Abstract
Ubiquitination is emerging as an important post-translational modification (PTM) for numerous cellular functions including protein degradation, DNA damage repair and tolerance, and cell cycle progression. Compared with other small-molecule modifiers found in phosphorylation, acetylation and glycosylation, ubiquitin is a small protein modifier that exists as either a single ubiquitin or a polyubiquitin chain. Furthermore, the polyubiquitin chains are formed via various linkages imparting an additional layer of specificity in cellular signaling. In order to adequately study ubiquitin signaling and particularly deubiquitination, a number of ubiquitin activity-based probes (ABPs) were developed and utilized in understanding the deubiquitinase (DUBs) function. Here, we focus on the current state of the DUB ABP development and their application in understanding DUB function and specificity for polyubiquitin chains and ubiquitinated proteins.
Keywords: PCNA, α-Globin, Deubiquitination, Cell-permeable ubiquitin probes, Hybrid triubiquitin probes, Monoubiquitin probes, Post-translational modification, Ubiquitination
4.23.1. Ubiquitination
Ubiquitin (Ub) is a small (8.6 kDa) regulatory protein of 76 amino acids that adopts a β-grasp fold. Ub is highly conserved in eukaryotic organisms. The conjugation of ubiquitin to a target protein is called ubiquitination or ubiquitylation.1 Typically, Ub is attached to proteins through an isopeptide linkage between the C-terminal carboxylate of ubiquitin (glycine 76) and an ε-amino group of a lysine residue in the acceptor proteins. Ubiquitination is an important, reversible post-translational modification (PTM) in eukaryotic cells. Since its discovery in the late 1970s and early 1980s, the modification by ubiquitin has emerged as an essential regulatory mechanism in almost all cellular processes in eukaryotes. Ubiquitination affects substrate proteins in many different ways including signaling, proteasomal degradation, altering cellular localization, modulating catalytic activity, and promoting or preventing protein interactions.2, 3 The cellular processes regulated by ubiquitination include cell cycle, transcription, trafficking, inflammation and DNA repair. Notably, many of the processes are independent of proteasome-mediated protein degradation.4
Ubiquitination involves three main enzymatic steps catalyzed by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s) (see Fig. 1 ).5 First, ubiquitin is activated in a two-step reaction by an E1 (ubiquitin-activating enzyme) with the consumption of ATP, forming a ubiquitin adenylate intermediate and subsequently a thioester bond between the C-terminal carboxyl group of ubiquitin and the active site cysteine of E1.6, 7 The human genome contains two E1s, i.e. UBA1 and UBA6.8 E2 catalyzes the transfer of Ub from the Ub-E1 conjugate to the active site cysteine of E2 and forms the Ub-E2 conjugate through a transthioesterification reaction. The human genome possesses more than 30 different E2 enzymes.9 The E3 ubiquitin ligase catalyzes the final step of the ubiquitination cascade by transferring Ub from the Ub-E2 conjugate to the substrate protein. E3s have substrate specificity for the E2 enzymes. The cullin-RING ligases, which constitute the largest group of E3s (around 600 members), do not form a covalent bond with Ub. Two smaller groups of E3s, the HECT ligases (around 30 members) and RBR ligases (around 12 members), form a Ub-thioester intermediate with the E3 active site cysteine.10
Fig. 1.
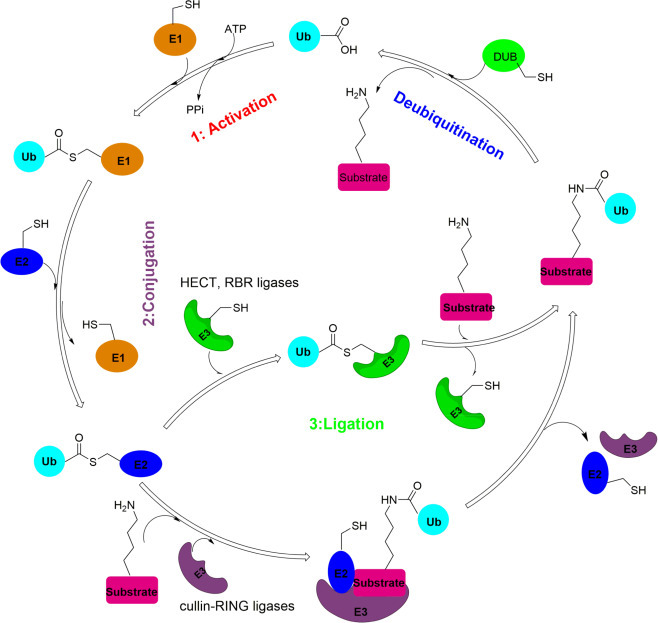
Overview of ubiquitination and deubiquitination process. Ubiquitination involves three main steps: activation, conjugation, and ligation, catalyzed by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s) respectively. Deubiquitination is catalyzed by deubiquitinating enzymes (DUBs).
Notably, substrate proteins can be ubiquitinated at different lysine residues resulting in multi-monoubiquitination. Ub itself can be further ubiquitinated, namely polyubiquitination, giving rise to Ub chains.11 This can occur through one of seven Ub Lys residues (K6, K11, K27, K29, K33, K48, and K63) or the Ub N-terminal methionine residue (M1).3 Furthermore, mixed and branched Ub chains also exist in cells.12 Different Ub chain linkages are associated with various cellular functions. For instance, the K48-linked polyubiquitination is the best studied Ub modification and it targets proteins to the proteasome for degradation.13 The K63-linked polyubiquitination is mostly related to non-proteolytic processes such as endocytic trafficking, inflammation, translation, and DNA repair.1, 14 M1-linked (also called linear) Ub chains play a very important roles in immune response.15
4.23.2. Deubiquitinases
Deubiquitinating enzymes or deubiquitinases (DUBs) are isopeptidases that hydrolyze the isopeptide bond between a Ub C-terminal carboxyl group and a side-chain amine group of a lysine residue in the acceptor proteins. The receptor proteins can be a ubiquitinated target protein or a receptor Ub in a polyubiquitin chain. In the human genome there are close to 100 DUBs, which can be classified into two main categories, i.e. cysteine proteases and metalloproteases.16 The cysteine proteases can be further divided into six families: ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), ovarian tumor proteases (OTU), Machado-Josephin domain DUBs (MJD), motif interacting with Ub-containing novel DUB family (MINDY) and the newly characterized zinc finger with UFM1-specific peptidase (ZUFSP). In the USP family, there are around 56 known DUBs (Fig. 2 ). The UCH family includes BAP1, UCHL1, UCHL3, UCHL5. The OTU family includes A20, Cezannes, and OTUs (Fig. 2). The MJD family has four members, Ataxin-3, Ataxin-3 like, JosD1, JosD2. The MINDY family contains four members MINDY-1 to -4. ZUFSP is the most recently reported DUB with no homology to any known DUBs.17 The metalloprotease group contains only the Jab1/Mov34/Mpr1, Pad1 N-terminal+, (MPN+), (JAMM) domain proteases. JAMM family contains eight members: BRCC36, CSN5, PSMD14, AMSH, AMSH-LP, MPND, MYSM1 and PRPF8.
Fig. 2.
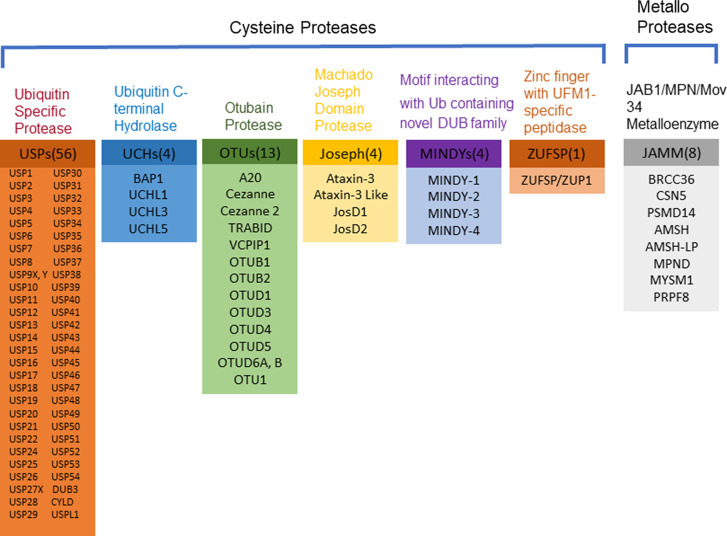
Overview of human deubiquitinating enzymes (DUBs). Human DUBs can be classified into two main categories: cysteine proteases and metalloproteases. The cysteine proteases are categorized into ubiquitin specific proteases (USPs), ubiquitin carboxyl-terminal hydrolases (UCHs), otubain (OTU) proteases, Machado Joseph Domain (MJD) proteases, motif interacting with Ub containing novel DUB family (MINDYs), zinc finger with UFM1-specific peptidase (ZUFSP). The metalloprotease family is JAB1/MPN/Mov34 metalloenzyme (JAMM). The human genome encodes more than 100 DUBs, each belonging to one of the above seven families.
DUBs often contain a catalytic domain surrounded by one or more additional domains, some of which contribute to target recognition.18 These additional domains include Ub-specific protease (DUSP) domain; ubiquitin-like (UBL) domain; meprin and TRAF homology (MATH) domain; zinc-finger ubiquitin-specific protease (ZnF-UBP) domain; zinc-finger myeloid, nervy and DEAF1 (ZnF-MYND) domain; ubiquitin-associated (UBA) domain; CHORD-SGT1 (CS) domain; microtubule-interacting and trafficking (MIT) domain; rhodenase-like domain; TBC/RABGAP domain; and the B-box domain.19
DUBs process polyubiquitin chains through different modes of action. Some DUBs can completely disassemble Ub chains from substrate protein, whereas others may be involved in chain editing, in which a chain is partially trimmed. These types of DUBs cleave Ub modules in a chain and have specific Ub-binding pockets on adjacent sides of the active site: one that binds the Ub moiety preceding (S1) and one following (S1′) the scissile bond shown in Fig. 3 .20 Other DUBs can cleave monoUb or Ub chains from protein substrates. These DUBs have a S1 site where the Ub proximal to the substrate protein would bind, but lack an S1′ Ub-binding site. Instead, these DUBs may have a specific S1′ substrate-binding site (Fig. 3). An S2 or even S3 site following the S1 Ub-binding site may accommodate more distal Ub modules of a chain to enhance specificity of DUBs.
Fig. 3.
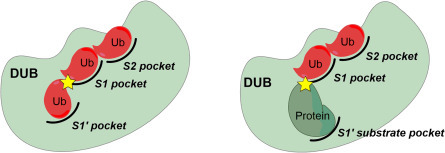
Possible modes of action in deubiquitination by DUBs. The specificity of DUBs for differently linked Ub chains and ubiquitinated substrates is governed by ubiquitin and target protein binding sites.
The Ub system regulates a variety of cellular processes. Abnormality in the Ub system is known to cause cancers, neurological disorders and other human diseases.21, 22, 23 The human DUBs are responsible for deubiquitination of over 5000 human proteins.24 DUBs are known to interact with a large number of partner proteins which further increase the diversity and specificity of DUBs.25 Human DUBs have recently garnered increasing attention as drug targets for pharmacological intervention due to better druggability than Ub ligases.26
4.23.3. Activity-Based Probes for DUBs
Activity-based probes (ABPs) have been developed for different families of enzymes,27 including serine hydrolases,28 cysteine protease29 and others. Although ABPs often mimic substrate, they usually are not processed by the target enzyme, instead forming a covalent bond with the active site residue of the target enzyme.30 Thus, the extent of probe labeling is an indirect measure of enzyme activity. ABPs are particularly well suited for studying enzymes with nucleophilic catalytic residues, as a reactive electrophile can be conveniently incorporated into the probe to form a covalent bond with the enzyme active-site residue. Cysteine, threonine, and serine proteases have been extensively studied with ABPs.
ABP usually contains three components, i.e. reactive group, recognition element and reporter tag, as shown in Fig. 4A.31 For those enzymes with an active site nucleophile, the reactive group in the ABP usually is an electrophile, often referred to as a “warhead”. The choice of warhead will affect both the reactivity and selectivity of the probe. The recognition element of ABPs confers selectivity toward the target enzyme, which can be a small molecule, a short peptide, or a full-length protein. A reporter tag (also referred to as a handle or label), can be used to detect probe labeled proteins. There are several generally used reporter tags. While the fluorescent tag can facilitate rapid and sensitive detection of labeled proteins, affinity tags (such as biotin or HA) also allow for isolation and enrichment of labeled proteins. Alternatively, a small biorthogonal group such as an alkyne or azide may be incorporated into the probe to allow subsequent attachment of a reporter through click chemistry.32 This is often referred to as “two-step” labeling, and may be particularly advantageous when larger tags interfere with reactivity, selectivity, or physiochemical properties of the probe. For larger ABPs, such as the DUB probes, substantial increase in the molecular weight of the labeled protein, as detected on a SDS-PAGE gel, can be used as an indication of probe labeling.
Fig. 4.
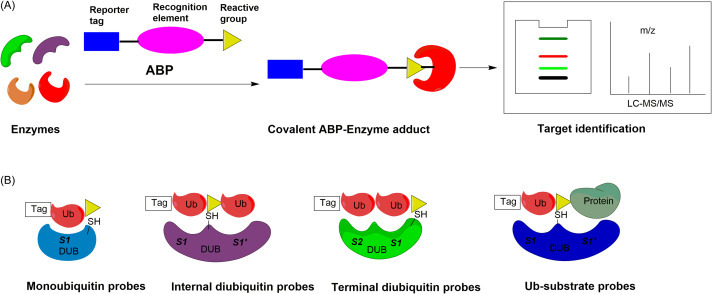
(A) Overview of activity-based protein profiling. (B) Probes used to study the DUB specificities and functions targeting the various Ub-binding sites. Monoubiquitin probes targeting the S1 pocket; internal diubiquitin probes targeting S1-S1′ pockets; terminal diubiquitin probes targeting S1-S2 pockets; Ub-substrate probes targeting S1-S1′ pockets.
The DUBs are well suited for study using Ub-based ABPs, due to the prevalence of enzyme active site nucleophiles (Fig. 4B). ABPs bearing cysteine-targeting electrophiles have been successfully developed for proteases.31 DUBs are mainly cysteine proteases. Hence, Ub-based DUB probes have been developed and used to identify new DUBs, assess the potency and selectivity of DUB inhibitors, characterize DUB enzymatic activity, and uncover the physiological roles of DUBs.33
In order to profile the activity of DUBs, several types of Ub probes were developed during the last decade.34 The most widely used DUB activity-based probes contain a mono-Ub recognition element with an electrophilic group conjugated to its C-terminus, exemplified by Ub propargylamide (Ub-PA) or Ub vinyl methyl ester (Ub-VME). Recently, the Ub-based DUB ABPs have expanded to include internal and terminal diUb-based ABPs,20, 35, 36, 37, 38 ubiquitin photoaffinity probes,39, 40 ubiquitin interactor affinity probes,41, 42 Ub-substrate protein probes,43, 44 and a reactive-site-centric DUB probe.45 The above-mentioned DUB ABPs have greatly enhanced our knowledge of DUBs through elucidation of linkage specificity. In this review, we will discuss newly developed activity-based DUB probes and their utility in understanding DUB linkage and target protein specificities.
4.23.4. Monoubiquitin Probes
The first generation of ABPs developed for DUBs contain a single Ub (1–75) with an electrophile (warhead) in place of the C-terminal glycine residue (G76), exemplified by the ubiquitin aldehyde (Ub-Al) and ubiquitin-nitrile (Ub-CN).46, 47 These two probes were important tools in the early mechanistic studies of DUBs. However, the modification of DUBs by Ub-Al and Ub-CN is reversible and is not compatible with the strongly reducing conditions of SDS/PAGE gel analysis. To overcome the limitations of these probes, Borodovsky et al. developed irreversible DUB ABPs by introducing a C-terminal reactive group using the intein-based chemical ligation method.48, 49 To generate the desired DUB ABPs, N-terminal HA-tagged Ub (HA-Ub) lacking Gly76 was expressed in E. coli as a fusion protein with an intein and a chitin binding domain. Purification using chitin beads, followed by a transthioesterification reaction with sodium 2-mercaptoethanesulfonate (MESNA) led to the isolation of the desired thioester. The desired irreversible DUB ABPs were synthesized by chemical ligation of the reactive groups (methyl (E)-4-aminobut-2-enoate et al.) with HA-Ub1–75-MESNA and purified by cation exchange chromatography. The covalent ABPs labeled DUBs in cell lysates and could be detected following SDS/PAGE immunoblotting due to the presence of an HA tag on the probe. Following similar chemical ligation methods, related Ubl-VS probes for ISG-15, SUMO-1, GATE-16, GABARAP, MAP1-LC3, and Apg8L were reported shortly thereafter.50
To date, a wide range of monoubiquitin probes have been developed with different electrophiles at the C-terminal of Ub, as summarized in the Fig. 5 .34 Electrophiles are classified according to the nature of the reaction with the catalytic cysteine, i.e. nucleophilic substitution, direct 1,2- addition and 1,4-conjugation addition. Surprisingly, propargyl amides also react with DUB active site cysteine residue, forming a vinyl thioether.51 The reaction appears to proceed via direct nucleophilic attack on the internal alkyne carbon, facilitated by stabilization of the developing carbanion by the “oxyanion hole” of the active site. The resulting adduct is stable to denaturing and reducing conditions, but could be cleaved in acid, which is useful for proteomic studies.
Fig. 5.
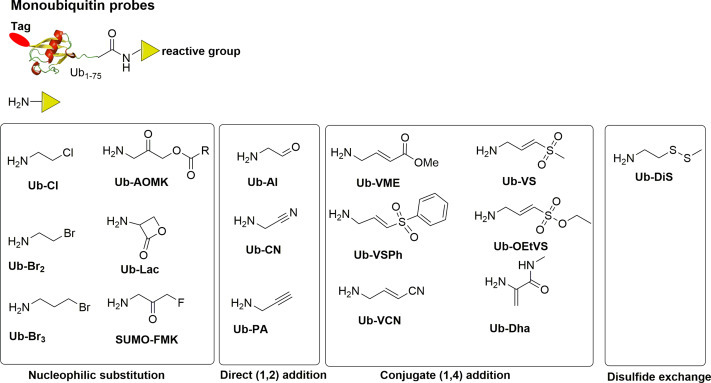
Monoubiquitin probe structures and warheads. Monoubiquitin probes usually contain a reporter tag (red ellipse), ubiquitin and a reactive group at the C-terminus of ubiquitin (yellow triangle). Based on the warheads, monoubiquitin probes can be categorized into three families: nucleophilic substitution, direct 1,2-addition, 1,4-addition and disulfide exchange.
Recently, Jong et al. reported a reversible disulfide Ub probe.52 Solid phase peptide synthesis was used to generate an N-terminal biotin and rhodamine labeled Ub disulfide probe. These probes can bind DUBs covalently by forming a disulfide bond between the active site cysteine residue and the Ub-based probe. The in-vitro labeling result showed that disulfide ubiquitin probes can label OTUB2, UCHL1, USP7 efficiently. Moreover, the disulfide bond probe can label DUBs in cell lysate. Most importantly, the disulfide bridges can be easily broken by the addition of a reducing agent such as DTT or TCEP. These probes can be used to capture and subsequently release catalytically active DUBs, whereas existing capturing agents bind irreversibly.
4.23.5. Diubiquitin Probes
Although the monoubiquitin DUB probes have proven to be useful tools for profiling DUBs, they rely solely on the interaction between Ub and the S1 site of DUBs for affinity and specificity. Thus, they provide little information about the chain linkage and target specificity of DUBs. While some DUBs, notably those of the UCH family, prefer to process Ub with only short C-terminus extensions, most DUBs cleave Ub from ubiquitinated substrates. These substrates may be Ub itself, or other proteins. DUBs can display specificity for position in the Ub chain (exo, endo, or base cleavage) or linkage type using additional Ub binding sites, which can recognize Ub on the distal or proximal side. DiUb APBs have therefore been developed in order to study DUB activity and linkage specificity.
The first diUb mimetic ABPs was reported by Iphofer et al., who used short peptides conjugated through Lys isopeptide bonds to mimic the proximal Ub, as shown in Fig. 6 .53 The authors first synthesized a ubiquitin peptide containing a lysine with its ε-amino group modified by 4-aminobut-2-enoic acid using solid phase peptide synthesis. The modified peptide was then used to react with HA-Ub1–75-thioester to afford the final probe. Compared with HA-Ub-VME, probes mimicking K48 and K63 linked diUb showed the expected reactivity toward a small number of recombinant DUBs, and labeled different sets of DUBs in cell lysate, as determined by LC/MS based protein profiling and immunoblotting experiments.
Fig. 6.
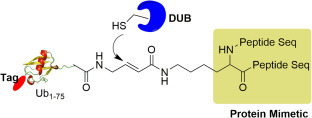
Internal diubiquitin mimetic ABP. The probe contains a reporter tag shown as red ellipse, ubiquitin (1–75) and a Michael acceptor reactive group between the C-terminal of ubiquitin and peptide sequence. Dubs can react covalently with the probe by a Michael addition.
It has become clear that DUBs likely make extensive contacts with proximal Ub in the DUB S1′ site.20 Thus, probes containing short Ub peptides may not recapitulate the interaction between DUB and full length Ub. Subsequent efforts have therefore been directed to making diUb probes containing two intact mono Ubs. McGouran et al. generated activity-based diUb probes with eight different Ub linkages (M1, K6, K11, K27, K29, K33, K48, K63) using azide-alkyne click reaction (Fig. 7 ).35 To generate the desired diUb probe, the Cu(I)-catalyzed click reaction was used to link the proximal and distal Ub. In the distal Ub, an intein-based method was used to introduce a Michael acceptor warhead and an alkyne group at the C-terminus of HA-Ub1–75. For the proximal ubiquitin, an unnatural amino acid, azidohomoalanine (Aha), was introduced to substitute the target lysine residue using the methionine analog incorporation approach.54 Subsequently, the proximal Ub was linked to the C-terminus of the distal Ub by the Cu(I)-catalyzed click reaction forming a 1,4-triazole linker. With the diUb probes in hand, 29 DUBs in total including 18 USPs, 4 UCHs, 5 OTUs, 1 MJD, and one SUMO deconjugating enzyme were captured by the probes from HEK293T cell lysate using LC-MS/MS.
Fig. 7.
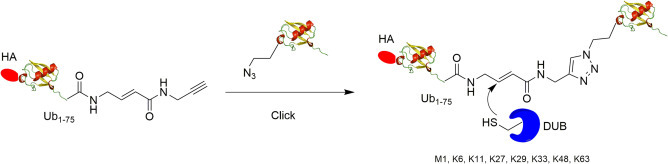
Generation of internal diubiquitin probe using azide-alkyne click reaction.
Li et al. generated the full length K48 and K63 diUb probes using intein-based chemical ligation method as shown in Fig. 8 .36 Chemical ligation reaction between the newly introduced cysteine to replace the selected lysine residue in proximal Ub and an α-bromoketone modified distal Ub yields the final diUb probe with a Michael acceptor warhead between the distal and proximal Ub. The resulting diUb probes closely mimic the native linkage of diUb in terms of linkage length. The K48 and K63 diUb probes were demonstrated to label DUBs from different families. A HA tag was introduced into the N-terminus of the proximal ubiquitin to facilitate its detection by the anti-HA antibody. Different labeling activity was observed between K48 and K63 internal diUb probes in both purified DUBs and DUBs in HEK293T cell lysate.
Fig. 8.
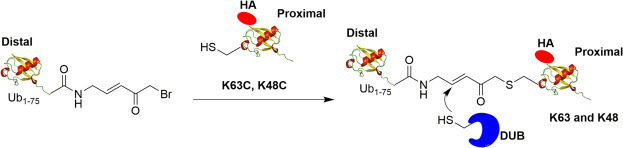
Generation of K63, K48 internal diUb probe. Distal Ub with C-terminus Michael acceptor linker can react with proximal ubiquitin mutant (K63C or K48C) to generate internal diUb probe. DUBs can form a covalent bond with the probe through Michael addition between the active site cysteine and the α,β-unsaturated ketone.
Haj-Yahya et al. generated dehydroalanine (DHA)-containing diUb (M1, K48, K63 linkage) probes using solid phase peptide synthesis and native chemical ligation as shown in Fig. 9 .37 For the synthesis of the proximal Ub, orthogonally protected Fmoc-Lys-(Dde)-OH was used in position 48 or 63 of native Ub. Subsequent to chain assembly, the ε-amine of the Lys residue was unmasked and coupled with Boc-Cys(Trt)-OH. After peptide cleavage and purification, the intermediate was ligated with the Ub1–75-thioester to afford a diUb intermediate. Next, the 1,4-dibromobutane was used to convert the cysteine in diUb to the DHA and the desired diUb product was obtained by HPLC purification.
Fig. 9.
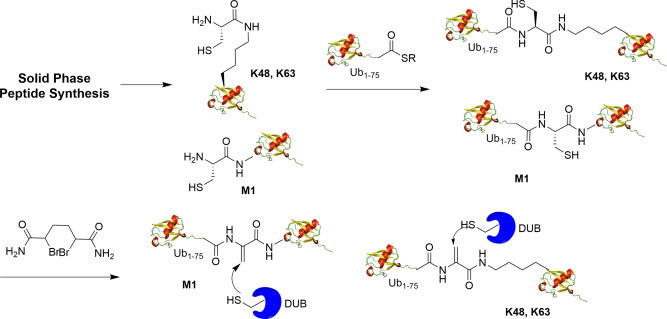
Generation of M1, K48, K63 internal dehydroalanine based diubiquitin probe using solid phase peptide synthesis and native chemical ligation.
Mulder et al. reported a method of generating diUb probes (K6, K11, K27, K29, K33, K48, K63) using solid phase peptide synthesis coupled with native chemical ligation as shown in Fig. 10 .38 To generate the desired probes, a lysine residue of interest in proximal Ub was replaced with a diaminobutyric acid residue. Using linear Fmoc-based solid phase peptide synthesis of the Ub polypeptide, seven Ub mutants were synthesized. Then, native chemical ligation of the proximal Ub mutant and distal Ub1–75-SEt thioester was performed to yield the diUb intermediate. Finally, 2,5-dibromohexanediamide was used for desulfurization to generate the final diUb probe with a reactive Michael acceptor between the proximal and distal Ub. The authors further investigated diUb reactivity toward DUBs and demonstrated the ability of these probes to capture active DUBs selectively with distinct target preferences. Fluorescent versions of the K11 and K48 diUb ABPs were also generated, allowing a more sensitive and faster read-out of labeling experiments.
Fig. 10.
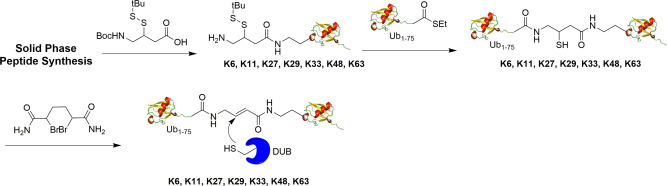
Generation of K6, K11, K27, K29, K33, K48, K63 linkage-specific internal diubiquitin probe using solid phase peptide synthesis.
The above described diUb probes all contain a warhead between the distal and proximal Ub for reaction with the catalytic cysteine of DUBs. This class of diUb probes allows the interrogation of the DUB S1 and potential S1′Ub-binding sites. To interrogate the S1 and S2 Ub-binding sites, Dennis et al. designed a set of diUb probes with a reactive warhead at the C-terminus of the proximal Ub (Fig. 11 ).20 The two Ubs are connected through a triazole linker which is resistant to DUBs cleavage. Solid phase peptide synthesis was used to generate distal ubiquitin containing a N-terminal tetramethyl rhodamine as a reporter tag and a propargylamide warhead at C-terminus of the proximal Ub. For the proximal Ub, the unnatural amino acid azidohomoalanine (Aha) was introduced at the specific position using a methionine analog incorporation approach.54 Then, the distal ubiquitin mutant can react with proximal ubiquitin mutant through a Cu(I)-catalyzed click reaction to generate a diUb intermediate. Finally, the diUb intermediate can react with propargylamine to generate the final terminal diUb probes. Seven linkage specific (K6, K11, K27, K29, K33, K48, K63) terminal diUb probes were generated by this method. With the probes, OTUD2 was shown to possess specificity for K11 and K33-linked terminal diUb probes due to engagement of S1-S2 sites on OTUD2. Additionally, an S2 site on OTUD3 interacting with K11-linked terminal diUb probes was found to provide the linkage specificity.
Fig. 11.
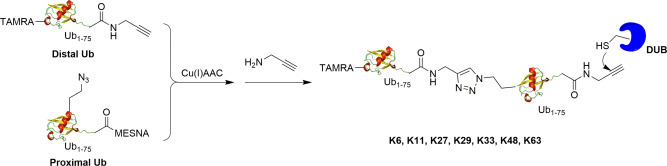
Synthesis of triazole-linked activity-based terminal diUb probes using Cu(I)AAC catalyzed click reaction.
4.23.6. Triubiquitin Probes
Polyubiquitin probes to address multiple auxiliary ubiquitin binding sites (beyond S2/S1 or S1/S1′binding modes) are highly desirable in investigating how DUBs process linear, mixed and branched chain.55 Another focus is understanding endo/exo chain cleavage activity of DUBs, which has remained poorly defined due to the lack of suitable tools. To date several branched chain triubiquitin substrates were synthesized utilizing thiol-ene reaction, chemoenzymatic methods, and solid phase synthesis.56, 57 Moreover, free polyUb or polyUb substrate protein have been generated using chemical and semisynthetic approaches.58, 59, 60, 61, 62, 63
Given that multiple Ub binding sites may exist on DUBs, polyUb probes are desirable in assessing the binding and processing of polyUb chains by DUBs. Paudel et al. recently reported hybrid triubiquitin probes with a Michael acceptor or a non-cleavable linker introduced at selected isopeptide linkage in a triUb.64 In brief, the synthesis of the probes were accomplished via a two-step method (Fig. 12 ). The first step utilized specific E1 and E2 combinations to generate K11, K48 and K63-linked native diUb containing a lysine to cysteine mutation. In parallel, HA-Ub1–75-MESNA was either reacted with a Michael acceptor (MA) or non-cleavable (NC) linker molecule to generate HA-Ub-MA and HA-Ub-NC, respectively. In the second step, HA-Ub-NC or HA-Ub-MA was reacted with the mutant diUb species to yield triUb-MA1-CL2 or triUb-NC1-CL2. The triUb probes can be used to interrogate the endo/exo cleavage modes of DUBs by simultaneously assessing the binding of all three Ub moieties to the potential S1, S2 and S1′ sites in DUBs. Using hybrid triUb probes, the USP9X catalytic domain (USP9X CD) was shown to process triUb chains in a linkage-specific way. Specifically, USP9X CD cleaves K48-linked polyUb chain in an exo cleavage mode, as demonstrated by labeling with K48 triUb-MA1-CL2, while no cleavage activity of K48 triUb-NC1-CL2. In contrast, USP9X CD displayed endo recognition of K11 polyUb chains by efficiently cleaving K11 triUb-NC1-CL2 and K11 triUb-MA1-CL2 probes while showing very low level labeling by K11 triUb-MA1-CL2 probe. These triUb probes can be used to investigate other DUBs in processing polyUb chains of different linkages.
Fig. 12.
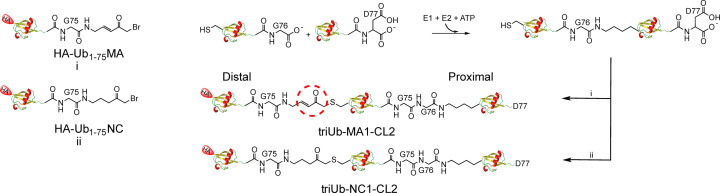
Generation of hybrid triubiquitin probes. Chemoenzymatic synthetic routes to develop triubiquitin probes are illustrated.
4.23.7. Ubiquitinated Substrate Protein Activity-Based Probes
In addition to the chain linkage specificity, the DUB's specificity toward target proteins is thought to be important for the physiological function of DUBs.3 In humans, around 5000 proteins have been found to be modified by Ub and 19,000 different lysine residues were identified.24 Therefore, the development of ubiquitinated substrate protein ABPs is of importance for a better understanding of DUBs’ target protein specificity. Challenges exist in making this type of probe synthetically or semi-synthetically given that many target proteins are prone to denaturation under harsh reaction conditions.
Roman et al. report a Ub α-Globin activity-based probe by applying a sequential dehydroalanine formation strategy to expressed proteins as shown in Fig. 13 .43 α-Globin was reported to undergo ubiquitination at various sites, including Lys100 in the C-terminus region. α-Globin contains a single Cys104 residue in the C-terminus region, close to the native ubiquitination site Lys100. The authors took advantage of the single cysteine (104) residue in α-Globin to generate a dehydroalanine α-Globin species using 2,5-dibromohexanediamide. Then a thiol-ene reaction and Pd-catalyzed reaction were used to generate α-Globin mutant, which can undergo native chemical ligation with a biotin-containing Ub thioester to generate ubiquitinated α-Globin through an amide bond. Finally, 2,5-dibromohexanediamide was used to generate the dehydroalanine ubiquitin α-Globin probe. After HPLC purification and buffer exchange, the ubiquitin α-globin probe can be refolded. The ubiquitin α-Globin probe mimics the native ubiquitinated α-Globin and cognate DUBs can be trapped by the probe through a Michael addition reaction. As a control, the α-Globin dehydroalanine probe without ubiquitin was also generated.
Fig. 13.
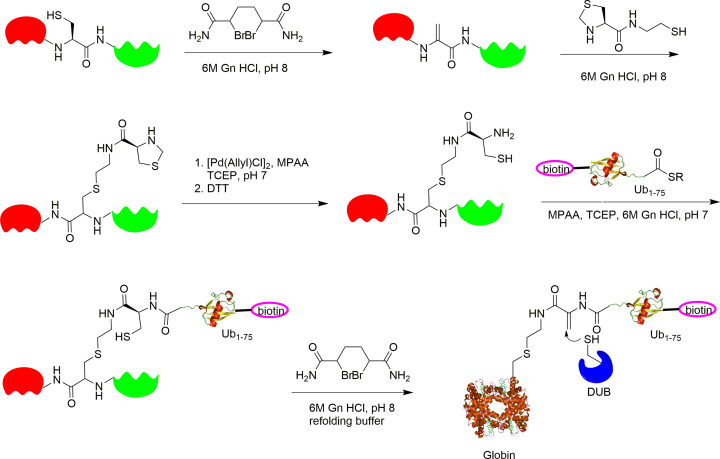
General synthetic strategy for the preparation of ubiquitinated protein ABPs on the basis of sequential DHA formation. DTT, dithiothreitol; Gn, guanidine; MPAA, 4-mercaptophenylacetic acid; TCEP, tris(2-carboxyethyl)phosphine.
The in vitro labeling results showed that the ubiquitin α-Globin probe labels a number of purified DUBs. Also, the ubiquitinated α-globin probe was used to identify potential DUBs from erythrocyte lysate capable of deubiquitinating α-globin using MS/MS-based proteomics. USP15, USP14, USP5, and UCHL3 were significantly enriched by the ubiquitin-α-Globin probe. An in vitro cleavage assay using native α-globin-Ub as a substrate was used to validate the identified DUBs. α-globin-Ub cleavage activity was observed for USP7 and USP15.
Another protein-based DUB probe was reported by Gong et al. using an expressed protein ligation strategy (Fig. 14 ).44 The target protein is proliferating cell nuclear antigen (PCNA), a nuclear protein essential for DNA replication, repair and damage tolerance.65 It has been shown that yeast PCNA can be ubiquitinated at multiple sites.66 While monoubiquitination of yeast PCNA at Lys164 plays an important role in DNA translesion synthesis (TLS) across DNA lesions such as cyclobutane pyrimidine dimer (CPD) induced by UV irradiation in eukaryotes, monoubiquitination of PCNA at Lys107 also exists in S. cerevisiae and has been linked to DNA ligase I deficiency in yeast.65, 67, 68, 69 In order to investigate the PCNA deubiquitination at different sites, activity-based Ub-PCNA probes were generated and used to identify the DUB in yeast S. cerevisiae responsible for PCNA deubiquitination. A major challenge in generating Ub-PCNA DUB probe is the labile nature of PCNA and the requirement of a mild reaction condition in probe preparation.
Fig. 14.
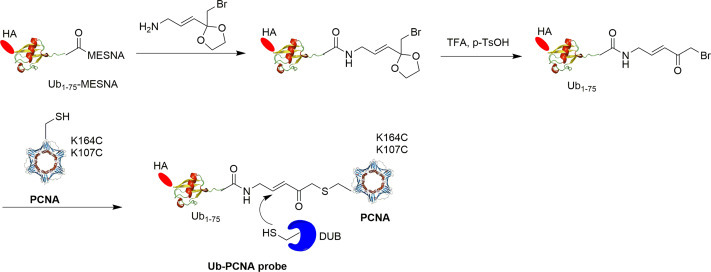
Scheme illustrating the generation of Ub-PCNA activity-based DUB probes.
To prepare the Ub-PCNA probes, a warhead was introduced between the Ub moiety and PCNA for covalent trapping of the catalytic cysteine in the DUB active site. First, Ub1–75 with an α-bromide introduced at its C-terminus was generated using an intein-based method.36 Then, ligation between the cysteine residue introduced at position 164 or 107 in the cysteine-light PCNA mutant with the α-bromide at the C-terminus of Ub yielded a Ub-PCNA probe with a stable thioether linkage under native reaction condition. Using the similar ligation method, Ub-PCNA probes containing a noncleavable linker were also generated.
The availability of the above described Ub-PCNA DUB probes coupled with pulldown and proteomics allowed the identification of yeast DUB, particularly Ubp10, that recognizes monoubiquitinated PCNA at K164 and catalyzes its deubiquitination. The result showed that Ubp10 is captured by the K164C Ub-PCNA probe specifically and revealed position-specific deubiquitination of a target protein substrate by DUB.
4.23.8. Cell-Permeable Activity-Based Probes for Human DUBs
Activity-based DUB probes are widely used in investigating DUB's function and activity. However, most early developed Ub-based DUB ABPs are limited to purified proteins and cell lysates due to poor cell permeability. One disadvantage of cell lysate-based study is the dilution of cytoplasm and disruption of cellular organelles, which may lead to altered activity of cellular DUBs and DUB complexes. To better understand the cellular functions of DUBs, cell-permeable DUB ABPs are needed for the interrogation of DUB activity and regulation in live cells. This class of DUB probes will also find use in drug development against DUBs.
Previously, several strategies have been developed to enable the in-cell profiling of DUBs, including catch-and-release probes and the use of pore-forming toxins (PFOs).70 These strategies facilitate the entry of Ub-based ABPs into live cells. Also, Ward et al. reported a small molecule cell-permeable DUB probe.71 With this probe, 12 USPs were identified using LC-MS/MS methods. Mulder et al. used electroporation to deliver the Ub cascade probe (Ub-Dha) into human cells.72 In addition to the labeling of enzymes in the ubiquitin cascade (2 E1s, 19 E2s, 2 E2/E3s, and 2E3s), the authors also identified four DUBs using a proteomics approach.
Recently, Gui et al. reported cell-permeable activity-based Ub probes that enable intracellular profiling of human DUBs.73 In this new class of DUB probes, such as HA-Cys(cR10)-Ub-PA, a cyclic polyarginine (cR10) peptide was linked to HA-Ub-PA through a disulfide bond as shown in Fig. 15A. The cR10 peptide was shown to enhance the cellular uptake of protein cargo, compared to linear and cyclic TAT peptides.74, 75, 76 The disulfide bond is cleaved in cells due to a reducing environment. Two other cR10-containing cell-permeable probes, HA-Cys(cR10)-Ub-VME and HA-Cys(cR10)-Ub-EA, were also prepared using similar methods as shown in Fig. 15B.
Fig. 15.
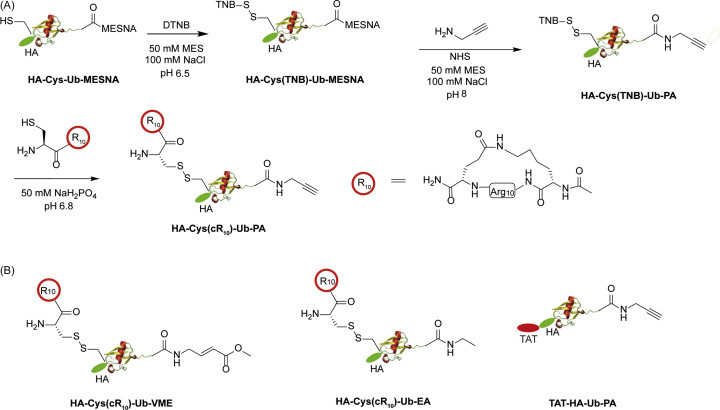
Generation of cell-permeable DUB ABPs. (A) Chemical steps of generating cell-permeable HA-Cys(cR10)-Ub-PA probe. (B) Structure of cell-permeable probe HA-Cys(R10)-Ub-VME, HA-Cys(R10)-Ub-EA and TAT-Ub-PA.
The HA-Cys(cR10)-Ub-PA probe was used in in-cell DUB labeling. A clear difference between HeLa cell lysate and in-cell labeling was observed. Twenty nine DUBs were significantly enriched by HA-Cys(cR10)-Ub-PA probe in HeLa cells using label-free quantitative mass spectrometry analysis. Additionally, the authors demonstrated that the cell-permeable DUB ABPs can be used in assessing the inhibition of DUBs by small molecule inhibitors.
4.23.9. Structure Studies Using Diubiquitin ABPs
Recently, diubiquitin ABPs have been utilized in co-crystallization with DUBs. The breadth of information from the structures has allowed for a careful analysis of binding interactions as well as preferential binding sites in DUBs. Although active-site cysteine mutant DUBs with native diubiquitin co-crystal structures have been solved for CYLD(Lys63),77 AMSH(Lys63),78 OTUD2(Lys11),79 and USP30(Lys6),80 the DUB-di-Ub ABP co-crystal structure captures DUB in action and provides further information about DUB catalysis. To date co-crystal structures using diUb-ABPs have been reported for USP21,81 Cezanne/OTUD7B82 and SARS PLpro83 interrogating endo/exo binding as well as the contribution of the auxiliary sites for Met1, K11, and K48-linked diubiquitin linkages (Fig. 16 ). These structural studies showcased the utility of activity-based diUb probes in understanding how DUBs recognize ubiquitin chains through multiple Ub binding sites and provided an in-depth understanding of their catalytic mechanisms.
Fig. 16.
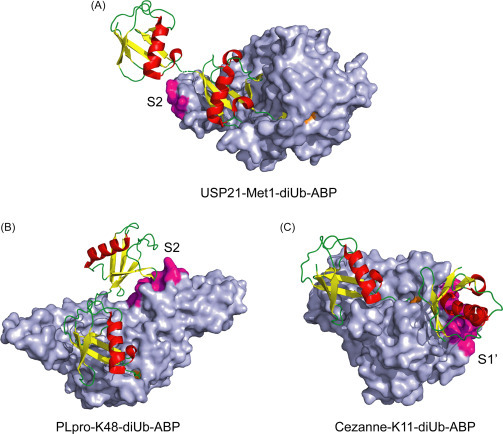
Co-crystal DUB structures using diUb-ABPs. (A) USP21 bound to Met1 diUb ABP that contains a terminal aldehyde warhead (PDB: 2Y5B). (B) SARS PLpro in complex with a triazole linked K48 diUb ABP that contains a terminal propargyl warhead (PDB: 5E6J). (C) Cezanne/OTUD7B in complex with K11 diUb-ABP that contains an internal Michael acceptor warhead (PDB: 5LRV). Auxiliary sites next to S1 sites are colored in magenta while catalytic cysteine is colored orange.
In the case of the structure of USP21 with linear diubiquitin aldehyde, a relatively small area (387 Å2) was revealed for the potential S2 site (Fig. 16a).81 The S2 site is located in the ZnF region of USP21 CD. Notably, the Ile36 hydrophobic patch of distal ubiquitin was found to contact the S2 site. However, further mutation of key residues in the USP21 S2 site failed to decrease the enzyme activity with linear diUb and tetraUb chains, which may be due to the exo-cleavage activity of USP21. Subsequent diUb- and triUb FlAsH anisotropy experiments were used to demonstrate the contribution of S2 site to Ub binding.
Human SARS coronavirus PLpro (papain like protease) specifically hydrolyzes K48 linked polyubiquitin chains. Structure elucidation with PLpro bound with terminal K48-linked diUb ABP revealed a potential S2 site in addition to the canonical S1 site in PLpro accommodating both ubiquitin moieties (Fig. 16b).83 The authors found that the Ile44 patch in the S2 site-bound ubiquitin interacts with an α-helix between the PLpro palm domain and N-terminal Ub-like domain. Mutation of these contacting residues resulted in lowered catalytic activity of PLpro against polyubiquitin chains. The buried area for the S2 site was determined to be 540 Å2, which is smaller than that in the S1 site (890 Å2). Furthermore, mutations in the S2 site using K48-linked diUb-AMC substrates showed a drastic reduction in hydrolysis supporting the importance of S2 site binding of distal Ub.
A detailed analysis of internal K11-linked diUb ABP binding to Cezanne revealed that recognition of S1′ site ubiquitin leads to a large dynamic conformational change and subsequent catalysis in Cezanne. The S1′ was determined to be transiently formed upon binding of diubiquitin, as ascertained by comparing monoUb and K11-diUb ABPs through co-crystallization, HDX, fluorescent polarization, and NMR experiments. In particular, a “Cys-loop” near the active site occupies the catalytic groove rendering the enzyme inactive in its apo state. In the co-crystal structure, binding of the proximal ubiquitin to the S1′ site resulted in movement of the Cys-loop out of the catalytic site, thus enabling the catalytic Cys194 for reaction with the diUb ABP. The interaction regions of the S1′ site Ub were found to be the α-helix of ubiquitin as well as its Thr12 and Glu16 residues. In Cezanne, the S1′ site comprises the α1, α2 helices and α3-α4 linker region.82 Mutation of residues in S1′ site did not drastically reduce polyubiquitin cleavage activity, likely due to the weak interaction in S1′ site as compared to the S1 site. The structural changes upon K11 diubiquitin binding supports the importance of S1′ site ubiquitin binding, which imparts the selectivity toward K11 linked polyubiquitin chains by Cezanne/OTUD7B.82
4.23.10. Conclusion and Future Direction
Activity-based ubiquitin probes are among the most crucial and versatile tools to understand specificity and activity of interacting proteins and DUBs. Currently, several methods of synthesizing monoubiquitin, diubiquitin and triubiquitin ABPs have been developed including solid phase synthesis and chemoenzymatic reactions. These probes have already yielded valuable information on how DUBs recognize and process polyubiquitin chains and ubiquitinated proteins. The molecular and structural diversity of polyubiquitin chains makes understanding ubiquitin signaling a challenging yet exciting undertaking.
The future ubiquitin research will benefit from the advent and continued development of ubiquitin-based probes. Ubiquitin ABPs are becoming more sophisticated to include polyubiquitin chains of mixed and branched linkages as well as polyubiquitinated proteins. Additionally, these ABPs allow for interrogation of auxiliary sites useful for development of inhibitors targeting DUBs and ubiquitin binding proteins. They will help to deepen our understanding of complex ubiquitin cellular signaling pathways and develop new therapies targeting the ubiquitin system.
References
- 1.Ikeda F., Crosetto N., Dikic I. What Determines the Specificity and Outcomes of Ubiquitin Signaling? Cell. 2010;143(5):677–681. doi: 10.1016/j.cell.2010.10.026. [DOI] [PubMed] [Google Scholar]
- 2.Glickman M.H., Ciechanover A. The Ubiquitin-Proteasome Proteolytic Pathway: Destruction for the Sake of Construction. Physiol. Rev. 2002;82(2):373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 3.Komander D., Rape M. The Ubiquitin Code. Annu. Rev. Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 4.Mukhopadhyay D., Riezman H. Proteasome-Independent Functions of Ubiquitin in Endocytosis and Signaling. Science. 2007;315(5809):201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 5.Pickart C.M., Eddins M.J. Ubiquitin: Structures, Functions, Mechanisms. Biochim. Biophys. Acta. 2004;1695(1–3):55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 6.Pickart C.M. Mechanisms Underlying Ubiquitination. Annu. Rev. Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 7.Schulman B.A., Harper J.W. Ubiquitin-Like Protein Activation by E1 Enzymes: The Apex for Downstream Signalling Pathways. Nat. Rev. Mol. Cell Biol. 2009;10(5):319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groettrup M., Pelzer C., Schmidtke G., Hofmann K. Activating the Ubiquitin Family: UBA6 Challenges the Field. Trends Biochem. Sci. 2008;33(5):230–237. doi: 10.1016/j.tibs.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 9.van Wijk S.J., Timmers H.T. The Family of Ubiquitin-Conjugating Enzymes (E2s): Deciding between Life and Death of Proteins. FASEB J. 2010;24(4):981–993. doi: 10.1096/fj.09-136259. [DOI] [PubMed] [Google Scholar]
- 10.Metzger M.B., Hristova V.A., Weissman A.M. HECT and RING Finger Families of E3 Ubiquitin Ligases at a Glance. J. Cell Sci. 2012;125(Pt 3):531–537. doi: 10.1242/jcs.091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng J., Schwartz D., Elias J.E., Thoreen C.C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S.P. A Proteomics Approach to Understanding Protein Ubiquitination. Nat. Biotechnol. 2003;21(8):921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 12.Yau R., Rape M. The Increasing Complexity of the Ubiquitin Code. Nat. Cell Biol. 2016;18(6):579–586. doi: 10.1038/ncb3358. [DOI] [PubMed] [Google Scholar]
- 13.Chau V., Tobias J.W., Bachmair A., Marriott D., Ecker D.J., Gonda D.K., Varshavsky A. A Multiubiquitin Chain Is Confined to Specific Lysine in a Targeted Short-Lived Protein. Science. 1989;243(4898):1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 14.Zhao S., Ulrich H.D. Distinct Consequences of Posttranslational Modification by Linear Versus K63-Linked Polyubiquitin Chains. Proc. Natl. Acad. Sci. U. S. A. 2010;107(17):7704–7709. doi: 10.1073/pnas.0908764107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zinngrebe J., Montinaro A., Peltzer N., Walczak H. Ubiquitin in the Immune System. EMBO Rep. 2014;15(1):28–45. doi: 10.1002/embr.201338025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reyes-Turcu F.E., Ventii K.H., Wilkinson K.D. Regulation and Cellular Roles of Ubiquitin-Specific Deubiquitinating Enzymes. Annu. Rev. Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwasna D., Abdul Rehman S.A., Natarajan J., Matthews S., Madden R., De Cesare V., Weidlich S., Virdee S., Ahel I., Gibbs-Seymour I., Kulathu Y. Discovery and Characterization of ZUFSP/ZUP1, a Distinct Deubiquitinase Class Important for Genome Stability. Mol. Cell. 2018;70(1):150–164.e6. doi: 10.1016/j.molcel.2018.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komander D., Clague M.J., Urbé S. Breaking the Chains: Structure and Function of the Deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009;10(8):550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 19.Nijman S.M., Luna-Vargas M.P., Velds A., Brummelkamp T.R., Dirac A.M., Sixma T.K., Bernards R. A Genomic and Functional Inventory of Deubiquitinating Enzymes. Cell. 2005;123(5):773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Flierman D., van der Heden van Noort G.J., Ekkebus R., Geurink P.P., Mevissen T.E., Hospenthal M.K., Komander D., Ovaa H. Non-hydrolyzable Diubiquitin Probes Reveal Linkage-Specific Reactivity of Deubiquitylating Enzymes Mediated by S2 Pockets. Cell Chem. Biol. 2016;23(4):472–482. doi: 10.1016/j.chembiol.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoeller D., Dikic I. Targeting the Ubiquitin System in Cancer Therapy. Nature. 2009;458(7237):438–444. doi: 10.1038/nature07960. [DOI] [PubMed] [Google Scholar]
- 22.Edelmann M.J., Kessler B.M. Ubiquitin and Ubiquitin-like Specific Proteases Targeted by Infectious Pathogens: Emerging Patterns and Molecular Principles. Biochim. Biophys. Acta. 2008;1782(12):809–816. doi: 10.1016/j.bbadis.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ciechanover A., Brundin P. The Ubiquitin Proteasome System in Neurodegenerative Diseases: Sometimes the Chicken, Sometimes the Egg. Neuron. 2003;40(2):427–446. doi: 10.1016/s0896-6273(03)00606-8. [DOI] [PubMed] [Google Scholar]
- 24.Kim W., Bennett E.J., Huttlin E.L., Guo A., Li J., Possemato A., Sowa M.E., Rad R., Rush J., Comb M.J., Harper J.W., Gygi S.P. Systematic and Quantitative Assessment of the Ubiquitin-Modified Proteome. Mol. Cell. 2011;44(2):325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sowa M.E., Bennett E.J., Gygi S.P., Harper J.W. Defining the Human Deubiquitinating Enzyme Interaction Landscape. Cell. 2009;138(2):389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrigan J.A., Jacq X., Martin N.M., Jackson S.P. Deubiquitylating Enzymes and Drug Discovery: Emerging Opportunities. Nat. Rev. Drug Discov. 2018;17(1):57–78. doi: 10.1038/nrd.2017.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jessani N., Cravatt B.F. The Development and Application of Methods for Activity-Based Protein Profiling. Curr. Opin. Chem. Biol. 2004;8(1):54–59. doi: 10.1016/j.cbpa.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y., Patricelli M.P., Cravatt B.F. Activity-Based Protein Profiling: The Serine Hydrolases. Proc. Natl. Acad. Sci. U. S. A. 1999;96(26):14694–14699. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato D., Boatright K.M., Berger A.B., Nazif T., Blum G., Ryan C., Chehade K.A., Salvesen G.S., Bogyo M. Activity-Based Probes that Target Diverse Cysteine Protease Families. Nat. Chem. Biol. 2005;1(1):33–38. doi: 10.1038/nchembio707. [DOI] [PubMed] [Google Scholar]
- 30.Sanman L.E., Bogyo M. Activity-Based Profiling of Proteases. Annu. Rev. Biochem. 2014;83:249–273. doi: 10.1146/annurev-biochem-060713-035352. [DOI] [PubMed] [Google Scholar]
- 31.Cravatt B.F., Wright A.T., Kozarich J.W. Activity-Based Protein Profiling: From Enzyme Chemistry to Proteomic Chemistry. Annu. Rev. Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 32.Prescher J.A., Bertozzi C.R. Chemistry in Living Systems. Nat. Chem. Biol. 2005;1(1):13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- 33.Gopinath P., Ohayon S., Nawatha M., Brik A. Chemical and Semisynthetic Approaches to Study and Target Deubiquitinases. Chem. Soc. Rev. 2016;45(15):4171–4198. doi: 10.1039/c6cs00083e. [DOI] [PubMed] [Google Scholar]
- 34.Hewings D.S., Flygare J.A., Bogyo M., Wertz I.E. Activity-Based Probes for the Ubiquitin Conjugation-Deconjugation Machinery: New Chemistries, New Tools, and New Insights. FEBS J. 2017;284(10):1555–1576. doi: 10.1111/febs.14039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGouran J.F., Gaertner S.R., Altun M., Kramer H.B., Kessler B.M. Deubiquitinating Enzyme Specificity for Ubiquitin Chain Topology Profiled by Di-ubiquitin Activity Probes. Chem. Biol. 2013;20(12):1447–1455. doi: 10.1016/j.chembiol.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li G., Liang Q., Gong P., Tencer A.H., Zhuang Z. Activity-Based Diubiquitin Probes for Elucidating the Linkage Specificity of Deubiquitinating Enzymes. Chem. Commun. 2014;50(2):216–218. doi: 10.1039/c3cc47382a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haj-Yahya N., Hemantha H.P., Meledin R., Bondalapati S., Seenaiah M., Brik A. Dehydroalanine-Based Diubiquitin Activity Probes. Org. Lett. 2014;16(2):540–543. doi: 10.1021/ol403416w. [DOI] [PubMed] [Google Scholar]
- 38.Mulder M.P., El Oualid F., ter Beek J., Ovaa H. A Native Chemical Ligation Handle that Enables the Synthesis of Advanced Activity-Based Probes: Diubiquitin as a Case Study. ChemBioChem. 2014;15(7):946–949. doi: 10.1002/cbic.201402012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang J., Zhang L., Tan X.L., Qi Y.K., Feng S., Deng H., Yan Y., Zheng J.S., Liu L., Tian C.L. Chemical Synthesis of Diubiquitin-Based Photoaffinity Probes for Selectively Profiling Ubiquitin-Binding Proteins. Angew. Chem. Int. Ed. Engl. 2017;56(10):2744–2748. doi: 10.1002/anie.201611659. [DOI] [PubMed] [Google Scholar]
- 40.Tan X.D., Pan M., Gao S., Zheng Y., Shi J., Li Y.M. A Diubiquitin-Based Photoaffinity Probe for Profiling K27-Linkage Targeting Deubiquitinases. Chem. Commun. 2017;53(73):10208–10211. doi: 10.1039/c7cc05504h. [DOI] [PubMed] [Google Scholar]
- 41.Zhang X., Smits A.H., van Tilburg G.B., Jansen P.W., Makowski M.M., Ovaa H., Vermeulen M. An Interaction Landscape of Ubiquitin Signaling. Mol. Cell. 2017;65(5):941–955.e8. doi: 10.1016/j.molcel.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 42.Zhang X., Smits A.H., van Tilburg G.B., Ovaa H., Huber W., Vermeulen M. Proteome-Wide Identification of Ubiquitin Interactions Using UbIA-MS. Nat. Protoc. 2018;13(3):530–550. doi: 10.1038/nprot.2017.147. [DOI] [PubMed] [Google Scholar]
- 43.Meledin R., Mali S.M., Kleifeld O., Brik A. Activity-Based Probes Developed by Applying a Sequential Dehydroalanine Formation Strategy to Expressed Proteins Reveal a Potential α-Globin-Modulating Deubiquitinase. Angew. Chem. Int. Ed. Engl. 2018;57(20):5645–5649. doi: 10.1002/anie.201800032. [DOI] [PubMed] [Google Scholar]
- 44.Gong P., Davidson G.A., Gui W., Yang K., Bozza W.P., Zhuang Z. Activity-Based Ubiquitin-Protein Probes Reveal Target Protein Specificity of Deubiquitinating Enzymes. Chem. Sci. 2018;9(40):7859–7865. doi: 10.1039/c8sc01573b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hewings D.S., Heideker J., Ma T.P., AhYoung A.P., El Oualid F., Amore A., Costakes G.T., Kirchhofer D., Brasher B., Pillow T., Popovych N., Maurer T., Schwerdtfeger C., Forrest W.F., Yu K., Flygare J., Bogyo M., Wertz I.E. Reactive-Site-Centric Chemoproteomics Identifies a Distinct Class of Deubiquitinase Enzymes. Nat. Commun. 2018;9(1):1162. doi: 10.1038/s41467-018-03511-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pickart C.M., Rose I.A. Mechanism of Ubiquitin Carboxyl-Terminal Hydrolase. Borohydride and Hydroxylamine Inactivate in the Presence of Ubiquitin. J. Biol. Chem. 1986;261(22):10210–10217. [PubMed] [Google Scholar]
- 47.Hershko A., Rose I.A. Ubiquitin-Aldehyde: A General Inhibitor of Ubiquitin-Recycling Processes. Proc. Natl. Acad. Sci. U. S. A. 1987;84(7):1829–1833. doi: 10.1073/pnas.84.7.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borodovsky A., Kessler B.M., Casagrande R., Overkleeft H.S., Wilkinson K.D., Ploegh H.L. A Novel Active Site-Directed Probe Specific for Deubiquitylating Enzymes Reveals Proteasome Association of USP14. EMBO J. 2001;20(18):5187–5196. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Borodovsky A., Ovaa H., Kolli N., Gan-Erdene T., Wilkinson K.D., Ploegh H.L., Kessler B.M. Chemistry-Based Functional Proteomics Reveals Novel Members of the Deubiquitinating Enzyme Family. Chem. Biol. 2002;9(10):1149–1159. doi: 10.1016/s1074-5521(02)00248-x. [DOI] [PubMed] [Google Scholar]
- 50.Hemelaar J., Borodovsky A., Kessler B.M., Reverter D., Cook J., Kolli N., Gan-Erdene T., Wilkinson K.D., Gill G., Lima C.D., Ploegh H.L., Ovaa H. Specific and Covalent Targeting of Conjugating and Deconjugating Enzymes of Ubiquitin-like Proteins. Mol. Cell. Biol. 2004;24(1):84–95. doi: 10.1128/MCB.24.1.84-95.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ekkebus R., van Kasteren S.I., Kulathu Y., Scholten A., Berlin I., Geurink P.P., de Jong A., Goerdayal S., Neefjes J., Heck A.J., Komander D., Ovaa H. On Terminal Alkynes that Can React with Active-Site Cysteine Nucleophiles in Proteases. J. Am. Chem. Soc. 2013;135(8):2867–2870. doi: 10.1021/ja309802n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Jong A., Witting K., Kooij R., Flierman D., Ovaa H. Release of Enzymatically Active Deubiquitinating Enzymes upon Reversible Capture by Disulfide Ubiquitin Reagents. Angew. Chem. Int. Ed. Engl. 2017;56(42):12967–12970. doi: 10.1002/anie.201706738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iphöfer A., Kummer A., Nimtz M., Ritter A., Arnold T., Frank R., van den Heuvel J., Kessler B.M., Jänsch L., Franke R. Profiling Ubiquitin Linkage Specificities of Deubiquitinating Enzymes with Branched Ubiquitin Isopeptide Probes. ChemBioChem. 2012;13(10):1416–1420. doi: 10.1002/cbic.201200261. [DOI] [PubMed] [Google Scholar]
- 54.Kiick K.L., Saxon E., Tirrell D.A., Bertozzi C.R. Incorporation of Azides into Recombinant Proteins for Chemoselective Modification by the Staudinger Ligation. Proc. Natl. Acad. Sci. U. S. A. 2002;99(1):19–24. doi: 10.1073/pnas.012583299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ohtake F., Saeki Y., Ishido S., Kanno J., Tanaka K. The K48-K63 Branched Ubiquitin Chain Regulates NF-kappaB Signaling. Mol. Cell. 2016;64(2):251–266. doi: 10.1016/j.molcel.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 56.Valkevich E.M., Guenette R.G., Sanchez N.A., Chen Y.C., Ge Y., Strieter E.R. Forging Isopeptide Bonds Using Thiol-Ene Chemistry: Site-Specific Coupling of Ubiquitin Molecules for Studying the Activity of Isopeptidases. J. Am. Chem. Soc. 2012;134(16):6916–6919. doi: 10.1021/ja300500a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dixon E.K., Castañeda C.A., Kashyap T.R., Wang Y., Fushman D. Nonenzymatic Assembly of Branched Polyubiquitin Chains for Structural and Biochemical Studies. Bioorg. Med. Chem. 2013;21(12):3421–3429. doi: 10.1016/j.bmc.2013.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Castañeda C., Liu J., Chaturvedi A., Nowicka U., Cropp T.A., Fushman D. Nonenzymatic Assembly of Natural Polyubiquitin Chains of any Linkage Composition and Isotopic Labeling Scheme. J. Am. Chem. Soc. 2011;133(44):17855–17868. doi: 10.1021/ja207220g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang K., Gong P., Gokhale P., Zhuang Z. Chemical Protein Polyubiquitination Reveals the Role of a Noncanonical Polyubiquitin Chain in DNA Damage Tolerance. ACS Chem. Biol. 2014;9(8):1685–1691. doi: 10.1021/cb500133k. [DOI] [PubMed] [Google Scholar]
- 60.Rösner D., Schneider T., Schneider D., Scheffner M., Marx A. Click Chemistry for Targeted Protein Ubiquitylation and Ubiquitin Chain Formation. Nat. Protoc. 2015;10(10):1594–1611. doi: 10.1038/nprot.2015.106. [DOI] [PubMed] [Google Scholar]
- 61.Singh S.K., Sahu I., Mali S.M., Hemantha H.P., Kleifeld O., Glickman M.H., Brik A. Synthetic Uncleavable Ubiquitinated Proteins Dissect Proteasome Deubiquitination and Degradation, and Highlight Distinctive Fate of Tetraubiquitin. J. Am. Chem. Soc. 2016;138(49):16004–16015. doi: 10.1021/jacs.6b09611. [DOI] [PubMed] [Google Scholar]
- 62.Bondalapati S., Jbara M., Brik A. Expanding the Chemical Toolbox for the Synthesis of Large and Uniquely Modified Proteins. Nat. Chem. 2016;8(5):407–418. doi: 10.1038/nchem.2476. [DOI] [PubMed] [Google Scholar]
- 63.Tang S., Liang L.J., Si Y.Y., Gao S., Wang J.X., Liang J., Mei Z., Zheng J.S., Liu L. Practical Chemical Synthesis of Atypical Ubiquitin Chains by Using an Isopeptide-Linked Ub Isomer. Angew. Chem. Int. Ed. Engl. 2017;56(43):13333–13337. doi: 10.1002/anie.201708067. [DOI] [PubMed] [Google Scholar]
- 64.Paudel P., Zhang Q., Leung C., Greenberg H.C., Guo Y., Chern Y.-H., Dong A., Li Y., Vedadi M., Zhuang Z., Tong Y. Crystal Structure and Activity-Based Labeling Reveal the Mechanisms for Linkage-Specific Substrate Recognition by Deubiquitinase USP9X. PNAS. 2019;116(15):7288–7297. doi: 10.1073/pnas.1815027116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moldovan G.L., Pfander B., Jentsch S. PCNA, the Maestro of the Replication Fork. Cell. 2007;129(4):665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 66.Chen J., Bozza W., Zhuang Z. Ubiquitination of PCNA and Its Essential Role in Eukaryotic Translesion Synthesis. Cell Biochem. Biophys. 2011;60(1–2):47–60. doi: 10.1007/s12013-011-9187-3. [DOI] [PubMed] [Google Scholar]
- 67.Stelter P., Ulrich H.D. Control of Spontaneous and Damage-Induced Mutagenesis by SUMO and Ubiquitin Conjugation. Nature. 2003;425(6954):188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 68.Hoege C., Pfander B., Moldovan G.L., Pyrowolakis G., Jentsch S. RAD6-Dependent DNA Repair Is Linked to Modification of PCNA by Ubiquitin and SUMO. Nature. 2002;419(6903):135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 69.Das-Bradoo S., Nguyen H.D., Wood J.L., Ricke R.M., Haworth J.C., Bielinsky A.K. Defects in DNA Ligase I Trigger PCNA Ubiquitylation at Lys 107. Nat. Cell Biol. 2010;12(1):74–79. doi: 10.1038/ncb2007. sup pp 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Claessen J.H.L., Witte M.D., Yoder N.C., Zhu A.Y., Spooner E., Ploegh H.L. Catch-and-Release Probes Applied to Semi-Intact Cells Reveal Ubiquitin-Specific Protease Expression in Chlamydia Trachomatis Infection. ChemBioChem. 2013;14(3):343–352. doi: 10.1002/cbic.201200701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ward J.A., McLellan L., Stockley M., Gibson K.R., Whitlock G.A., Knights C., Harrigan J.A., Jacq X., Tate E.W. Quantitative Chemical Proteomic Profiling of Ubiquitin Specific Proteases in Intact Cancer Cells. ACS Chem. Biol. 2016;11(12):3268–3272. doi: 10.1021/acschembio.6b00766. [DOI] [PubMed] [Google Scholar]
- 72.Mulder M.P., Witting K., Berlin I., Pruneda J.N., Wu K.P., Chang J.G., Merkx R., Bialas J., Groettrup M., Vertegaal A.C., Schulman B.A., Komander D., Neefjes J., El Oualid F., Ovaa H. A Cascading Activity-Based Probe Sequentially Targets E1-E2-E3 Ubiquitin Enzymes. Nat. Chem. Biol. 2016;12(7):523–530. doi: 10.1038/nchembio.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gui W., Ott C.A., Yang K., Chung J.S., Shen S., Zhuang Z. Cell-Permeable Activity-Based Ubiquitin Probes Enable Intracellular Profiling of Human Deubiquitinases. J. Am. Chem. Soc. 2018;140(39):12424–12433. doi: 10.1021/jacs.8b05147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Herce H.D., Garcia A.E., Cardoso M.C. Fundamental Molecular Mechanism for the Cellular Uptake of Guanidinium-Rich Molecules. J. Am. Chem. Soc. 2014;136(50):17459–17467. doi: 10.1021/ja507790z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nischan N., Herce H.D., Natale F., Bohlke N., Budisa N., Cardoso M.C., Hackenberger C.P. Covalent Attachment of Cyclic TAT Peptides to GFP Results in Protein Delivery into Live Cells with Immediate Bioavailability. Angew. Chem. Int. Ed. Engl. 2015;54(6):1950–1953. doi: 10.1002/anie.201410006. [DOI] [PubMed] [Google Scholar]
- 76.Herce H.D., Schumacher D., Schneider A.F.L., Ludwig A.K., Mann F.A., Fillies M., Kasper M.A., Reinke S., Krause E., Leonhardt H., Cardoso M.C., Hackenberger C.P.R. Cell-Permeable Nanobodies for Targeted Immunolabelling and Antigen Manipulation in Living Cells. Nat. Chem. 2017;9(8):762–771. doi: 10.1038/nchem.2811. [DOI] [PubMed] [Google Scholar]
- 77.Sato Y., Goto E., Shibata Y., Kubota Y., Yamagata A., Goto-Ito S., Kubota K., Inoue J., Takekawa M., Tokunaga F., Fukai S. Structures of CYLD USP with Met1- or Lys63-Linked Diubiquitin Reveal Mechanisms for Dual Specificity. Nat. Struct. Mol. Biol. 2015;22(3):222–229. doi: 10.1038/nsmb.2970. [DOI] [PubMed] [Google Scholar]
- 78.Sato Y., Yoshikawa A., Yamagata A., Mimura H., Yamashita M., Ookata K., Nureki O., Iwai K., Komada M., Fukai S. Structural Basis for Specific Cleavage of Lys 63-Linked Polyubiquitin Chains. Nature. 2008;455(7211):358. doi: 10.1038/nature07254. [DOI] [PubMed] [Google Scholar]
- 79.Mevissen T.E., Hospenthal M.K., Geurink P.P., Elliott P.R., Akutsu M., Arnaudo N., Ekkebus R., Kulathu Y., Wauer T., El Oualid F., Freund S.M., Ovaa H., Komander D. OTU Deubiquitinases Reveal Mechanisms of Linkage Specificity and Enable Ubiquitin Chain Restriction Analysis. Cell. 2013;154(1):169–184. doi: 10.1016/j.cell.2013.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sato Y., Okatsu K., Saeki Y., Yamano K., Matsuda N., Kaiho A., Yamagata A., Goto-Ito S., Ishikawa M., Hashimoto Y., Tanaka K., Fukai S. Structural Basis for Specific Cleavage of Lys6-Linked Polyubiquitin Chains by USP30. Nat. Struct. Mol. Biol. 2017;24(11):911. doi: 10.1038/nsmb.3469. [DOI] [PubMed] [Google Scholar]
- 81.Ye Y., Akutsu M., Reyes-Turcu F., Enchev R.I., Wilkinson K.D., Komander D. Polyubiquitin Binding and Cross-Reactivity in the USP Domain Deubiquitinase USP21. EMBO Rep. 2011;12:350–357. doi: 10.1038/embor.2011.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mevissen T.E.T., Kulathu Y., Mulder M.P.C., Geurink P.P., Maslen S.L., Gersch M., Elliott P.R., Burke J.E., von Tol B.D.M., Akutsu M., Oualid F.E., Kawasaki M., Freund S.M.V., Ovaa H., Komander D. Molecular Basis of Lys11-Polyubiquitin Specificity in the Deubiquitinase Cezanne. Nature. 2016;538(7625):402. doi: 10.1038/nature19836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bekes M., van der Heden van Noort G.J., Ekkebus R., Ovaa H., Huang T.T., Lima C.D. Recognition of Lys48-Linked Di-Ubiquitin and Deubiquitinating Activities of the SARS Coronavirus Papain-Like Protease. Mol. Cell. 2016;62(4):572–585. doi: 10.1016/j.molcel.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]


