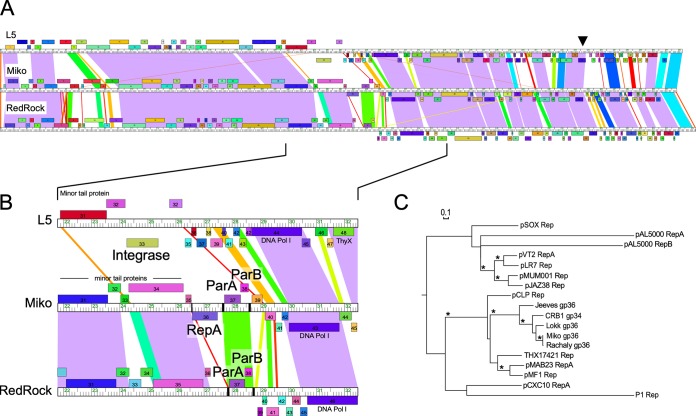
FIG 2.
Alignment of parABS phages used in this study. (A) Representative RepA (Miko) and non-RepA (RedRock) phage genomes were aligned to that of the integrating phage L5. The ruler shows genome length in kilobases, while ORFs shown above and below the ruler are transcribed rightward and leftward, respectively. ORFs of the same color have been assigned to the same gene phamily. A black arrowhead indicates the location of the immunity repressor gene in all three genomes. Shared nucleotide sequence similarity is represented as spectrum-colored shading, with violet representing the most similar and red the least similar above a BLASTN E-value threshold of 10−4. (B) Genome segments containing genes relevant for prophage replication and maintenance. Where present, integrase (L5), homologues to repA (Miko), parA and parB (Miko and RedRock) are labeled. The locations of centromere-like parS sites are indicated in the Miko and RedRock genome rulers with black bars. DNA Pol I, DNA polymerase I. (C) Relatedness of the phage RepA proteins to replication initiator proteins found in actinobacterial plasmids and the E. coli phage P1, shown as a phylogeny generated by maximum likelihood (PhyML). Bootstrap values greater than 70% are indicated with an asterisk, and the bar indicates the number of substitutions per site.