Bacteriophages are the most abundant biological entities in the biosphere and are a source of uncharacterized biological mechanisms and genetic tools. Here, we identify segments of phage genomes that are used for stable extrachromosomal replication in the prophage state. Autonomous replication of some of these phages requires a RepA-like protein, although most lack repA and use RNA-based systems for replication initiation. We describe a suite of plasmids based on these prophage replication functions that vary in copy number, stability, host range, and compatibility. These plasmids expand the toolbox available for genetic manipulation of Mycobacterium and other Actinobacteria, including Gordonia terrae.
KEYWORDS: Mycobacterium, bacteriophage genetics, bacteriophages
ABSTRACT
Temperate bacteriophages are common and establish lysogens of their bacterial hosts in which the prophage is stably inherited. It is typical for such prophages to be integrated into the bacterial chromosome, but extrachromosomally replicating prophages have been described also, with the best characterized being the Escherichia coli phage P1 system. Among the large collection of sequenced mycobacteriophages, more than half are temperate or predicted to be temperate, most of which code for a tyrosine or serine integrase that promotes site-specific prophage integration. However, within the large group of 621 cluster A temperate phages, ∼20% lack an integration cassette, which is replaced with a parABS partitioning system. A subset of these phages carry genes coding for a RepA-like protein (RepA phages), which we show here is necessary and sufficient for autonomous extrachromosomal replication. The non-RepA phages appear to replicate using an RNA-based system, as a parABS-proximal region expressing a noncoding RNA is required for replication. Both RepA and non-RepA phage-based plasmids replicate at one or two copies per cell, transform both Mycobacterium smegmatis and Mycobacterium tuberculosis, and are compatible with pAL5000-derived oriM and integration-proficient plasmid vectors. Characterization of these phage-based plasmids offers insights into the variability of lysogenic maintenance systems and provides a large suite of plasmids for actinobacterial genetics that vary in stability, copy number, compatibility, and host range.
INTRODUCTION
Bacteriophages are the most abundant biological entities in the biosphere and are a source of vast genetic diversity (1). Mainly through the Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES) program, more than 17,000 bacteriophages infecting hosts of the phylum Actinobacteria have been isolated, of which more than 3,000 have been sequenced (https://phagesdb.org). These bacteriophages can be sorted into related groups (clusters A, B, C, etc.) according to their overall relatedness (2, 3), and ∼50% of these contain phages that are likely to be temperate, coding for predicted repressor and integrase genes (4). Two classes of integrases have been described—tyrosine and serine integrases—that are used to integrate the phage genome into the host chromosome when establishing lysogeny. This enables the prophage to be passively replicated with the host genome and ensures that a prophage is present in each of the daughter cells after division. Although integration systems are well studied and common among temperate phages (5–7), some temperate phages, including the prototype Escherichia coli phage P1, maintain their prophages extrachromosomally, and carry genes coding for components of partitioning and recombination systems that ensure prophage maintenance (8). “Plasmidial” prophages (9) are relatively uncommon but have been reported for diverse bacteria, including Bacillus anthracis (10), Borrelia burgdorferi (11), Chlamydia pneumoniae (12), and Staphylococcus aureus (9), in addition to P1-family phages (13) and the linearly replicating phage N15 (14). Extrachromosomal prophages are likely underrepresented in genome sequencing projects (15).
Partitioning systems have been reported for cluster A temperate mycobacteriophages, including CRB1 and RedRock (16, 17), that presumably facilitate prophage maintenance (18). RedRock lacks an integrase gene, but carries genes coding for a parABS system and replicates extrachromosomally with a prophage average copy number of 2.4 copies/cell (16). RedRock ParB is a DNA-binding protein and recognizes two parS loci (parS-L and parS-R), each of which contains eight directly repeated copies of an 8-bp motif. RedRock parA and parB are expressed lysogenically as expected, and the parABS cassette stabilizes extrachromosomally replicating shuttle plasmids such as those based on oriM from Mycobacterium fortuitum plasmid pAL5000 (16).
Extrachromosomal maintenance requires a system for initiation of DNA replication. In phage P1, an initiator protein, RepA and an origin of replication is required, as well as ParA and ParB (reviewed in reference 19); related systems have been described for prophages of ΦHAP-1 of Halomonas aquamarina (20), pVv01 of Vibrio vulnificus (21) and lcp3 of Leptospira interrogans (22). Replication initiator protein genes are commonly found in plasmids, and putative replication initiator protein open reading frames (ORFs) have been identified in several naturally occurring mycobacterial plasmids, including plasmids pLR7 (Mycobacterium avium) (23), pJAZ38 (Mycobacterium fortuitum) (24), pCLP (Mycobacterium celatum) (25), and pMF1 (M. fortuitum) (26). The replication cassette commonly used in plasmids for genetic manipulation of mycobacteria (oriM) derives from M. fortuitum plasmid pAL5000, which has a copy number in Mycobacterium smegmatis of about 23 (27). The replication cassette genes encode two proteins that are required for replication, RepA and the DNA-binding protein, RepB (28). Mycobacteriophage DNA replication systems are not well-characterized for either lytic growth or extrachromosomal replication.
Many different plasmid replication systems have been described (29), and although most require an initiator protein, some require only RNA products for initiation and copy number regulation. The most common example is the replication cassette of E. coli plasmid ColE1, the basis for many plasmids used in recombinant DNA, including pBR322, pUC18/19, and their derivatives (30). In these systems, an RNA molecule (RNA II) acts as a primer for DNA replication by the host DNA polymerase I (Pol I), and a second RNA (RNA I) modulates its activity to determine copy number (31). We previously suggested that the parABS mycobacteriophage RedRock might use an RNA-based replication system for prophage replication, as a noncoding RNA is expressed adjacent to the parABS cassette, and no repA homologue or similar gene in the genome was identified (16). However, we were not able to demonstrate autonomous replication by a DNA cassette containing this region. In contrast, mycobacteriophage CRB1 codes for a putative RepA protein (17).
Here, we characterize the prophage origins of replication for eight temperate mycobacteriophages: Miko, Rachaly, Jeeves, RedRock, Alma, Gladiator, Et2Brutus, and LadyBird. Miko, Rachaly, and Jeeves prophages initiate replication with a RepA-like replication initiator protein, but RedRock, Alma, Gladiator, Et2Brutus, and LadyBird use an initiator RNA, and to our knowledge, these are first RNA-based prophage replication systems. Plasmids that include these origins vary in copy number, retention without selection, and compatibility in M. smegmatis mc2155, and differ in functionality in Mycobacterium tuberculosis and Gordonia terrae. Plasmids based on these prophage origins broaden the suite of tools available for genetic manipulation of Actinobacteria.
RESULTS
Spectrum of extrachromosomally replicating actinobacteriophage prophages.
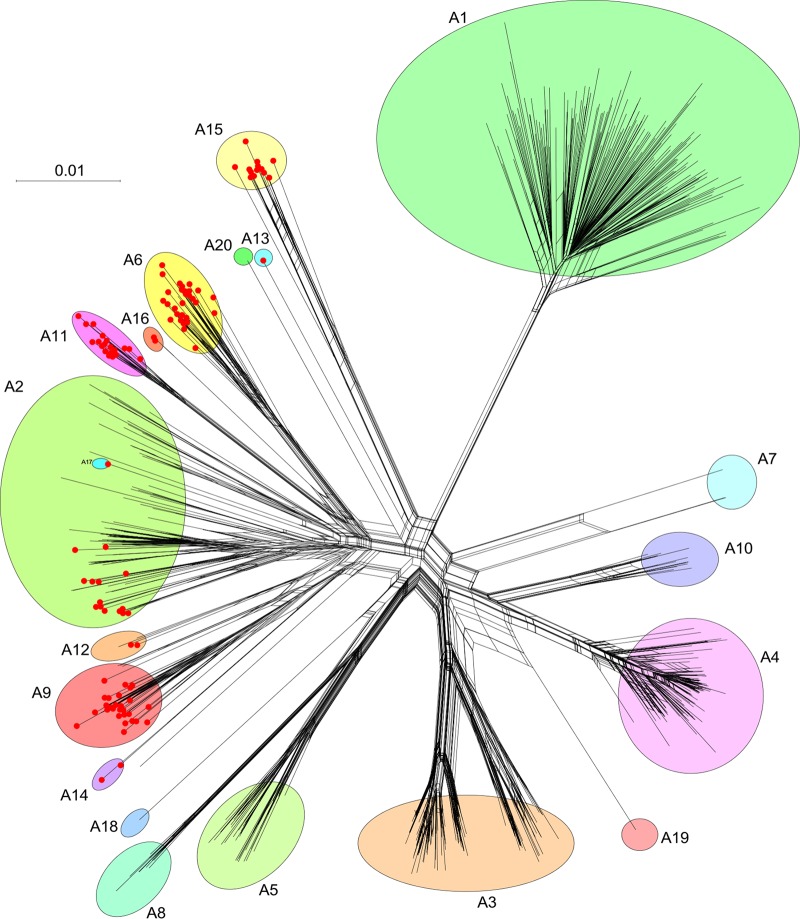
The number of sequenced actinobacteriophages has increased substantially over the past 5 years, including a 3.5-fold increase in the number of cluster A phages (32). Reexamination of the sequenced genomes reveals an expanded set of 110 “parABS” phages (see Table S1 in the supplemental material) grouped in the large 621-member cluster A. However, the cluster A phages are a highly diverse group, and they can be divided into 20 subclusters (Fig. 1). The largest subcluster, A1, is devoid of parABS phages, and all phages contain either a tyrosine or serine integrase (33); there are also no parABS phages in subclusters A3, A4, A5, A7, A8, A10, A18, A19, or A20 (Fig. 1). In contrast, all of the members of subclusters A6, A11, A13, A14, A15, A16, and A17 have a parABS system, together with 27 of the 31 subcluster A9 phages, two of the four subcluster A12 phages, and 15 of the 90 subcluster A2 phages (Fig. 1). Of the 110 parABS phages, 97 infect Mycobacterium smegmatis mc2155, and 13 infect Gordonia terrae 3612 (https://phagesdb.org), although all of the Gordonia parABS phages are in subcluster A15 (Fig. 1). In all 110 phages, the parABS cassette is centrally located in the viral genome, in a colinear position to the integration cassettes of closely related genomes (Fig. 2A). All of the parA proteins appear to be homologues and are grouped into a single protein “phamily” (3). However, there is considerable diversity among the parB proteins, which fall into at least four distinct protein phamilies. Three of these are represented in the 15 subcluster A2 parAB phages, and it is likely that parB is under selection to diversify to avoid incompatibility.
FIG 1.
Network phylogeny of cluster A mycobacteriophages. A network phylogeny of 621 cluster A mycobacteriophages was constructed based on gene content and represented using Splitstree (58). A database “Actino_Draft” dated 9 December 2019 was used in which predicted gene products were sorted into groups (phamilies) of related sequences as described previously (1, 59) (C. Gauthier and G. F. Hatfull, unpublished data), and a nexus-formatted file generated using the custom script “PhamNexus.” Colored circles illustrate the 20 subclusters (A1 to A20), and red dots at nodes indicate phages carrying parABS systems. The bar indicates the number of substitutions per site.
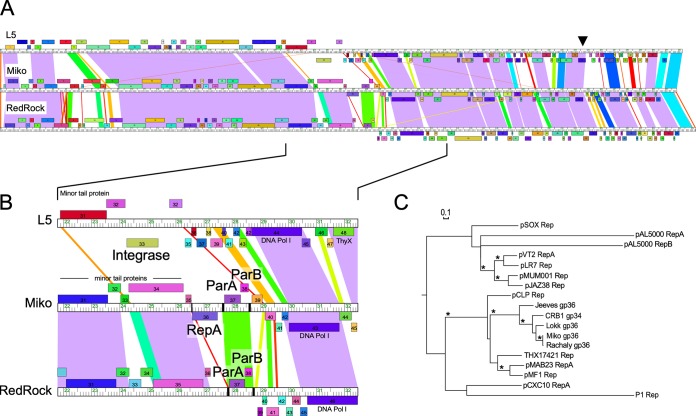
FIG 2.
Alignment of parABS phages used in this study. (A) Representative RepA (Miko) and non-RepA (RedRock) phage genomes were aligned to that of the integrating phage L5. The ruler shows genome length in kilobases, while ORFs shown above and below the ruler are transcribed rightward and leftward, respectively. ORFs of the same color have been assigned to the same gene phamily. A black arrowhead indicates the location of the immunity repressor gene in all three genomes. Shared nucleotide sequence similarity is represented as spectrum-colored shading, with violet representing the most similar and red the least similar above a BLASTN E-value threshold of 10−4. (B) Genome segments containing genes relevant for prophage replication and maintenance. Where present, integrase (L5), homologues to repA (Miko), parA and parB (Miko and RedRock) are labeled. The locations of centromere-like parS sites are indicated in the Miko and RedRock genome rulers with black bars. DNA Pol I, DNA polymerase I. (C) Relatedness of the phage RepA proteins to replication initiator proteins found in actinobacterial plasmids and the E. coli phage P1, shown as a phylogeny generated by maximum likelihood (PhyML). Bootstrap values greater than 70% are indicated with an asterisk, and the bar indicates the number of substitutions per site.
Cluster A actinobacteriophages that contain parABS. Download Table S1, DOCX file, 0.02 MB (23KB, docx) .
Copyright © 2020 Wetzel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Identification of putative prophage origins of replication.
In the parABS phages, substitution of the integrase functionality requires not only parABS partitioning functions but also the functions needed to promote the initiation of extrachromosomal DNA replication and copy number control. Genome alignments suggest that the replication and partitioning functions of the parABS phages must be closely linked and centrally located, downstream of the virion structure and assembly genes (Fig. 2). Using genome alignments, we identified two subsets of parABS phages. First, the “RepA” phages which carry genes encoding homologues of RepA plasmid initiation proteins located to the left of the parABS cassette, and second, the “non-RepA” phages for which RepA homologues have not been identified (Table S1). The non-RepA phages, including RedRock, represent the vast majority of parABS phages (95%; 105/110) and have representatives in each of the subclusters containing parABS phages. Five phages code for a RepA-like protein, four in subcluster A2 (Miko, Rachaly, Lokk, and CRB1), and one (Jeeves) of the two subcluster A14 parABS phages (Fig. 1; Table S1). In each of these, repA is transcribed in the opposite direction to both parABS and the virion structure and assembly genes (Fig. 2).
The five phage-encoded RepA-like proteins are 320 to 350 residues long and share ∼40% conserved amino acid residues. Miko and Rachaly RepA are very closely related (98% amino acid identity), but distantly related to Jeeves RepA, to which they share only ∼50% amino acid identity. Database searches strongly support the functional assignment of these phage-encoded RepA proteins. For example, Jeeves RepA is related to a plasmid-encoded RepA in Mycobacterium abscessus plasmid pMAB23 (34) (46% identity over 250 residues), as well as RepA proteins found in mycobacterial plasmids of the pMSC262 family, such as plasmid pCLP (Mycobacterium celatum) and plasmid pMF1 (M. fortuitum) (25, 26). Related proteins are also present in genome assemblies of several Mycobacterium species, including M. abscessus, Mycobacterium cosmeticum, and Mycobacterium tusciae (e.g., NCBI:protein accession number TXH17421; Fig. 2C). The mycobacteriophage RepA proteins are not closely related to RepA proteins of pMSC262 family members pLR7 and pJAZ38 (23, 24, 35) or to RepA and RepB encoded by pAL5000 (36). The phage RepA proteins contain an N-terminal predicted helix-turn-helix DNA-binding domain (Miko residues 95 to 132), a common feature of plasmid replication initiator proteins (31).
Well-characterized plasmid replication proteins typically bind to either direct repeat sequences (“iterons,” as in RepA of phage P1 [37]), palindromic sequences (e.g., RepB of pAL5000 oriM), or conserved sequence motifs (e.g., plasmids pMF1, pCLP, pJAZ38, pLR7, and pMSC262) at the origin of replication; these are typically tightly linked to the repA gene (26, 31). We have not identified repeated sequence motifs in the regions between the virion tail genes and parS-L but note that nucleotide sequence conservation extends ∼50 bp upstream of the repA gene in Rachaly, Miko, Lokk, and CRB1. It thus seems likely that the origin of replication lies either immediately upstream of repA or within the repA gene itself, similar to the position of the phage lambda origin within the o gene (38, 39). We note that the RepA phages Miko and Rachaly differ from the previously characterized non-RepA parABS phages (RedRock, Alma, Et2Brutus, Gladiator, and LadyBird [16]) in having an additional parS locus containing five direct repeats, immediately downstream of the repA gene (see Fig. 5). In phages Lokk and CRB1, there is no intergenic space between the 3′ ends of repA and the adjacent rightward-transcribed gene and no additional parS site. We note that Jeeves, Lokk, and CRB1 appear to lack parS-R (see Fig. 5), which had been observed for several other parABS phages (16).
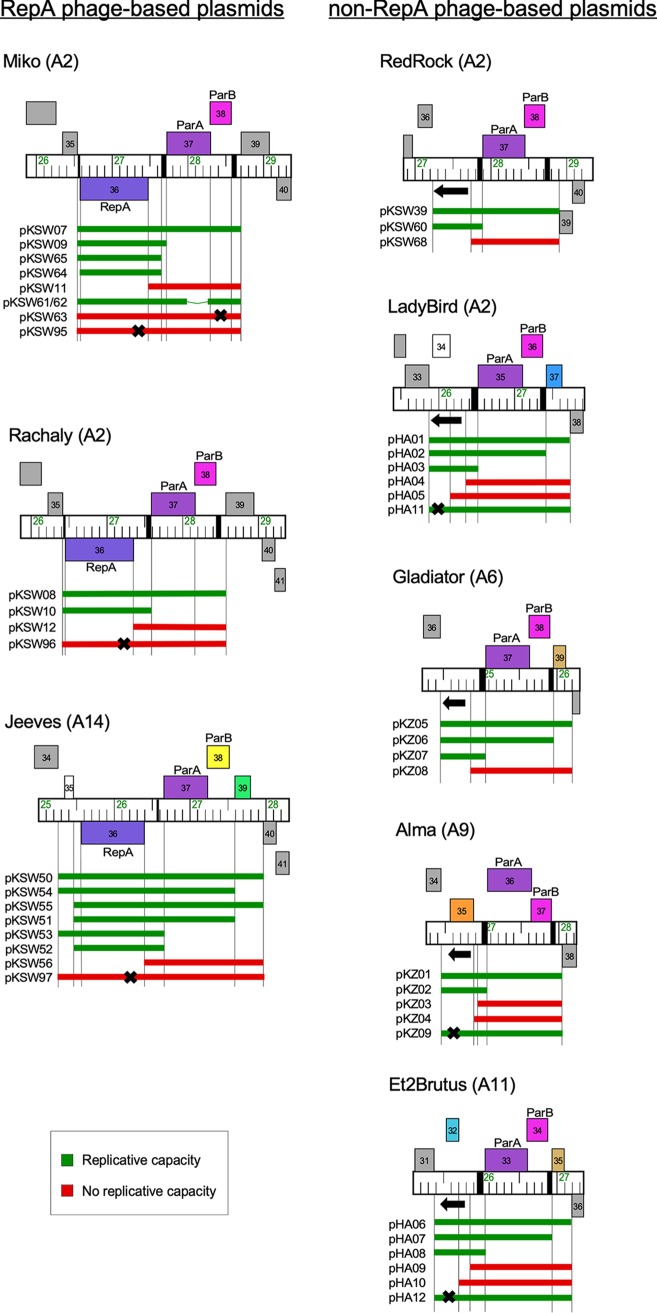
FIG 5.
Phage genome segments that support autonomous replication. Segments of eight phage genome maps are shown with relevant genes labeled; the locations of parS repeats are indicated by black boxes on the genome ruler. A black arrow indicates the location and transcription direction of the noncoding RNA implicated in replication initiation. Bars underneath each map indicate a genome segment inserted into the nonreplicating vector pMOS-Hyg and then electroporated into M. smegmatis mc2155. Replication-proficient plasmids efficiently transforming M. smegmatis (>104 CFU/μg DNA) are shown in green, and those that fail to transform are shown in red. A black X indicates the position of a stop codon introduced by mutagenesis.
Copy numbers of non-RepA prophages.
We previously determined that the prophage copy number in a lysogen of the non-RepA phage RedRock was 2.4 copies/cell (16); here we extended this analysis to include additional non-RepA lysogens of Alma, Et2Brutus, and LadyBird. DNA was extracted from lysogens and sequenced, and the ratio of sequence reads mapping to the bacterial chromosome and the prophage genome was calculated (Table 1). Because the sample also contains phage DNA from spontaneous lytic induction, the proportion of prophage-derived reads was derived from the ratio of sequence reads traversing the cohesive ends (i.e., from prophages) relative to those corresponding to genome cleaved at cos during packaging (i.e., from packaged genomes). The adjusted prophage copy numbers were 4.8, 3.7, and 2.5 for Alma, Et2Brutus, and LadyBird, respectively (Table 1). These copy numbers may be slightly overestimated, because some reads across genome ends could be derived from unpackaged concatemers during lytic replication. However, ratios of ligated to cleaved cos sites as high as 3:1 (Et2Brutus; Table 1) are unlikely to solely indicate lytic growth and are more consistent with extrachromosomally replicating prophages. We were not able to measure copy numbers for prophages of Miko, Rachaly, and Jeeves due to somewhat higher levels of spontaneous lytic induction.
TABLE 1.
Copy numbers of extrachromosomal prophages
| Strain(phage) | Phage readsa |
Precise end readsb |
End- spanning readsc |
Total end reads |
End- spanning reads: total end reads |
Coverage |
Phage/host coverage |
||
|---|---|---|---|---|---|---|---|---|---|
| Phage | mc2155 | Raw | Correctedd | ||||||
| mc2155(Alma) | 59,386 | 41 | 46 | 87 | 0.53 | 167.5 | 18.5 | 9.1 | 4.80 |
| mc2155(Et2Brutus) | 32,465 | 11 | 35 | 46 | 0.76 | 92.8 | 19.2 | 4.8 | 3.69 |
| mc2155(LadyBird) | 27,265 | 11 | 19 | 30 | 0.63 | 77.0 | 19.1 | 4.0 | 2.55 |
Sequence reads mapping to the phage genome out of a total of ∼1 million per sample.
Sequence reads beginning at precisely the terminus of viral genomic DNA.
Sequence reads that span the predicted 5′ and 3′ ends of the genomes.
The ratio of phage/host genome coverage multiplied by the end-spanning reads:total end reads.
Miko repA is required for prophage replication.
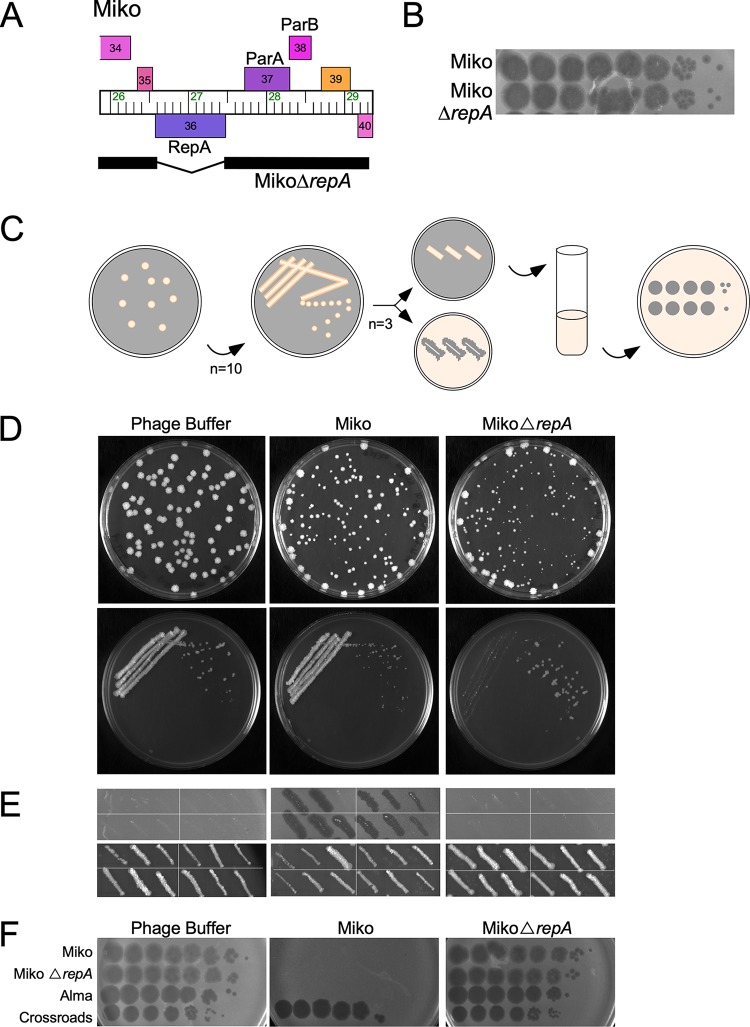
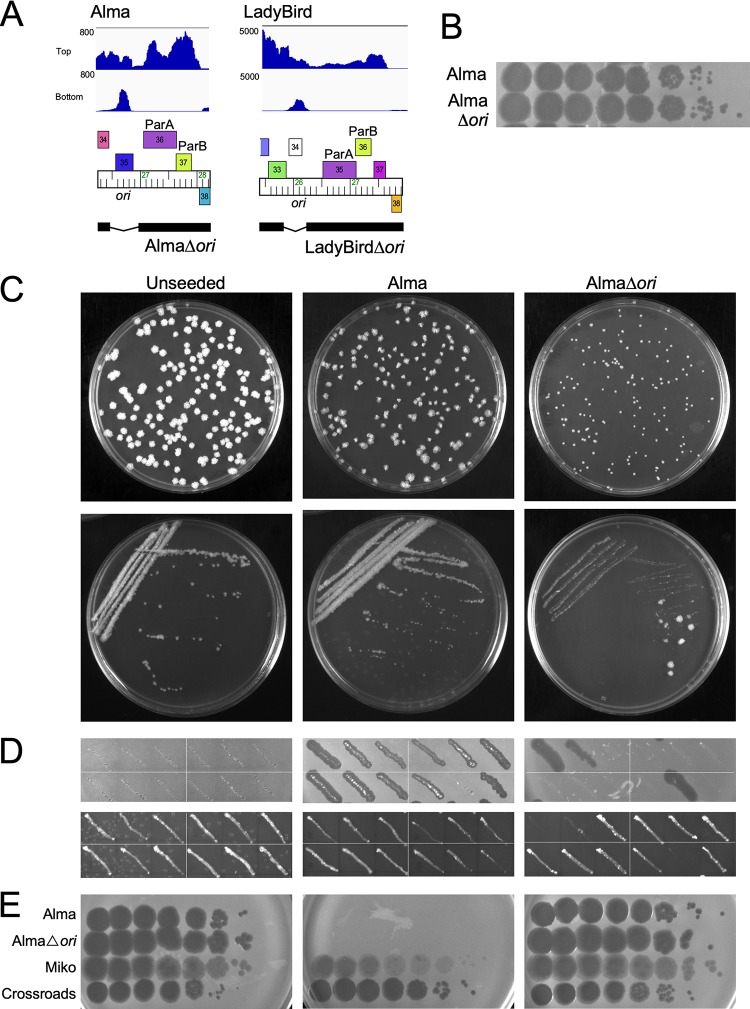
To determine whether repA of phage Miko is required for prophage replication, we reasoned that repA deletion would have little or no effect on lytic growth but would lead to reduced lysogenic stability. A deletion derivative (MikoΔrepA) was constructed using an adaptation of bacteriophage recombineering of electroporated DNA (BRED) engineering (40) (Fig. 3A) and appears unaltered in its lytic properties; it can be readily propagated to high titer and has a plaque morphology similar to that of its parent phage (Fig. 3B). To test for lysogeny, a liquid culture of M. smegmatis mc2155 was diluted, and colonies were recovered on solid media seeded with Miko or MikoΔrepA (Fig. 3C). Similar numbers of colonies were recovered on Miko-seeded medium as with a buffer control, reflecting a high lysogenization frequency under these experimental conditions (Table 2). To confirm that the recovered derivatives are lysogenic for Miko, 10 individual colonies were restreaked to remove phage particles carried over from the selection plate and tested for phage release and superinfection immunity (Fig. 3D); nearly all (28/30) of the individual colonies picked from the restreaks spontaneously released phage (Fig. 3E; Table 2), and a tested subset were all immune to superinfection (Fig. 3F). Interestingly, a similar number of survivors was recovered on MikoΔrepA-seeded plates consistent with a high rate of lysogenization, although the colonies were smaller than those on the Miko-seeded plates, reflecting slowing growth (Table 2). When restreaked, the pattern of growth is distinctly different from those taken from the Miko-seeded plates (Fig. 3D); the densest part of the streak fails to grow well—presumably due to phage carryover and lytic phage replication—and the single colonies recovered are not lysogens when retested for phage release and immunity (Fig. 3E and F; Table 2). These data suggest that Miko repA is not required for establishment of lysogeny and immunity, but that it is required for stable lysogeny and prophage inheritance.
FIG 3.
RepA is necessary to form stable lysogens of Miko. (A) Genome of MikoΔrepA mutant phage. Miko was engineered to remove repA, and the black bar shows the retained region (deletion coordinates 26597 to 27466). (B) Titer and plaque morphology of MikoΔrepA mutant phage. Lysates of Miko and MikoΔrepA were 10-fold serially diluted and plated onto a lawn of M. smegmatis mc2155. (C) Scheme to characterize MikoΔrepA lysogens. Lysogens were recovered by plating exponentially growing M. smegmatis mc2155 onto solid media seeded with phage buffer, Miko, or MikoΔrepA. Ten individual colonies were streaked onto solid media to remove phage particles carried over from the selection plate. Three colonies from each streak plate were patched onto solid media and M. smegmatis lawns to test for spontaneous phage release. Liquid cultures were grown from these patches to test for phage superinfection. (D) Plates seeded with phage buffer, Miko, or MikoΔrepA (top) and representative streaks from colonies grown on the plates (bottom). Larger colonies were recovered at the edges of the seeded plates where phage particles are likely less abundant were avoided. (E) Spontaneous phage release from four representative colonies grown in the presence of phage buffer, Miko, or MikoΔrepA. None of the colonies recovered on media with phage buffer or MikoΔrepA released phages and are not stably lysogenic; at least two of the three purified streaks from colonies recovered on Miko-seeded plates are lysogenic and release phage particles. (F) Susceptibility of colonies to phage superinfection. Liquid cultures were grown from patches of 6 of the 10 colonies from the seeded plates and tested for their susceptibility to Miko, MikoΔrepA, Alma, and a control phage Crossroads (L2). A representative example of each is shown.
TABLE 2.
Lysogens of MikoΔrepA and AlmaΔori
| Phage or control | No. of colonies/plate |
% frequency of colony formationa |
Phage release from patch (3 per original colony)b |
Colonies yielding ≥1 patch with phage releasec |
|---|---|---|---|---|
| Miko | 91 | 128 | 28 | 10 |
| Miko△repA | 100 | 140 | 0 | 0 |
| Phage buffer | 71 | 100 | 0 | 0 |
| Alma | 154 | 112 | 29 | 10 |
| Alma△ori | 158 | 114 | 11 | 7 |
| Unseeded | 138 | 100 | 0 | 0 |
Relative to the colony recovery using only phage buffer.
Ten colonies were picked from the seeded plates and restreaked, and three colonies of each were tested for phage release and immunity. The numbers of colonies of the 30 total colonies releasing phage are shown.
The proportions of each of the original 10 colonies restreaked from seeded plates of which at least one of the three retested colonies released phage.
Role of the putative replication origin of phages Alma and LadyBird in lysogeny.
Phages Alma and LadyBird lack a repA gene, but transcriptome sequencing (RNA-Seq) data for both show expression of RNA immediately upstream of parA, but in the reverse direction (Fig. 4A; see also Fig. S1 in the supplemental material). This RNA does not correspond to a predicted ORF, as we reported previously for several other non-RepA phages (16). We note that some of these phages have predicted ORFs in the forward direction that overlap with the noncoding RNA, a subset of which (e.g., Alma 35) are also present in integrase-encoding cluster A phages and are unlikely to be involved in extrachromosomal replication (Fig. 4A). To explore whether these regions are required for prophage maintenance, we constructed deletion derivatives of Alma and LadyBird (AlmaΔori and LadyBirdΔori, respectively) in which these transcribed regions (defined here as ori) are removed (Fig. 4A). Both derivatives have normal lytic growth and amplify to high titer (Fig. 4B; data not shown). Using a similar approach to that described above for Miko (Fig. 3C), AlmaΔori appears unaltered in its lysogenic establishment, and similar numbers of M. smegmatis colonies were recovered on Alma- and AlmaΔori-seeded plates (Table 2); however, the AlmaΔori-derived colonies are small and very slow growing compared to Alma-derived colonies (Fig. 3C). When the AlmaΔori-derived colonies were restreaked, the densest part of the streak failed to grow (as seen for Miko [Fig. 3]), but a mixture of very small and larger isolated colonies were observed (Fig. 4C). Upon further testing, most of the large colonies were nonlysogenic, whereas the small colonies released phage and appeared to be lysogens (Fig. 4D and E; Table 2); nonlysogenic derivatives were recovered only rarely using wild-type Alma. These data suggest that all or part of the deleted region is required for prophage replication. Attempts to recover lysogens from regions where Alma and AlmaΔori phages were spotted on M. smegmatis lawns support similar conclusions (Fig. S2). Although neither LadyBird nor LadyBirdΔori lysogens could be recovered from phage-seeded plates, streaking from infected areas of M. smegmatis lawns yielded results similar to the results with Alma, and the LadyBirdΔori survivors are not stably lysogenic (Fig. S2). We conclude that these small transcribed regions are required for prophage stability.
FIG 4.
ori is necessary to form stable lysogens of Alma. (A) Genomes of Δori mutant phages. Phages were engineered to delete the noncoding RNA region (labeled ori) expressed by the prophage (Alma, LadyBird). The black bar below the genome map segment shows the retained regions. The coordinates of the deleted regions are 26463 to 26959 and 25900 to 26299 for Alma and LadyBird, respectively. Strand-specific RNA-Seq reads are aligned above the Alma and LadyBird maps. (B) Titer and plaque morphology of AlmaΔori. Lysates of Alma and AlmaΔori were 10-fold serially diluted and plated onto a lawn of M. smegmatis mc2155. (C) Characterization of AlmaΔori lysogens (akin to Fig. 3C). Lysogens were recovered by plating serial dilutions of exponentially growing M. smegmatis mc2155 onto unseeded solid media or solid media seeded with Alma or AlmaΔori (Top). Ten (Alma, AlmaΔori) or six (unseeded) individual colonies were streaked onto solid media to remove phage particles carried over from the selection plate (bottom) (representative shown). (D) Spontaneous phage release from colonies grown in the presence of AlmaΔori. Three colonies from each streak plate were patched onto solid media and M. smegmatis lawns to test for spontaneous phage release. None of the colonies recovered on unseeded media released phage, but at least two of the three colonies from purified streaks from colonies recovered on Alma-seeded plates are lysogenic and release phage particles. Some patches originating from AlmaΔori-seeded plates released phage, while others did not. Patches that did not release phage grew well on solid media, while patches that did release phage grew poorly on solid media. (E) Susceptibility of colonies to phage superinfection. Liquid cultures were grown from patches of 6 of the 10 colonies from the seeded plates and tested for their susceptibilities to Alma, AlmaΔori, Miko and a control phage Crossroads (L2). A representative example of each is shown.
Transcriptomic profile of the LadyBird lysogen. RNA was isolated from a LadyBird lysogen in the log phase of growth, and libraries were generated and sequenced using the Illumina platform. The reads per base are plotted along a Phamerator map of LadyBird, with reads aligning to the top strand shown in blue and reads mapping to the bottom strand shown in red. Robust expression of the immunity repressor gene (79) and parAB (35 and 36) is observed, as well as the opposite strand RNA proximal to parAB (labeled “ori”). Low expression of left-arm structural genes indicates some spontaneous induction. Download FIG S1, PDF file, 0.2 MB (181.2KB, pdf) .
Copyright © 2020 Wetzel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ori is necessary to form stable lysogens of Alma and LadyBird. (A) Representative streaks (n = 3) from areas of infection of Alma (right) and AlmaΔori (left) on an M. smegmatis lawn. (B) Spontaneous phage release from three individual colonies per streak. Individual colonies were patched onto solid media (left) and an M. smegmatis lawn (center). The original infecting phage per streak plate is shown as a grid (right). (C) Susceptibility of colonies to phage superinfection. Liquid cultures were grown from one patch per original area of infection and tested for its susceptibility to Alma, AlmaΔori, Miko (A2), and a control phage Crossroads (L2). A representative example of each is shown. (D) Representative streaks (n = 3) from areas of infection of LadyBird (right) and LadyBirdΔori (left) on an M. smegmatis lawn. (E) Spontaneous phage release from three individual colonies per streak. Individual colonies were patched onto solid media (left) and an M. smegmatis lawn (center). The original infecting phage per streak plate is shown as a grid (right). (F) Susceptibility of colonies to phage superinfection. Liquid cultures were grown from one patch per original area of infection and tested for its susceptibility to LadyBird, LadyBirdΔori, Alma (A9), and a control phage Crossroads (L2). A representative example of each is shown. Download FIG S2, PDF file, 0.9 MB (897.4KB, pdf) .
Copyright © 2020 Wetzel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phage RepA and non-RepA origins support extrachromosomal autonomous replication.
To further characterize the phage components required for autonomous replication, we constructed a series of recombinant plasmids carrying segments of RepA phages Miko, Rachaly, and Jeeves and segments of the non-RepA phages, RedRock, LadyBird, Gladiator, Alma, and Et2Brutus, into a vector (pMOS-Hyg) incapable of replicating in M. smegmatis. For the RepA phages, initial recombinant plasmids contained the regions encompassing repA, parA, parB, and included the parS sites (Fig. 5). For the non-RepA phages, the initial recombinants included parA, parB, the parS sites, as well as the ∼600-bp region upstream of parA carrying the putative ori, and one or two of the closely linked ORFs (Fig. 5). All of the plasmids (pKSW07, pKSW08, pKSW50, pKSW39, pHA01, pKZ05, pKZ01, and pHA06, carrying segments from phages Miko, Rachaly, Jeeves, RedRock, LadyBird, Gladiator, Alma, and Et2Brutus, respectively) were able to transform M. smegmatis mc2155 with efficiencies similar to those for a control plasmid (pCCK38) containing oriM (Table 3). We note that the efficient transformation of M. smegmatis with the RedRock-derived plasmid pKSW39 (Fig. 5) differs from prior reports that a similar phage DNA fragment did not support extrachromosomal replication (16). The primary differences between these constructs is the orientation of the RedRock insert in relation to distinct antibiotic resistance genes (against hygromycin instead of kanamycin), suggesting that the juxtaposition of vector sequences can have a strong impact on replicon functionality.
TABLE 3.
Transformation efficiencies of phage-based plasmids in M. smegmatis
| Plasmid | Phagea | Feature(s) | Transformation efficiency (CFU/μg of DNA) |
|---|---|---|---|
| pCCK38 | N/A | oriM | 1.2 × 105 |
| pHA06 | Et2Brutus | ori + parABS | 1.1 × 105 |
| pHA08 | Et2Brutus | ori | 3.2 × 104 |
| pKZ05 | Gladiator | ori + parABS | 9.6 × 104 |
| pKZ07 | Gladiator | ori | 1.2 × 105 |
| pHA01 | LadyBird | ori + parABS | 6.5 × 104 |
| pHA03 | LadyBird | ori | 1.2 × 105 |
| pKZ01 | Alma | ori + parABS | 1.0 × 105 |
| pKZ02 | Alma | ori | 1.3 × 105 |
| pKSW39 | RedRock | ori + parABS | 8.9 × 104 |
| pKSW60 | RedRock | ori | 6.6 × 104 |
| pKSW07 | Miko | repA + parABS | 9.0 × 104 |
| pKSW09 | Miko | repA | 1.0 × 105 |
| pKSW08 | Rachaly | repA + parABS | 9.7 × 104 |
| pKSW10 | Rachaly | repA | 8.1 × 104 |
| pKSW50 | Jeeves | repA + parABS | 1.1 × 105 |
| pKSW52 | Jeeves | repA | 8.4 × 104 |
| pMOS-Hyg | N/A | None | 0 |
N/A, not applicable.
To determine whether the parABS partitioning systems are required for autonomous replication, we constructed deletion derivatives of the parental plasmids in which parA and parB are removed (Fig. 5). All three of the RepA phage-derived plasmids and all five of the non-RepA phage-derived plasmids lacking parABS efficiently transformed M. smegmatis, and the parABS cassettes are clearly not required for autonomous replication (Fig. 5). These and similar plasmids in which one or more of the flanking ORFs are removed similarly show that these are also not required for replication (Fig. 5).
For the three RepA phages, repA and the flanking intergenic regions are sufficient for replication of plasmids pKSW09, pKSW10, and pKSW52, derived from Miko, Rachaly, and Jeeves, respectively (Fig. 5). We further characterized these by constructing additional derivatives and testing their ability to transform M. smegmatis. These experiments showed both that the parS sites are not required (e.g., pKSW64 for Miko) and that interruption of the repA open reading frame by introduction of an early translation termination codon (in plasmids pKSW95, pKSW96, and pKSW97 for Miko, Rachaly, and Jeeves, respectively) eliminates the replication capacity (Fig. 5); reversion to the wild-type repA sequence restored transformation ability (data not shown). The parABS systems alone in the absence of repA do not support replication, as expected (Fig. 5) (16). For Miko, the minimum segment shown to support replication contains repA and 171 bp of upstream sequence (Fig. 5).
For the non-RepA phages, plasmids carrying ∼600 bp to the left of parA transform M. smegmatis efficiently (Table 3) and autonomously replicate. In two of these (plasmids pKSW60 and pKZ07 from RedRock and Gladiator, respectively), there are no predicted ORFs, whereas phages LadyBird, Alma, and Et2Brutus have a predicted rightward-transcribed ORF in this region (genes 34, 35, and 32, respectively). Removal of regions containing these ORFs results in loss of transformation (Fig. 5), but the ORFs themselves are not required, because introduction of early translation termination codons does not prevent replication, although the transformants grow somewhat slower than their parental counterparts (data not shown). The reason for reduced growth of these mutant plasmids is unclear, but reversion back to the wild-type sequence restored normal transformation and colony growth. Together, these observations are consistent with the conclusion that the non-RepA phages do not require protein products for replication and that they use RNAs to initiate autonomous replication.
The five non-RepA phage origin of replication regions (defined by the ∼600-bp regions upstream of parA sufficient for autonomous replication) vary in relatedness at the nucleotide level (Table 4). Alma and LadyBird are the most similar with ∼71% average nucleotide identity (ANI), and additional pairwise comparisons between RedRock, Alma, Gladiator, and LadyBird range from 62% to 65% ANI (Table 4). The region in Et2Brutus is more distantly related and has between 41% (RedRock) and 49% (Gladiator) ANI. Moreover, there is no open reading frame shared between these phages that is conserved and could potentially be involved in replication.
TABLE 4.
Percent nucleotide identity of putative replication origins of non-RepA phages
| Phage (subcluster) | Coordinates | % nucleotide identity |
||||
|---|---|---|---|---|---|---|
| Et2Brutus | Gladiator | RedRock | LadyBird | Alma | ||
| Et2Brutus (A11) | 25381−26052 | 100 | ||||
| Gladiator (A6) | 24464−25062 | 49.0 | 100 | |||
| RedRock (A2) | 27232−27897 | 41.4 | 62.3 | 100 | ||
| LadyBird (A2) | 25875−26522 | 48.9 | 62.5 | 64.4 | 100 | |
| Alma (A9) | 26448−27060 | 44.0 | 62.5 | 65.5 | 71.0 | 100 |
Roles of parABS in plasmid maintenance.
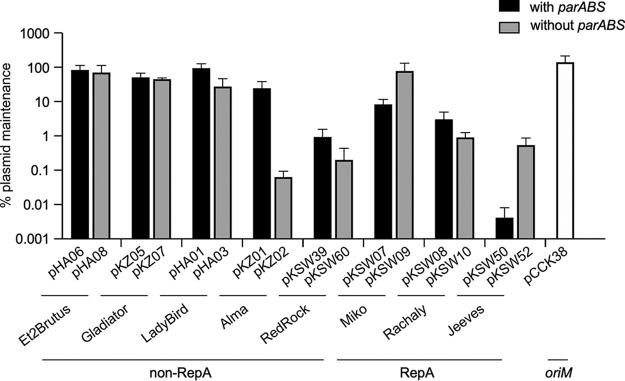
The parABS cassette is not necessary for autonomous replication of any of the plasmids tested here, but it is likely required for plasmid maintenance as described for the RedRock parABS cassette (16); it also could play a regulatory role in replication. To further explore the roles of parABS, we measured the stability of autonomously replicating plasmids and the impact of removal of parABS (Fig. 6). Somewhat surprisingly, the stability of plasmids containing parABS varied substantially in the absence of selection, varying from being well-maintained (LadyBird) to very unstable (Jeeves). For two of the non-RepA phage derivatives (from Ladybird and Alma), removal of parABS resulted in increased plasmid loss as expected, but there was little impact on those derived from Et2Brutus or Gladiator (Fig. 6).
FIG 6.
Maintenance of plasmids without selection. M. smegmatis transformants with plasmids (as indicated) were grown in liquid culture with selection to saturation and then serially passaged for a total of ∼40 generations without selection. The percentage of plasmid maintenance was determined by plating serial dilutions of culture on solid media with and without selection. Data represent the mean values from four independent cultures, and error bars represent one standard deviation.
Surprisingly, plasmids pKSW07 and pKSW50 (derived from RepA phages Miko and Jeeves, respectively) are not only very poorly maintained, but removal of parABS results in substantial increases in plasmid retention (Fig. 6). We reasoned that a plausible explanation for this paradox is that plasmid copy numbers may have changed from regulatory consequences of removing parABS. To determine this, the plasmid copy numbers were measured by whole-genome sequencing of bacterial cultures grown with antibiotic selection (Table 5). We found that the phage-based plasmids with the parABS cassettes had copy numbers ranging from 0.4 to 2.1, with the RepA phage-based plasmids having 0.4 to 0.8 copies/cell, and the non-RepA phage-based plasmids having one or two copies/cell (Table 5). Cultures carrying plasmid pKSW50 grow notably slower than other cultures (data not shown), which is likely related to its low average copy number (0.4 copies/cell), reflecting a substantial proportion of nonviable cells when growing in the presence of antibiotic. When parABS is removed from the plasmids, copy numbers vary widely, from 0.8/cell (RedRock) to 16.4/cell (Et2Brutus) (Table 5).
TABLE 5.
Copy number of phage-based plasmids
| Plasmid | Phage | Feature(s) | Copy no. |
|---|---|---|---|
| pCCK38 | N/A | oriM | 15.6 |
| pHA06 | Et2Brutus | ori + parABS | 2.1 |
| pHA08 | Et2Brutus | ori | 16.4 |
| pKZ05 | Gladiator | ori + parABS | 1.8 |
| pKZ07 | Gladiator | ori | 7.6 |
| pHA01 | LadyBird | ori + parABS | 1.9 |
| pHA03 | LadyBird | ori | 5.4 |
| pKZ01 | Alma | ori + parABS | 1.2 |
| pKZ02 | Alma | ori | 1.8 |
| pKSW39 | RedRock | ori + parABS | 1 |
| pKSW60 | RedRock | ori | 0.8 |
| pKSW07 | Miko | repA + parABS | 0.8 |
| pKSW09 | Miko | repA | 11.7 |
| pKSW08 | Rachaly | repA + parABS | 0.8 |
| pKSW10 | Rachaly | repA | 2 |
| pKSW50 | Jeeves | repA + parABS | 0.4 |
| pKSW52 | Jeeves | repA | 2.4 |
The basis for the change in copy number is unclear, but could result either from changes in RepA/ori expression, or from parS-associated handcuffing or other regulatory mechanisms (41). However, the copy number variation likely accounts for the observed patterns of plasmid stability. First, the most well-maintained plasmids lacking parABS have the highest copy numbers (Et2Brutus,16.4 copies/cell), and the least well-maintained have much lower copy numbers (RedRock, 0.8 copies/cell; Alma, 1.8 copies/cell; Table 5); with higher copy numbers, production of plasmid-less cells at division is reduced, especially without a partitioning system. Second, plasmids containing parABS typically have lower copy numbers than their cognate parental plasmids, with the exception of RedRock, where there is little difference (Table 5). In systems such as in Gladiator, where the parABS cassette is not evidently contributing to maintenance, it enables a lower-copy-number plasmid (pKZ05) to be maintained similarly to a higher-copy-number plasmid (pKZ07). Nonetheless, it is surprising that many of these phage-derived plasmids are not well-maintained even with inclusion of the parABS cassette (Table 5). Although additional regulation through the phage repressor, which is encoded by an unlinked gene, is a possibility, we note that the replication/partitioning regions are largely devoid of predicted repressor binding sites; only Rachaly and Jeeves have such sites within or flanking parABS (42). Because vector context appears to be important, as illustrated by the behaviors of RedRock-derived plasmids (see above), stabilities and copy numbers of the recombinant plasmids may not fully reflect their parent prophages. For RedRock, LadyBird, Alma, and Et2Brutus, the plasmid copy numbers (one or two copies/cell) are similar although modestly lower than the cognate prophage copy numbers (Tables 1 and 5).
The behaviors and stabilities of this series of plasmids raise the question as to whether the parABS cassettes, particularly for RepA phages Miko and Rachaly, are active in partitioning at all. To address this, we used a similar strategy to that described previously to characterize the RedRock parABS system, which dramatically stabilizes an oriM plasmid expressing mCherry (pLO87) that is very unstable in the absence of selection (16). Recombinant versions of plasmid pLO87 carrying the parABS cassettes of Miko and Rachaly confer stability similar to that observed for RedRock parABS (16), and increased plasmid retention from <1% to >80% retention over ∼40 generations of unselected growth (Table 6). The reason why the pLO87-derived plasmids are more stable than the plasmids carrying the phage-derived replication systems is unclear, but perhaps is influenced by the vector backbone and vector genes. We note that when the Miko and Rachaly phage cassettes (including repA and parABS cassettes) are inserted into a different nonreplication plasmid vector, pMD04 (43), we observed similar stabilities (16.6% and 8.6% retention over 40 generations) to their cognate plasmids pKSW07 and pKSW08 (Table 5).
TABLE 6.
Maintenance of plasmids containing mCherry, oriM, and phage parABS
| Plasmid | Phage | Feature(s) | % maintenancea |
|---|---|---|---|
| pLO87 | N/A | N/A | 0 |
| pMO01 | RedRock | parABS | 91.0 |
| pKSW35 | Miko | parABS | 96.1 |
| pKSW36 | Rachaly | parABS | 90.0 |
| pKSW37 | Miko | parABS + gp39 | 89.2 |
| pKSW38 | Rachaly | parABS + gp39 | 81.4 |
Percentage of colonies carrying plasmid as determined by fluorescence after 40 generations of unselected growth.
Further evidence for the functionality of parA and parB in the context of the Miko-derived plasmids is provided by additional plasmid derivatives in which the genes have been interrupted or inactivated (Fig. 5). An early translational termination mutation in Miko parB (plasmid pKSW63) results in a nontransformation phenotype, perhaps due to parA overexpression or mislocalization, a phenotype similar to that reported previously for RedRock (16). Two plasmid derivatives containing separate in-frame deletions of Miko parA (pKSW60 and pKSW61) are competent to transform M. smegmatis but form extremely small colonies, consistent with very poor plasmid retention (data not shown). These observations suggest that the Miko parABS system is functional in plasmid pKSW07, even though it does not fully promote plasmid maintenance.
Compatibility of prophage origin plasmids.
Plasmids with common replication systems are typically incompatible as they compete for the replication machinery; therefore, we tested compatibility of a subset of these systems with each other and with the commonly used oriM plasmids derived from pAL5000. For this assay, one replicon partner was plasmid pKSW09 (Miko), pKSW52 (Jeeves), pHA08 (Et2Brutus), pKZ07 (Gladiator), or control plasmid pCCK38 (containing oriM) or pJV39 (with the L5 attP-int integration apparatus), and the other was a prophage of Miko, Et2Brutus, or LeBron (an unrelated integrating phage) carried by a lysogenic M. smegmatis strain. The plasmids (which lack parABS systems that could independently influence compatibility) were transformed into the lysogens, liquid cultures were grown with plasmid selection, and prophage maintenance was determined by spontaneous phage release (Fig. 7A). If a given prophage and a particular plasmid are compatible, then we expected to see prophage loss that is no greater than in the absence of the plasmid or a vector control (Fig. 7). In contrast, incompatibility would lead to prophage loss and a greater proportion of nonlysogens.
FIG 7.
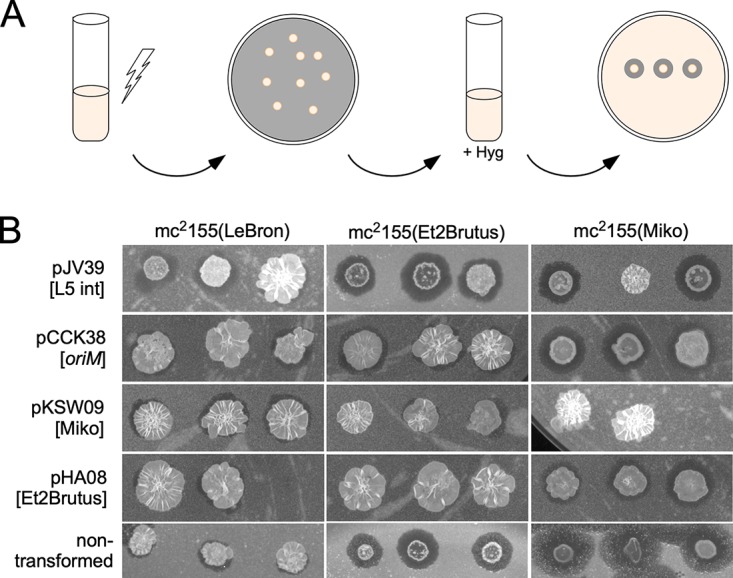
Compatibility of phage-based plasmids and prophages. (A) Scheme to test prophage origin compatibility. M. smegmatis mc2155 lysogens of phages LeBron, Et2Brutus, and Miko were transformed with various phage-based plasmids. The resulting colonies were grown in liquid culture with selection for the plasmid and then tested for prophage maintenance by spotting cultures onto lawns of M. smegmatis and observing spontaneous phage release. (B) The compatibilities of LeBron, Et2Brutus, and Miko prophages with plasmids pJV39, pCCK38, pKSW09, and pHA08, as measured by prophage maintenance. These data are a subset of data shown in Table 6. Three transformants were grown per transformation per experiment; missing spots in figure indicate transformant cultures that had not yet grown to saturation at the time of analysis.
Using a LeBron lysogen as a control, we observed stable prophage maintenance in transformants of all of the tested plasmids (Fig. 7B; Table 7), showing that all of the combinations of the LeBron prophage and plasmid are compatible. Transformants of an Et2Brutus lysogen, carrying the unrelated integrating vector pJV39 also maintain their prophage, but pHA08 transformants (Et2Brutus ori) efficiently lose the prophage due to incompatibility (Table 7). The Et2Brutus prophage is fully compatible with plasmids pKSW52 and pKSW09 from Jeeves and Miko, respectively, but is at least partially incompatible with pKZ07 (Gladiator) (Table 7). The Miko prophage is stable in nontransformed cells but is seemingly antagonized by the integrating vector pJV39—perhaps resulting from growth in the presence of hygromycin—leading to substantial prophage loss (Table 7). Transformants with pKSW09 (Miko) are more unstable, as anticipated, but HA08 (Et2Brutus) is relatively well-tolerated and is likely compatible; Miko prophage loss is also observed with pKZ07 (Gladiator) and pKSW02 (Jeeves), but at levels similar to that of the pJV39 control. Both the Miko and Et2Brutus prophages appear to be compatible with the oriM plasmid, and prophage loss is not evidently greater than with the control plasmids. These observations suggest that both Miko- and Et2Brutus-derived plasmids can be used compatibly with oriM plasmids in mycobacterial genetics and that at least Miko- and Et2Brutus-derived plasmids can be used together without interference.
TABLE 7.
Compatibility as measured by percent maintenance of prophage with plasmid selection
| Prophage | % maintenance of prophage with plasmid selection |
||||||
|---|---|---|---|---|---|---|---|
| pCCK38 | pJV39 | Jeeves (pKSW52) |
Miko (pKSW09) |
Et2Brutus (pHA08) |
Gladiator (pKZ07) |
No plasmid |
|
| LeBron | 83.3 | 100 | 100 | 100 | 100 | 100 | 100 |
| Miko | 58.3 | 41.6 | 33 | 22.2 | 83.3 | 33 | 100 |
| Et2Brutus | 100 | 100 | 100 | 91.6 | 0 | 66.67 | 100 |
Host range of prophage origin plasmids.
We tested plasmid derivatives for each of these systems (with the exception of Rachaly) for their ability to transform M. tuberculosis mc27000. Initially, we used plasmids lacking parABS (i.e., pKSW09, pKSW52, pKSW60, pHA03, pKZ07, pKZ02, and pHA08 from Miko, Jeeves, RedRock, LadyBird, Gladiator, Alma, and Et2Brutus, respectively), and all except pKSW09 and pKSW52 (Miko and Jeeves) successfully transformed with frequencies of >104 CFU/μg DNA (Table 8). Colony sizes varied, with pHA03 transformants (LadyBird) yielding the largest colonies and pHA08 (Et2Brutus) yielding the smallest (data not shown). Because pKSW09 and pKSW52 gave no transformants at all, we tested the parent parABS-containing plasmids pKSW07 and pKSW50, each of which efficiently transforms M. tuberculosis mc27000 (Table 8). Thus, for these two systems, the parABS partitioning system is required for M. tuberculosis transformation, a notable departure from their behaviors in M. smegmatis. We also tested the ability of the phage-based plasmids to transform G. terrae 3612. We observed robust transformation frequencies (>105) for plasmids pKSW09 (Miko), pKZ07 (Gladiator), and pHA08 (Et2Brutus), whereas pHA03 (LadyBird), pKZ02 (Alma), pKSW60 (RedRock), and pKSW52 (Jeeves) did not transform (Table 8); Rachaly-based plasmids were not tested. In contrast to our findings for M. tuberculosis, inclusion of the partitioning cassette did not confer the ability to transform G. terrae to the nontransforming phage-based plasmids (Table 8). We note that subcluster A15 contains 13 phages, all of which infect G. terrae, and all of which code for a parABS system and are in the non-RepA phage category.
TABLE 8.
Transformation efficiencies of phage-based plasmids in other Actinobacteria
| Phage | Feature(s) | Plasmid | Transformation efficiency (CFU/μg of DNA) |
|
|---|---|---|---|---|
|
M. tuberculosis mc27000 |
G. terrae
3612 |
|||
| Et2Brutus | ori | pHA08 | >104 | >105 |
| Gladiator | ori | pKZ07 | >104 | >105 |
| LadyBird | ori + parABS | pHA01 | Not tested | 0 |
| ori | pHA03 | >104 | 0 | |
| Alma | ori + parABS | pKZ01 | Not tested | 0 |
| ori | pKZ02 | >104 | 0 | |
| RedRock | ori + parABS | pKSW39 | Not tested | 0 |
| ori | pKSW60 | >104 | 0 | |
| Miko | repA + parABS | pKSW07 | >104 | Not tested |
| repA | pKSW09 | 0 | >105 | |
| Jeeves | repA + parABS | pKSW50 | >104 | 0 |
| repA | pKSW52 | 0 | 0 | |
| pMOS-Hyg | 0 | 0 | ||
DISCUSSION
We have described here the putative replication origins and partitioning functions of a series of temperate mycobacteriophages whose prophages are maintained extrachromosomally. Although more than 100 such phages have been reported, only a minority (5%) use a RepA-like initiator protein like that of the prototype P1 prophage. We have demonstrated that RepA is both required and sufficient for autonomous replication, and the cis-acting ori sequences presumably lie within or immediately adjacent to repA. However, most of the autonomously replicating phages do not have repA, there are no identifiable protein-coding genes, and it is likely that they use the transcribed RNA to initiate replication. A region expressing these RNAs is necessary and sufficient for autonomous replication.
Mapping and characterizing these prophage replication origins are confounded by differences in the behaviors of related systems derived from different phages, necessitating inclusion of many different systems in the analysis. This is especially notable in the variations in maintenance and copy numbers of recombinant plasmids, and it is likely that non-phage, vector-derived genes or transcripts influence these properties. Nonetheless, by analyzing a repertoire of RepA phages and non-RepA phages, we show that the prophages and prophage-derived plasmids replicate at low copy numbers and that parABS promotes plasmid maintenance. Furthermore, at least a subset of these systems provide a new suite of plasmid vectors for use in actinobacterial genetics that offer stably maintained low-copy-number plasmids capable of replicating in both fast- and slow-growing mycobacteria as well as other actinobacterial strains such as Gordonia. These are compatible with pAL5000 oriM plasmids, facilitating the construction of complex recombinant strains. Although different phages and plasmids may be optimal for specific applications, we note that the Et2Brutus plasmids have the desirable properties of good retention without selection, compatibility with a variety of origins, and broad host range.
The observation that phage-derived DNA segments containing the replication origin and the partitioning functions behave differently in different contexts is quite striking. There are several notable and informative observations. First, prior attempts to characterize the replication system of the non-RepA phage RedRock were discouraging, as insertion of an ori-parABS cassette into a vector did not promote replication and transformation of M. smegmatis (16). However, inserting the same RedRock DNA cassette into a similar vector backbone (to give pKSW39) but in a different orientation to a distinct antibiotic resistance gene (against hygromycin instead of kanamycin) results in efficient transformation and replication at a copy number similar to that of the prophage (Table 5) (16). Nonetheless, this plasmid is not stably maintained in the absence of selection, even though the parABS system should be fully functional (Table 6) (16), and removing it further reduces stability (Fig. 6). Second, it is notable that the Miko repA-parABS cassette facilitates efficient transformation of M. smegmatis and replicates at low copy number (Table 5), although it is quite unstable in the absence of selection (Fig. 6). Removal of parABS results in a striking increase in stability, seemingly facilitated by a 10-fold increase in plasmid copy number. In general, these behaviors suggest that replication is tightly regulated with a potential interaction between the partitioning and replication systems.
A further oddity is the unexpected dependence on the parABS cassette of Miko and Jeeves for transformation of M. tuberculosis. Both use a RepA-dependent replication system, and neither requires parABS for M. smegmatis transformation. In M. smegmatis, RepA plasmids lacking parABS have higher copy numbers than RepA plasmids that contain parABS do (Table 5) and are more stable. It is unclear why such changes would result in the inability to replicate in M. tuberculosis. However, we note that at least in some contexts, RepA expression is likely toxic, as plasmids expressing Miko RepA from the strong constitutive hsp60 promoter do not transform M. smegmatis (data not shown). Nonetheless, a variety of plasmids with different origins are available for use as M. tuberculosis plasmid vectors, and their compatibility with oriM plasmids, integrating, and parABS phage-based plasmids represents a substantial expansion of opportunities for M. tuberculosis genetics. The Gladiator-, Alma-, and Miko-derived plasmids also replicate in Gordonia, and it is likely all or many of the plasmids described here will be useful for genetic analysis of other actinobacterial strains, including nontuberculosis mycobacteria (NTM) pathogens such as M. abscessus and Mycobacterium avium. Additionally, the instability of some of these plasmids could be utilized to develop transient transposon delivery systems for various actinobacterial strains.
The extrachromosomally replicating temperate actinobacteriophages are almost exclusively found within cluster A; the exception, Streptomyces phage pZL12, is a singleton phage with no close relatives (44). Cluster A is exceptionally large (>600 individual phages), so we cannot exclude the possibility that other extrachromosomally replicating temperate phages will not be found in other less-well-sampled clusters of temperate actinobacteriophages, all of which are less than a quarter of the size of cluster A. Interestingly, although 50% of the cluster A subclusters have parABS phages (Fig. 1), most have only parABS phages, whereas subclusters A2, A9, and A12 have both integrating and parABS phage members. Because both the integration cassettes of subcluster A2 phages such as those in L5 and D29 (45, 46) as well as the replication partitioning system of subcluster A2 RepA phages are fully functional outside their phage contexts, it seems likely that they can be readily exchanged between the two, and comparison of cluster A2 genomes suggests this has likely occurred in their relatively recent evolutionary pasts. The genomes of phages Lokk and BobSwaget exemplify this, as they have 97% ANI and identical gene content, except for the additional RepA homologue found in Lokk (47). We are not aware of other sets of closely related genomes where this is observed, and note that this would likely not occur with the prototype lambda phage in which the integration apparatus is well-integrated into the overall regulatory circuitry, including dependence on the unlinked cII gene for integrase expression (48).
MATERIALS AND METHODS
Bacteria and plasmids.
M. smegmatis mc2155, M. tuberculosis mc27000, and Gordonia terrae 3612 were grown as described previously (49, 50). To construct phage-based plasmids, genome segments were PCR amplified from phage lysates using Q5 HiFi 2× MasterMix (NEB), and amplicons were inserted into the HindIII-digested vector pMOS-Hyg or XmnI-digested pMD04 (43) using the NEBuilder HiFi assembly kit (NEB); plasmids containing point mutations or deletions were constructed using the Q5 site-directed mutagenesis (SDM) kit (NEB). Transformations used electroporation protocols described previously for Mycobacterium (51) and Gordonia (49), and transformants were recovered on solid media with antibiotics (pMOS-Hyg, 50 μg/ml hygromycin; pMD04, 20 μg/ml kanamycin) and incubated at 37°C (M. smegmatis and M. tuberculosis) or 30°C (G. terrae). Plasmids used in this study have been compiled in Table S3 and S4 in the supplemental material.
Construction of mutant bacteriophages.
Phage genomic DNA was extracted from high-titer lysates of LadyBird, Alma, and Miko using phenol-chloroform extraction. Phage DNA was coelectroporated into recombineering M. smegmatis mc2155::pJV53 cells (51) with substrate DNA complementary to 250 bp flanking the deletion as previously described (40); genome coordinates and primer sequences are shown in Table S2. Cells were recovered for 3 to 4 h at 37°C and then combined with an M. smegmatis strain containing a plasmid with an anhydrotetracycline (ATc)-inducible CRISPR system targeting unmutated phage and plated on 7H11 with albumin-dextrose-catalase (ADC), kanamycin (Kan), CaCl2, and 300 ng/ml ATc. Plaques were screened for deletion via PCR and sequenced as described previously (52).
DNA substrates and primers used in phage engineering. Download Table S2, DOCX file, 0.01 MB (13.1KB, docx) .
Copyright © 2020 Wetzel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmids used in this study. Download Table S3, DOCX file, 0.02 MB (17.3KB, docx) .
Copyright © 2020 Wetzel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mutant plasmids used in this study. Download Table S4, DOCX file, 0.01 MB (13.6KB, docx) .
Copyright © 2020 Wetzel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The genome of the Miko parent phage used here varies slightly from the published sequence (GenBank accession number MN369748), in that a 153-bp repeat (coordinates 24718 to 24870) in gene 34 (minor tail protein) occurs once, whereas in the published sequence, it occurs twice. On sequencing of LadyBirdΔori, several base changes were observed in gene 31, coding for a predicted minor tail protein. These mutations did not alter the instability of the prophage (Fig. S3). AlmaΔori also had a single base change in gene 17, coding for a predicted major capsid protein.
Several point mutations in a minor tail protein gene gp31 do not alter prophage stability phenotypes. (A) Representative streaks from areas of infection of LadyBird G24044A (right) and LadyBirdΔori G24044A, C24885A (center) and LadyBirdΔori G24044A, G24980A (left) on an M. smegmatis lawn. (B) Spontaneous phage release from three individual colonies per streak, three streaks total. Individual colonies were patched onto an M. smegmatis lawn (top) and solid media (bottom). At least two patches/colony from LadyBird G24044A released phage, indicative of lysogens. Only one patch from LadyBirdΔori G24044A, C24885A released phage and 0 patches from LadyBirdΔori G24044A, G24980A released phage, indicative of unstable prophages or nonlysogens. Patches from a streak from a spot of phage buffer on an M. smegmatis lawn are shown for comparison. Download FIG S3, PDF file, 0.8 MB (789KB, pdf) .
Copyright © 2020 Wetzel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phenotyping mutant bacteriophages.
Serial dilutions of M. smegmatis mc2155 in log phase (optical density at 600 nm [OD600] of 0.8) were spread on plates seeded with 109 phage particles, or phage buffer as a control, and incubated for 4 days. Well-isolated colonies were streaked to remove phage, and three colonies per streak were patched onto lawns of M. smegmatis mc2155 to test for spontaneous phage release. Liquid cultures were grown from patches on solid media to test for susceptibility to the original infecting phage.
RNA-Seq.
Strand-specific transcription profiles of the LadyBird lysogen were measured by transcriptome sequencing (RNA-Seq) as previously described (53) and viewed using Integrated Genomics Viewer (IGV) (54) (Fig. S2).
Amino acid and nucleotide alignment and phylogeny.
Amino acid sequences of plasmid and phage RepA proteins (Fig. 2C) were aligned using Clustal Omega (55), and a phylogenetic tree using maximum likelihood (PhyML) was constructed with SeaView (56) and visualized using EvolView (57). Nucleotide sequences for the non-RepA phage origins (Table 3) were aligned using Clustal Omega.
Plasmid maintenance assays.
M. smegmatis transformants carrying phage-based plasmids were grown with selection to saturation, diluted 1:10,000, and regrown to saturation in antibiotic-free media; this was performed three times for a total of ∼40 generations without selection. The final culture was serially diluted, spotted (10 μl) onto solid media with and without selection, and incubated for 3 days at 37°C. The resulting colonies were counted, and maintenance was calculated as the number of colonies on the plate with selection divided by the number of colonies on the plate lacking antibiotics. This experiment was performed twice with duplicates. To determine maintenance of mCherry-expressing plasmids, the experiment was performed similarly, but retention was measured as the proportion of pink colonies on unselected plates.
Compatibility of prophage origins.
An M. smegmatis mc2155 lysogen of Et2Brutus was described previously (42) and lysogens of LeBron and Miko were made following the same protocol. Electrocompetent cells of the LeBron, Miko, and Et2Brutus lysogens were transformed with plasmids pJV39, pCCK38, pKSW52, pKSW09, pHA08, and pKZ07. The transformed lysogens were grown on hygromycin (Hyg) plates for 3 to 4 days at 37°C. Three colonies were picked from each of these plates, and liquid cultures were grown to saturation with hygromycin at 37°C to select for the plasmid. The liquid cultures were spotted onto lawns of M. smegmatis mc2155 and incubated at 37°C. Prophage maintenance was determined by observation of spontaneous phage release from spots of liquid culture. Compatibility was calculated as the percentage of transformed lysogen cultures that maintained the prophage after selection for the plasmid for at least six independent cultures.
Determination of plasmid copy number.
M. smegmatis transformants carrying phage-based plasmids were grown with selection to log phase (OD600 of ∼0.8) and DNA was extracted using phenol-chloroform. The DNA was sequenced using the Illumina platform as previously described (52), and copy number was calculated as the ratio of the average coverage of the plasmid sequence to the M. smegmatis chromosome. Prophage copy number was calculated similarly. For several plasmids, two transformants were evaluated (pCCK38, pKSW50, pKSW52, pKZ05, and pKZ07) showing good repeatability (<5% variation except for the low-copy-number vector pKSW50, which varied by 25%, or 0.1, between replicates); therefore, the remaining transformants were evaluated once.
Data availability.
All phage genome sequences are available at phagesdb.org. RNA-Seq data for the LadyBird lysogen have been deposited in the Gene Expression Omnibus (GEO) with accession number GSE145724.
ACKNOWLEDGMENTS
We thank Carlos Guerrero, Matthew Montgomery, and Lexi Marx for excellent technical assistance, Daniel Russell for genome sequencing and assembly, Travis Mavrich, Matthew Montgomery, and Janine LeBlanc-Straceski for comments on the manuscript, and the other members of the Hatfull Lab for valuable discussions. We also thank PHIRE and SEA-PHAGES students of the following universities who isolated the phages used in this study: University of Pittsburgh, Virginia Commonwealth University, Washington University in St. Louis, St. Edward’s University, Gonzaga University, and Worcester Polytechnic Institute.
This work was supported by NIH grants AI128082 and R35GM131729 and Howard Hughes Medical Institute award 54308198.
Footnotes
This article is a direct contribution from Graham F. Hatfull, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Vincent Fischetti, Rockefeller University, and Hector Morbidoni, Facultad de Ciencias Médicas-Universidad Nacional de Rosario.
Citation Wetzel KS, Aull HG, Zack KM, Garlena RA, Hatfull GF. 2020. Protein-mediated and RNA-based origins of replication of extrachromosomal mycobacterial prophages. mBio 11:e00385-20. https://doi.org/10.1128/mBio.00385-20.
REFERENCES
- 1.Pope WH, Bowman CA, Russell DA, Jacobs-Sera D, Asai DJ, Cresawn SG, Jacobs WR, Hendrix RW, Lawrence JG, Hatfull GF, Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Science, Phage Hunters Integrating Research and Education, Mycobacterial Genetics Course. 2015. Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of phage genetic diversity. Elife 4:e06416. doi: 10.7554/eLife.06416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hatfull GF, Jacobs-Sera D, Lawrence JG, Pope WH, Russell DA, Ko C-C, Weber RJ, Patel MC, Germane KL, Edgar RH, Hoyte NN, Bowman CA, Tantoco AT, Paladin EC, Myers MS, Smith AL, Grace MS, Pham TT, O’Brien MB, Vogelsberger AM, Hryckowian AJ, Wynalek JL, Donis-Keller H, Bogel MW, Peebles CL, Cresawn SG, Hendrix RW. 2010. Comparative genomic analysis of 60 mycobacteriophage genomes: genome clustering, gene acquisition, and gene size. J Mol Biol 397:119–143. doi: 10.1016/j.jmb.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hatfull GF, Pedulla ML, Jacobs-Sera D, Cichon PM, Foley A, Ford ME, Gonda RM, Houtz JM, Hryckowian AJ, Kelchner VA, Namburi S, Pajcini KV, Popovich MG, Schleicher DT, Simanek BZ, Smith AL, Zdanowicz GM, Kumar V, Peebles CL, Jacobs WR Jr, Lawrence JG, Hendrix RW. 2006. Exploring the mycobacteriophage metaproteome: phage genomics as an educational platform. PLoS Genet 2:e92. doi: 10.1371/journal.pgen.0020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hatfull GF. 2018. Mycobacteriophages. Microbiol Spectr 6(5):GPP3-0026-2018. doi: 10.1128/microbiolspec.GPP3-0026-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith MCM. 2015. Phage-encoded serine integrases and other large serine recombinases. Microbiol Spectr 3(4):MDNA3-0059-2014. doi: 10.1128/microbiolspec.MDNA3-0059-2014. [DOI] [PubMed] [Google Scholar]
- 6.Murphy KC. 2012. Phage recombinases and their applications. Adv Virus Res 83:367–414. doi: 10.1016/B978-0-12-394438-2.00008-6. [DOI] [PubMed] [Google Scholar]
- 7.Landy A. 2015. The lambda integrase site-specific recombination pathway. Microbiol Spectr 3(2):MDNA3-0051-2014. doi: 10.1128/microbiolspec.MDNA3-0051-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sternberg N, Hoess R. 1983. The molecular genetics of bacteriophage P1. Annu Rev Genet 17:123–154. doi: 10.1146/annurev.ge.17.120183.001011. [DOI] [PubMed] [Google Scholar]
- 9.Utter B, Deutsch DR, Schuch R, Winer BY, Verratti K, Bishop-Lilly K, Sozhamannan S, Fischetti VA. 2014. Beyond the chromosome: the prevalence of unique extra-chromosomal bacteriophages with integrated virulence genes in pathogenic Staphylococcus aureus. PLoS One 9:e100502. doi: 10.1371/journal.pone.0100502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sozhamannan S, McKinstry M, Lentz SM, Jalasvuori M, McAfee F, Smith A, Dabbs J, Ackermann HW, Bamford JK, Mateczun A, Read TD. 2008. Molecular characterization of a variant of Bacillus anthracis-specific phage AP50 with improved bacteriolytic activity. Appl Environ Microbiol 74:6792–6796. doi: 10.1128/AEM.01124-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casjens S, Palmer N, van Vugt R, Huang WM, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson RJ, Haft D, Hickey E, Gwinn M, White O, Fraser CM. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol 35:490–516. doi: 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- 12.Read TD, Brunham RC, Shen C, Gill SR, Heidelberg JF, White O, Hickey EK, Peterson J, Utterback T, Berry K, Bass S, Linher K, Weidman J, Khouri H, Craven B, Bowman C, Dodson R, Gwinn M, Nelson W, DeBoy R, Kolonay J, McClarty G, Salzberg SL, Eisen J, Fraser CM. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res 28:1397–1406. doi: 10.1093/nar/28.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sergueev K, Dabrazhynetskaya A, Austin S. 2005. Plasmid partition system of the P1par family from the pWR100 virulence plasmid of Shigella flexneri. J Bacteriol 187:3369–3373. doi: 10.1128/JB.187.10.3369-3373.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ravin NV. 2015. Replication and maintenance of linear phage-plasmid N15. Microbiol Spectr 3(1):PLAS-0032-2014. doi: 10.1128/microbiolspec.PLAS-0032-2014. [DOI] [PubMed] [Google Scholar]
- 15.Deutsch DR, Utter B, Verratti KJ, Sichtig H, Tallon LJ, Fischetti VA. 2018. Extra-chromosomal DNA sequencing reveals episomal prophages capable of impacting virulence factor expression in Staphylococcus aureus. Front Microbiol 9:1406. doi: 10.3389/fmicb.2018.01406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dedrick RM, Mavrich TN, Ng WL, Cervantes Reyes JC, Olm MR, Rush RE, Jacobs-Sera D, Russell DA, Hatfull GF. 2016. Function, expression, specificity, diversity and incompatibility of actinobacteriophage parABS systems. Mol Microbiol 101:625–644. doi: 10.1111/mmi.13414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stella EJ, Franceschelli JJ, Tasselli SE, Morbidoni HR. 2013. Analysis of novel mycobacteriophages indicates the existence of different strategies for phage inheritance in mycobacteria. PLoS One 8:e56384. doi: 10.1371/journal.pone.0056384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerdes K, Howard M, Szardenings F. 2010. Pushing and pulling in prokaryotic DNA segregation. Cell 141:927–942. doi: 10.1016/j.cell.2010.05.033. [DOI] [PubMed] [Google Scholar]
- 19.Łobocka MB, Rose DJ, Plunkett G, Rusin M, Samojedny A, Lehnherr H, Yarmolinsky MB, Blattner FR. 2004. Genome of bacteriophage P1. J Bacteriol 186:7032–7068. doi: 10.1128/JB.186.21.7032-7068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mobberley JM, Authement RN, Segall AM, Paul JH. 2008. The temperate marine phage PhiHAP-1 of Halomonas aquamarina possesses a linear plasmid-like prophage genome. J Virol 82:6618–6630. doi: 10.1128/JVI.00140-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammerl JA, Klevanskaa K, Strauch E, Hertwig S. 2014. Complete nucleotide sequence of pVv01, a P1-like plasmid prophage of Vibrio vulnificus. Genome Announc 2:e00135-14. doi: 10.1128/genomeA.00135-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu W, Wang J, Zhu Y, Tang B, Zhang Y, He P, Zhang Y, Liu B, Guo X, Zhao G, Qin J. 2015. Identification of three extra-chromosomal replicons in Leptospira pathogenic strain and development of new shuttle vectors. BMC Genomics 16:90. doi: 10.1186/s12864-015-1321-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beggs ML, Crawford JT, Eisenach KD. 1995. Isolation and sequencing of the replication region of Mycobacterium avium plasmid pLR7. J Bacteriol 177:4836–4840. doi: 10.1128/jb.177.17.4836-4840.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gavigan JA, Ainsa JA, Perez E, Otal I, Martin C. 1997. Isolation by genetic labeling of a new mycobacterial plasmid, pJAZ38, from Mycobacterium fortuitum. J Bacteriol 179:4115–4122. doi: 10.1128/jb.179.13.4115-4122.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Picardeau M, Le Dantec C, Vincent V. 2000. Analysis of the internal replication region of a mycobacterial linear plasmid. Microbiology 146:305–313. doi: 10.1099/00221287-146-2-305. [DOI] [PubMed] [Google Scholar]
- 26.Bachrach G, Colston MJ, Bercovier H, Bar-Nir D, Anderson C, Papavinasasundaram KG. 2000. A new single-copy mycobacterial plasmid, pMF1, from Mycobacterium fortuitum which is compatible with the pAL5000 replicon. Microbiology 146:297–303. doi: 10.1099/00221287-146-2-297. [DOI] [PubMed] [Google Scholar]
- 27.Huff J, Czyz A, Landick R, Niederweis M. 2010. Taking phage integration to the next level as a genetic tool for mycobacteria. Gene 468:8–19. doi: 10.1016/j.gene.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stolt P, Stoker NG. 1996. Functional definition of regions necessary for replication and incompatibility in the Mycobacterium fortuitum plasmid pAL5000. Microbiology 142:2795–2802. doi: 10.1099/13500872-142-10-2795. [DOI] [PubMed] [Google Scholar]
- 29.Nossal NG. 1983. Prokaryotic DNA replication systems. Annu Rev Biochem 52:581–615. doi: 10.1146/annurev.bi.52.070183.003053. [DOI] [PubMed] [Google Scholar]
- 30.Brantl S. 2014. Plasmid replication control by antisense RNAs. Microbiol Spectr 2(4):PLAS-0001-2013. doi: 10.1128/microbiolspec.PLAS-0001-2013. [DOI] [PubMed] [Google Scholar]
- 31.del Solar G, Giraldo R, Ruiz-Echevarría MJ, Espinosa M, Díaz-Orejas R. 1998. Replication and control of circular bacterial plasmids. Microbiol Mol Biol Rev 62:434–464. doi: 10.1128/MMBR.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Russell DA, Hatfull GF. 2017. PhagesDB: the actinobacteriophage database. Bioinformatics 33:784–786. doi: 10.1093/bioinformatics/btw711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim AI, Ghosh P, Aaron MA, Bibb LA, Jain S, Hatfull GF. 2003. Mycobacteriophage Bxb1 integrates into the Mycobacterium smegmatis groEL1 gene. Mol Microbiol 50:463–473. doi: 10.1046/j.1365-2958.2003.03723.x. [DOI] [PubMed] [Google Scholar]
- 34.Ripoll F, Pasek S, Schenowitz C, Dossat C, Barbe V, Rottman M, Macheras E, Heym B, Herrmann JL, Daffe M, Brosch R, Risler JL, Gaillard JL. 2009. Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PLoS One 4:e5660. doi: 10.1371/journal.pone.0005660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin M, Taniguchi H, Mizuguchi Y. 1994. Analysis of the replication region of a mycobacterial plasmid, pMSC262. J Bacteriol 176:419–425. doi: 10.1128/jb.176.2.419-425.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Labidi A, Mardis E, Roe BA, Wallace RJ Jr.. 1992. Cloning and DNA sequence of the Mycobacterium fortuitum var fortuitum plasmid pAL5000. Plasmid 27:130–140. doi: 10.1016/0147-619x(92)90013-z. [DOI] [PubMed] [Google Scholar]
- 37.Abeles AL, Reaves LD, Youngren-Grimes B, Austin SJ. 1995. Control of P1 plasmid replication by iterons. Mol Microbiol 18:903–912. doi: 10.1111/j.1365-2958.1995.18050903.x. [DOI] [PubMed] [Google Scholar]
- 38.Zahn K, Blattner FR. 1985. Binding and bending of the lambda replication origin by the phage O protein. EMBO J 4:3605–3616. doi: 10.1002/j.1460-2075.1985.tb04124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alfano C, McMacken R. 1989. Ordered assembly of nucleoprotein structures at the bacteriophage lambda replication origin during the initiation of DNA replication. J Biol Chem 264:10699–10708. [PubMed] [Google Scholar]
- 40.Marinelli LJ, Piuri M, Swigonova Z, Balachandran A, Oldfield LM, van Kessel JC, Hatfull GF. 2008. BRED: a simple and powerful tool for constructing mutant and recombinant bacteriophage genomes. PLoS One 3:e3957. doi: 10.1371/journal.pone.0003957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Das N, Chattoraj DK. 2004. Origin pairing (‘handcuffing’) and unpairing in the control of P1 plasmid replication. Mol Microbiol 54:836–849. doi: 10.1111/j.1365-2958.2004.04322.x. [DOI] [PubMed] [Google Scholar]
- 42.Mavrich TN, Hatfull GF. 2019. Evolution of superinfection immunity in cluster A mycobacteriophages. mBio 10:e00971-19. doi: 10.1128/mBio.00971-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Donnelly-Wu MK, Jacobs WR Jr, Hatfull GF. 1993. Superinfection immunity of mycobacteriophage L5: applications for genetic transformation of mycobacteria. Mol Microbiol 7:407–417. doi: 10.1111/j.1365-2958.1993.tb01132.x. [DOI] [PubMed] [Google Scholar]
- 44.Zhong L, Cheng Q, Tian X, Zhao L, Qin Z. 2010. Characterization of the replication, transfer, and plasmid/lytic phage cycle of the Streptomyces plasmid-phage pZL12. J Bacteriol 192:3747–3754. doi: 10.1128/JB.00123-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee MH, Pascopella L, Jacobs WR Jr, Hatfull GF. 1991. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guerin. Proc Natl Acad Sci U S A 88:3111–3115. doi: 10.1073/pnas.88.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pena CE, Stoner J, Hatfull GF. 1998. Mycobacteriophage D29 integrase-mediated recombination: specificity of mycobacteriophage integration. Gene 225:143–151. doi: 10.1016/s0378-1119(98)00490-9. [DOI] [PubMed] [Google Scholar]
- 47.Butela KA, Gurney SMR, Hendrickson HL, LeBlanc-Straceski JM, Zimmerman AM, Conant SB, Freed NE, Silander OK, Thomson JJ, Berkes CA, Bertolez C, Davies CG, Elinsky A, Hanlon AJ, Nersesyan J, Patel P, Sherwood J, Ngo TT, Wisniewski KA, Yacoo K, Arendse PM, Bowlen NW, Cunmulaj J, Downs JL, Ferrenberg CA, Gassman AE, Gilligan CER, Gorkiewicz E, Harness C, Huffman A, Jones C, Julien A, Kupic AE, Latu SF, Manning TJ, Maxwell D, Merrimack College SEA-PHAGES Annotators 2016, Meyer CE, Reardon M, Slaughter M, Swasey R, Tennent RI, Torres V, Waller T, Worcester RM, Yost BL, Cresawn SG, Garlena RA, Jacobs-Sera D, Pope WH. 2017. Complete genome sequences of cluster A mycobacteriophages BobSwaget, Fred313, KADY, Lokk, MyraDee, Stagni, and StepMih. Genome Announc 5:e00182-17. doi: 10.1128/genomeA.01182-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Little JW. 2010. Evolution of complex gene regulatory circuits by addition of refinements. Curr Biol 20:R724–R734. doi: 10.1016/j.cub.2010.06.028. [DOI] [PubMed] [Google Scholar]
- 49.Montgomery MT, Guerrero Bustamante CA, Dedrick RM, Jacobs-Sera D, Hatfull GF. 2019. Yet more evidence of collusion: a new viral defense system encoded by Gordonia phage CarolAnn. mBio 10:e02417-18. doi: 10.1128/mBio.02417-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jacobs-Sera D, Marinelli LJ, Bowman C, Broussard GW, Guerrero Bustamante C, Boyle MM, Petrova ZO, Dedrick RM, Pope WH, Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Science (SEA-Phages) Program, Modlin RL, Hendrix RW, Hatfull GF. 2012. On the nature of mycobacteriophage diversity and host preference. Virology 434:187–201. doi: 10.1016/j.virol.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Kessel JC, Hatfull GF. 2007. Recombineering in Mycobacterium tuberculosis. Nat Methods 4:147–152. doi: 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]
- 52.Russell DA. 2018. Sequencing, assembling, and finishing complete bacteriophage genomes. Methods Mol Biol 1681:109–125. doi: 10.1007/978-1-4939-7343-9_9. [DOI] [PubMed] [Google Scholar]
- 53.Dedrick RM, Jacobs-Sera D, Bustamante CA, Garlena RA, Mavrich TN, Pope WH, Reyes JC, Russell DA, Adair T, Alvey R, Bonilla JA, Bricker JS, Brown BR, Byrnes D, Cresawn SG, Davis WB, Dickson LA, Edgington NP, Findley AM, Golebiewska U, Grose JH, Hayes CF, Hughes LE, Hutchison KW, Isern S, Johnson AA, Kenna MA, Klyczek KK, Mageeney CM, Michael SF, Molloy SD, Montgomery MT, Neitzel J, Page ST, Pizzorno MC, Poxleitner MK, Rinehart CA, Robinson CJ, Rubin MR, Teyim JN, Vazquez E, Ware VC, Washington J, Hatfull GF. 2017. Prophage-mediated defence against viral attack and viral counter-defence. Nat Microbiol 2:16251. doi: 10.1038/nmicrobiol.2016.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thorvaldsdottir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 57.Zhang H, Gao S, Lercher MJ, Hu S, Chen WH. 2012. EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res 40:W569–W572. doi: 10.1093/nar/gks576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huson DH. 1998. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- 59.Cresawn SG, Bogel M, Day N, Jacobs-Sera D, Hendrix RW, Hatfull GF. 2011. Phamerator: a bioinformatic tool for comparative bacteriophage genomics. BMC Bioinformatics 12:395. doi: 10.1186/1471-2105-12-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cluster A actinobacteriophages that contain parABS. Download Table S1, DOCX file, 0.02 MB (23KB, docx) .
Copyright © 2020 Wetzel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Transcriptomic profile of the LadyBird lysogen. RNA was isolated from a LadyBird lysogen in the log phase of growth, and libraries were generated and sequenced using the Illumina platform. The reads per base are plotted along a Phamerator map of LadyBird, with reads aligning to the top strand shown in blue and reads mapping to the bottom strand shown in red. Robust expression of the immunity repressor gene (79) and parAB (35 and 36) is observed, as well as the opposite strand RNA proximal to parAB (labeled “ori”). Low expression of left-arm structural genes indicates some spontaneous induction. Download FIG S1, PDF file, 0.2 MB (181.2KB, pdf) .
Copyright © 2020 Wetzel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ori is necessary to form stable lysogens of Alma and LadyBird. (A) Representative streaks (n = 3) from areas of infection of Alma (right) and AlmaΔori (left) on an M. smegmatis lawn. (B) Spontaneous phage release from three individual colonies per streak. Individual colonies were patched onto solid media (left) and an M. smegmatis lawn (center). The original infecting phage per streak plate is shown as a grid (right). (C) Susceptibility of colonies to phage superinfection. Liquid cultures were grown from one patch per original area of infection and tested for its susceptibility to Alma, AlmaΔori, Miko (A2), and a control phage Crossroads (L2). A representative example of each is shown. (D) Representative streaks (n = 3) from areas of infection of LadyBird (right) and LadyBirdΔori (left) on an M. smegmatis lawn. (E) Spontaneous phage release from three individual colonies per streak. Individual colonies were patched onto solid media (left) and an M. smegmatis lawn (center). The original infecting phage per streak plate is shown as a grid (right). (F) Susceptibility of colonies to phage superinfection. Liquid cultures were grown from one patch per original area of infection and tested for its susceptibility to LadyBird, LadyBirdΔori, Alma (A9), and a control phage Crossroads (L2). A representative example of each is shown. Download FIG S2, PDF file, 0.9 MB (897.4KB, pdf) .
Copyright © 2020 Wetzel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DNA substrates and primers used in phage engineering. Download Table S2, DOCX file, 0.01 MB (13.1KB, docx) .
Copyright © 2020 Wetzel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmids used in this study. Download Table S3, DOCX file, 0.02 MB (17.3KB, docx) .
Copyright © 2020 Wetzel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mutant plasmids used in this study. Download Table S4, DOCX file, 0.01 MB (13.6KB, docx) .
Copyright © 2020 Wetzel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Several point mutations in a minor tail protein gene gp31 do not alter prophage stability phenotypes. (A) Representative streaks from areas of infection of LadyBird G24044A (right) and LadyBirdΔori G24044A, C24885A (center) and LadyBirdΔori G24044A, G24980A (left) on an M. smegmatis lawn. (B) Spontaneous phage release from three individual colonies per streak, three streaks total. Individual colonies were patched onto an M. smegmatis lawn (top) and solid media (bottom). At least two patches/colony from LadyBird G24044A released phage, indicative of lysogens. Only one patch from LadyBirdΔori G24044A, C24885A released phage and 0 patches from LadyBirdΔori G24044A, G24980A released phage, indicative of unstable prophages or nonlysogens. Patches from a streak from a spot of phage buffer on an M. smegmatis lawn are shown for comparison. Download FIG S3, PDF file, 0.8 MB (789KB, pdf) .
Copyright © 2020 Wetzel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
All phage genome sequences are available at phagesdb.org. RNA-Seq data for the LadyBird lysogen have been deposited in the Gene Expression Omnibus (GEO) with accession number GSE145724.