Flagella and motility are widespread virulence factors among pathogenic bacteria. Motility enhances the initial host colonization, but the flagellum is a major antigen targeted by the host immune system. Here, we demonstrate that pathogenic E. coli strains employ a mechanosensory function of the flagellar motor to activate flagellar expression under high loads, while repressing it in liquid culture. We hypothesize that this mechanism allows pathogenic E. coli to regulate its motility dependent on the stage of infection, activating flagellar expression upon initial contact with the host epithelium, when motility is beneficial, but reducing it within the host to delay the immune response.
KEYWORDS: Escherichia coli, bacterial physiology, flagellar gene regulation, flagellar motility, mechanosensing, virulence
ABSTRACT
Bacterial flagellar motility plays an important role in many processes that occur at surfaces or in hydrogels, including adhesion, biofilm formation, and bacterium-host interactions. Consequently, expression of flagellar genes, as well as genes involved in biofilm formation and virulence, can be regulated by the surface contact. In a few bacterial species, flagella themselves are known to serve as mechanosensors, where an increased load on flagella experienced during surface contact or swimming in viscous media controls gene expression. In this study, we show that gene regulation by motility-dependent mechanosensing is common among pathogenic Escherichia coli strains. This regulatory mechanism requires flagellar rotation, and it enables pathogenic E. coli to repress flagellar genes at low loads in liquid culture, while activating motility in porous medium (soft agar) or upon surface contact. It also controls several other cellular functions, including metabolism and signaling. The mechanosensing response in pathogenic E. coli depends on the negative regulator of motility, RflP (YdiV), which inhibits basal expression of flagellar genes in liquid. While no conditional inhibition of flagellar gene expression in liquid and therefore no upregulation in porous medium was observed in the wild-type commensal or laboratory strains of E. coli, mechanosensitive regulation could be recovered by overexpression of RflP in the laboratory strain. We hypothesize that this conditional activation of flagellar genes in pathogenic E. coli reflects adaptation to the dual role played by flagella and motility during infection.
OBSERVATION
Sensing and rapid adaptation to changing environmental conditions are a common survival strategy of bacteria. One of the best-studied examples of a highly regulated cellular function is flagellum-mediated motility. In E. coli, which can exist in various environments and has adopted both commensal and pathogenic lifestyles, the expression of flagellar genes is organized in a hierarchical cascade of three classes that integrate a number of external and internal signals (1, 2). The class I transcriptional regulator FlhDC induces the expression of class II genes, which encode the components of the flagellar hook and basal body as well as a sigma factor, FliA. In turn, FliA is required for the expression of class III genes, which include those for the outer part of flagella and components of the chemotaxis pathway.
Flagella and motility have several functions in bacterial virulence (3, 4). In the early phase of infection, motility is important for moving through the mucus layer and establishing initial contact with the host epithelium, and flagella can serve as adhesins. However, flagella are also highly immunogenic, being rapidly recognized by the innate immune system (5). Known strategies to avoid this immune response used by such pathogens as Salmonella, Campylobacter, and Helicobacter rely on stochastic variation in the expression of flagellar genes, due either to genetic changes (6–10) or to changes at the transcriptional level (11–13). The latter regulation in Salmonella is mediated by RflP (formerly called YdiV) (14), which represses fliA expression by sequestering the FlhDC complex and targeting it for degradation in a subpopulation of cells (15, 16). Expression of RflP and hence of the fraction on nonmotile cells in the population depends on nutrient levels and cell envelope stress (15, 17).
Although flagellar systems of E. coli and Salmonella and their regulation are highly similar, only a few E. coli strains exhibit genetic phase variation in flagellar expression (18, 19). Furthermore, although the rflP gene is present in E. coli, it is inactive in laboratory strains because of attenuated translation (20). Thus, it remained unclear what mechanisms might be used by most pathogenic E. coli strains to control motility under conditions that resemble those encountered during infection.
In this work, we investigated the motility of several commensal and pathogenic strains of E. coli grown in liquid or in soft agar. We showed that pathogenic, but not commensal, E. coli strains activate motility only in a porous medium. The underlying regulation requires flagellar rotation as well as expression of RflP, and it is likely to represent a novel mechanism of motility-dependent mechanosensing.
Pathogenic isolates of E. coli downregulate flagellar gene expression in liquid.
We investigated the motility of a number of pathogenic, as well as a few commensal, natural isolates of E. coli obtained from the Leibniz Institute’s DSMZ collection (Table 1 and see Table S1 in the supplemental material). Surprisingly, while most tested strains (17 out of 19) appeared nonmotile or poorly motile when cultivated in liquid tryptone broth (TB), they became motile on soft (0.27% agar) TB (TBA 0.27%) motility plates, where E. coli cells can spread by swimming through agar pores (Table 1; Fig. 1A and B). Such dependence of motility on growth in agar was confirmed by reinoculation of liquid TB cultures from TBA 0.27% plates and vice versa. Interestingly, all E. coli strains following this motility pattern were implicated in pathogenicity, whereas both tested commensal strains from the DSMZ collection and the common laboratory strain MG1655 were motile under both conditions.
TABLE 1.
E. coli isolates used in this study
| E. coli straina | Source of isolation or type of pathogenicityb |
Motility on TBA 0.27% |
Motility in TBc |
|---|---|---|---|
| U5/41 | UPEC | – | – |
| DSM 50902 | NA | + | – |
| QST 40139 | Possibly commensal strain | – | – |
| AMC 198 | NA | – | – |
| E611 | EPEC | + | – |
| E2808 | EPEC | + | – |
| EW2129-54 | EPEC | + | +/– |
| C771 | EPEC | + | – |
| ICB 4004 | Urine | – | – |
| S13 | Septicemia patient | + | – |
| IHE3035 | Newborn with meningitis | + | +/– |
| M185/1-1 | Fecal isolate from healthy individual | + | + |
| B185/29-10 | Fecal isolate from healthy individual | + | + |
| D699(U5/41 Oac–) | Patient with urinary tract infection | + | – |
| IHE3043 | Newborn with meningitis | – | – |
| Z36 | Patient with urinary tract infection | + | – |
| T111 | Patient with urinary tract infection | + | – |
| PC 0886 | Possibly commensal strain | – | – |
| CDC 5624-50 | EPEC | – | – |
| MG1655 | Commensal lab strain | + | + |
Pathogenic and commensal strains that were motile on TBA 0.27% plates are indicated with boldface and italics, respectively.
UPEC, uropathogenic E. coli; EPEC, enteropathogenic E. coli; NA, not available.
+/– indicates that only a fraction of cells are motile.
FIG 1.
Motilities of pathogenic and commensal E. coli isolates in liquid and in 0.27% agar. (A) Cell trajectories of E. coli Z36 and MG1655, shown as their projected intensities during 20-s-long movies. Bacteria were grown in liquid TB at 37°C with shaking to an OD600 of 0.5 to 0.6, diluted to an OD600 of 0.05 to 0.1 in TB, and imaged at room temperature. Scale bars, 50 μm. (B) Motility halos formed by E. coli Z36 and MG1655 after 5 μl of the cells grown in TB (as for panel A) was spotted onto the surface of 0.27% TB agar (TBA 0.27%) and incubated at 37°C for 6 to 8 h. (C to H) Distributions of fluorescence levels of PflhD-gfp, PfliA-gfp, and PfliC-gfp reporters in E. coli E2808 (C), S13 (D), Z36 (E), T111 (F), M185/1-1 (G), and MG1655 (H) cells taken from the edges of spreading populations grown in TBA 0.27% or in liquid TB medium, as indicated. Fluorescence was measured using flow cytometry as described previously (46). AU, arbitrary units.
List of bacterial strains and plasmids used in this study. Download Table S1, DOCX file, 0.02 MB (25.2KB, docx) .
Copyright © 2020 Laganenka et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
For further analysis of the underlying regulatory mechanism, we transformed four pathogenic and two commensal E. coli strains with plasmids carrying green fluorescent protein (GFP) reporters for all three classes of promoters in a flagellar gene expression cascade. Whereas the activities of the flhD (class I) reporters were similar between cells grown in TBA 0.27% and in liquid TB for all strains (Fig. 1C to H and Fig. S1A to F), promoters of fliA (class II/class III) and fliC (flagellin, class III) were strongly downregulated in liquid TB in pathogenic strains (Fig. 1C to F and Fig. S1A to D). In contrast, no changes in flagellar gene expression were observed in commensal strains (Fig. 1G and H and Fig. S1E and F), consistent with their unchanged motility in liquid culture. Together, these results indicate the existence of a lifestyle-specific motility control in E. coli acting at the level of class II flagellar gene expression.
Expression of flagellar genes in pathogenic and commensal E. coli isolates in TBA and in liquid TB. Experiments were performed as described for Fig. 1. Percentages of GFP-positive cells in E2808 (A), S13 (B), Z36 (C), T111 (D), M185/1-1 (E), and MG1655 (F) populations are shown. Means from a minimum of four independent replicas are shown; error bars represent standard deviations. P values were calculated using the Mann-Whitney test. *, P < 0.05; ns, not significant. Download FIG S1, EPS file, 0.1 MB (100.2KB, eps) .
Copyright © 2020 Laganenka et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Rotation of the flagellar motor under a load is required for activation of flagellar gene expression in pathogenic E. coli.
Since the most apparent difference between growth in liquid and that in soft agar is the porous structure of agar, which interferes with flagellar rotation, we hypothesized that an increased mechanical load on the flagellar motor might signal flagellar gene expression in pathogenic E. coli. Such mechanosensing has indeed been described in several bacterial species (21, 22), although its molecular mechanisms remain poorly understood. In E. coli, the load is known to cause posttranslational remodeling of the flagellar motor, increasing the number of force-generating stator units (23), but no gene-regulatory mechanosensing mediated by flagella has been reported so far.
To test our hypothesis, we made E. coli Z36 knockout strains that lack flagellar filaments and therefore have largely reduced motor loads (ΔfliC mutant) or have paralyzed flagella due to the absence of the MotA stator protein (ΔmotA mutant). These strains were locked in the motility-off state at the level of gene expression in both liquid and agar (Fig. 2A to C; Fig. S2A to C), suggesting that in pathogenic E. coli, rotation of the flagellar motor under a load is indeed perceived as a signal activating the expression of class II and class III flagellar genes. These knockouts could be complemented for both motility and gene expression when respective proteins were produced from a plasmid (Fig. S3).
FIG 2.
Flagellar motor rotation under a load and RflP control motility in pathogenic E. coli. (A to D) Distributions of fluorescence levels of PfllhD-gfp, PfliA-gfp, and PfliC-gfp reporters in wild-type strain E. coli Z36 (A) and in its ΔmotA (B), ΔfliC (C), ΔrflP (D), ΔrflP ΔmotA (E), and ΔrflP ΔfliC (F) knockout mutants. Bacterial growth and measurements were as described for Fig. 1C to H. (G) Cell trajectories of E. coli Z36 and its ΔrflP mutant grown in liquid TB medium, acquired as described in the legend of Fig. 1A. Scale bars, 50 μm. (H) PfliC-gfp expression in WT Z36 and ΔmotA cells forced to the surfaces of microtiter plates (attached [ATT]) by centrifugation or statically incubated (planktonic [PL]), measured by flow cytometry. The mean fluorescence of each GFP-positive subpopulation is shown in arbitrary units (AU). Means from a minimum of four independent replicas are shown; error bars represent standard deviations. P values were calculated using the Mann-Whitney test. **, P < 0.005; ns, not significant.
Expression of flagellar genes in wild-type and mutant E. coli Z36. (A to D) The percentage of GFP-positive cells in each population and mean levels of fluorescence expressed in arbitrary units (AU) are shown for the wild-type (A), ΔmotA mutant (B), ΔfliC mutant (C), ΔrflP mutant (D), ΔrflP ΔmotA mutant (E), and ΔrflP ΔfliC mutant (F) strains grown and measured as described for Fig. 2. Means from a minimum of four independent replicas are shown; error bars represent standard deviations. P values were calculated using the Mann-Whitney test (**, P < 0.005; *, P < 0.05; ns, not significant). Download FIG S2, EPS file, 0.2 MB (161.7KB, eps) .
Copyright © 2020 Laganenka et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Complementation of ΔmotA and ΔfliC deletions. (A) Motility halos formed on TBA 0.27% by wild-type E. coli Z36, the ΔmotA mutant, and the ΔfliC mutant expressing motAB and fliC under the control of a leaky trc promoter without IPTG induction. (B) Distributions of fluorescence levels of PfllhD-gfp, PfliA-gfp, and PfliC-gfp reporters in the wild-type, ΔmotA, and ΔfliC strains expressing motAB and fliC under the control of a leaky trc promoter without IPTG induction. Bacterial growth and measurements were as described for Fig. 1C to H. Download FIG S3, PDF file, 0.1 MB (124.2KB, pdf) .
Copyright © 2020 Laganenka et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Besides swimming in porous (such as agar) or viscous media, the mechanical load on the flagellar motor is also expected to increase when bacteria swim toward or attach to a surface. Such surface sensing may indeed be the major function of flagellum-mediated mechanosensing (21, 24). Consistently, we observed that the expression of fliC became elevated when wild-type (WT) E. coli Z36 cells were either forced to the surface by centrifugation (Fig. 2H) or grown on the surface of a semisolid (0.5%) agar (Fig. S4). No fliC gene induction was observed in ΔmotA cells under these conditions, suggesting that induction is mediated by the same mechanosensing mechanism. Notably, when cells were grown on a 0.5% agar surface, the flhD promoter activity of ΔmotA cells was the same as that of WT E. coli Z36 cells (Fig. S4). This confirms that motA deletion has no direct effect on flhD activity and suggests that the lower expression of flhD in nonmotile knockout strains grown on TBA 0.27% than in TB (Fig. 2C and D) was rather due to a high cell density and a corresponding nutrient depletion within the nonspreading colony.
Class I and III flagellar gene expression on TBA 0.5%. Distributions of fluorescence levels of PflhD-gfp and PfliC-gfp in E. coli Z36 and its ΔmotA knockout strain grown on TBA 0.5%, quantified as described for Fig. 1C to H. Download FIG S4, EPS file, 0.1 MB (121.4KB, eps) .
Copyright © 2020 Laganenka et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Anti-FlhC2FlhD4 factor RflP is involved in mechanosensing.
Since fliA and fliC are known to be repressed by RflP in Salmonella, we tested the effect of rflP deletion on the observed regulation. Unlike in the laboratory E. coli strains where RflP had no effect on flagellar gene expression (20), rflP deletion in E. coli Z36 resulted in strong derepression of fliA and fliC expression, particularly apparent in liquid TB medium (Fig. 2D and Fig. S2D). Consequently, most E. coli Z36 ΔrflP cells remained motile in liquid (Fig. 2G). This effect could be complemented by expressing RflP from the plasmid (Fig. S5). Deletion of rflP also derepressed the flagellar gene expression in the ΔmotA and ΔfliC backgrounds, both in liquid and on TBA 0.27% (Fig. 2E and F and Fig. S2E and F). Although fliA and particularly fliC expression was still weakly upregulated on soft agar even in E. coli Z36 ΔrflP, it remains unclear whether this residual RflP-independent regulation is due to mechanosensing, since to some extent it was also observed in ΔrflP ΔmotA and ΔrflP ΔfliC double knockouts (Fig. 2E and F and Fig. S2E and F).
Complementation of rflP deletion in E. coli Z36. Distributions of fluorescence levels of PfliC-gfp in E. coli Z36 ΔrflP expressing an rflP-FLAG fusion from an IPTG-inducible promoter, without induction or induced with 10 μM IPTG. Cells were grown in liquid TB as described for Fig. 1. Download FIG S5, EPS file, 0.1 MB (118.9KB, eps) .
Copyright © 2020 Laganenka et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The interplay between RflP-mediated repression and mechanosensing remains to be elucidated (see below). Nevertheless, we observed that the activity of the rflP promoter was independent of the cultivation conditions (Fig. S6), suggesting that RflP is not regulated by mechanosensing at the transcriptional level. This is in contrast to the transcriptional regulation of rflP by cell envelope stress in Salmonella (17).
Expression of the PrflP-gfp promoter in E. coli Z36 in TBA and in liquid TB. Mean levels of fluorescence expressed in arbitrary units (AU) are shown for the wild type and the indicated mutant strains grown and measured as described for Fig. 1. Means from a minimum of four independent replicas are shown; error bars represent standard deviations. P values were calculated using the Mann-Whitney test (ns, not significant). Download FIG S6, EPS file, 0.1 MB (99.3KB, eps) .
Copyright © 2020 Laganenka et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
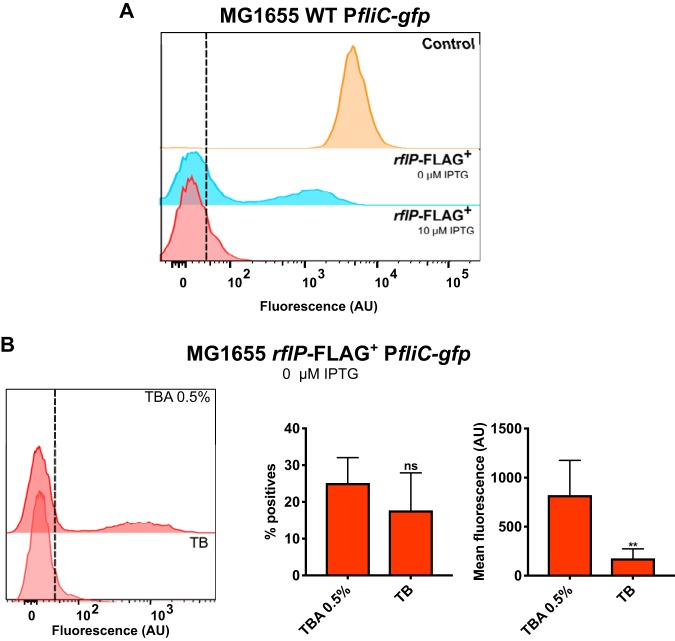
Importantly, our results also indicate that the determinants of mechanosensing other than RflP are present in E. coli MG1655. The overexpression of rflP not only led to the expected dose-dependent inhibition of fliC expression in MG1655 (Fig. 3A) but also apparently restored mechanosensing in this laboratory strain, with elevated fliC expression in soft agar compared to that in liquid (Fig. 3B). Since the inhibition of fliC expression by RflP was only partial in MG1655, this increase was particularly pronounced at the level of mean reporter activity rather than as a fraction of positive cells.
FIG 3.
The expression of RflP restores mechanosensing in E. coli MG1655. (A) Distribution of fluorescence levels of PfliC-gfp in E. coli MG1655 expressing an rflP-FLAG fusion from an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter, without induction or induced with 10 μM IPTG. Cells were grown in liquid TB as described for Fig. 1. (B) Distribution of fluorescence levels of PfliC-gfp in MG1655 cells expressing RflP, grown on TBA 0.5% or in liquid TB. Media were supplemented with ampicillin to prevent plasmid loss. Fluorescence was quantified as described for Fig. 1. Means from a minimum of seven independent replicas are shown; error bars represent standard deviations. P values were calculated using the Mann-Whitney test (**, P < 0.005; ns, not significant).
Proteome regulation by mechanosensing.
In order to identify nonflagellar proteins that could be regulated by mechanosensing, we performed comparative whole-proteome analysis of WT E. coli Z36 and ΔmotA cells grown either on 0.5% agar or in liquid TB medium. Apart from flagellar motor proteins, several metabolic enzymes and uncharacterized proteins were upregulated in a motA-dependent fashion on the agar surface, whereas several transporters and metabolic enzymes, as well as the sensor kinase QseC, were downregulated (Table 2). Since RflP could not be detected in these samples, its regulation at the protein level could not be assessed. Moreover, the expression of many other proteins, some of them involved in the acid stress response, was upregulated on the surface in a motA-independent manner (Table S2). Since QseC, a member of the QseB QseC two-component regulatory system, was among the most strongly downregulated proteins and it was previously shown to play a role in the regulation of flagellar expression (25, 26), we tested whether this system might have a regulatory role in mechanosensing. However, no effect of qseC deletion on fliC expression could be observed in E. coli Z36, grown either on soft agar or in TB (Fig. S7), suggesting that under our conditions and in this strain, this two-component system is inactive. Thus, QseC does not seem to have a mechanosensory role. Nevertheless, its mechanosensitive regulation by pathogenic E. coli might be important during infection, since QseC was shown to be important for virulence (26, 27).
TABLE 2.
Proteins regulated by motility-dependent surface sensing in E. coli Z36a
| Protein | Function | Fold change between: |
||
|---|---|---|---|---|
| ΔmotAsurf and WTsurf |
WTliq and WTsurf |
ΔmotAliq and ΔmotAsurf |
||
| YhdP | Uncharacterized protein | –21.18 | –177.91 | –1.13 |
| IroE | Uncharacterized protein | –5.49 | –78.78 | 1.60 |
| PrpC | Citrate synthase | –31.93 | –24.14 | 1.13 |
| PrpD | 2-Methylcitrate dehydratase | –4.96 | –5.62 | –1.03 |
| PrpE | Propionate-CoA ligase | –3.92 | –5.34 | –1.21 |
| FliH | Flagellar assembly protein | –3.71 | –2.22 | 1.77 |
| FliS | Flagellar protein | –3.20 | –2.10 | 1.88 |
| ElaB | Uncharacterized protein | 7.66 | –1.61 | –2.15 |
| MotB | Flagellar stator component | –5.02 | –1.27 | 2.75 |
| UbiB | Probable protein kinase | 29.56 | 1.00 | –1.25 |
| YfiQ | Uncharacterized protein | 7.61 | 2.32 | –1.52 |
| UidR | HTH-type transcriptional regulator | 3.65 | 2.51 | –1.63 |
| C2460 | Putative polyketide synthase | 3.56 | 3.42 | 1.64 |
| TdcF | Putative reactive intermediate deaminase | 3.34 | 3.54 | 1.09 |
| AnmK | Anhydro-N-acetylmuramic acid kinase | 3.10 | 3.75 | –1.15 |
| YffB | ArsC family protein | 4.44 | 4.07 | 1.81 |
| Gst | Glutathione S-transferase | 4.05 | 4.78 | 1.19 |
| Alr | Alanine racemase, biosynthetic | 4.60 | 5.11 | 1.19 |
| ThiQ | Thiamine import ATP-binding protein | 4.81 | 5.46 | 1.10 |
| NrdG | Anaerobic ribonucleoside-triphosphate reductase-activating protein |
5.57 | 6.79 | 1.19 |
| SdhE | FAD assembly factor | 5.20 | 7.30 | –1.88 |
| MobB | Molybdopterin-guanine dinucleotide biosynthesis protein |
4.95 | 7.76 | 2.42 |
| C3307 | Putative conserved protein | 5.65 | 10.05 | 1.15 |
| KefC | Glutathione-regulated potassium efflux system protein |
22.47 | 10.50 | –1.53 |
| YheV | Uncharacterized protein | 4.34 | 11.64 | 1.94 |
| C1760 | Putative transcriptional repressor | 19.74 | 13.14 | –1.12 |
| SapA | Peptide transport periplasmic protein | 19.54 | 18.54 | 1.46 |
| NusB | N utilization substance protein | 17.48 | 20.27 | 1.60 |
| YddE | Uncharacterized protein | 29.57 | 21.36 | –1.88 |
| C4017 | Putative ribose ABC transporter | 6.30 | 22.17 | 2.92 |
| YjjU | Uncharacterized protein | 77.64 | 37.40 | –1.68 |
| YbbL | Hypothetical ABC transporter ATP-binding protein |
26.50 | 43.96 | 2.04 |
| YcjZ | Hypothetical transcriptional regulator | 5.53 | 63.19 | 1.70 |
| IdnK | Gluconokinase | 33.19 | 84.34 | 2.27 |
| ThiJ | 4-Methyl-5(b-hydroxyethyl)-thiazole monophosphate biosynthesis enzyme |
163.82 | 111.05 | –1.09 |
| CyoC | Cytochrome bo3 ubiquinol oxidase subunit 3 | 69.54 | 130.08 | 1.47 |
| YgiY | Uncharacterized protein | 103.01 | 132.15 | 1.01 |
| QseC | Sensor protein | 103.01 | 132.15 | 1.01 |
| RlmC | 23S rRNA [uracil747-C5]methyltransferase | 92.80 | 166.17 | 2.17 |
Proteins were selected from the whole-proteome data based on the following criteria: differential expression between the WT and the ΔmotA mutant (fold change, ≥3) on TBA 0.5%, differential expression between the WT on TBA 0.5% (WTsurf) and in liquid TB (WTliq), and little or no difference (fold change, <3) between the ΔmotA mutant on TBA 0.5% (ΔmotAsurf) and in liquid TB (ΔmotAliq). The E. coli CFT073 protein annotation database was used as a reference. CoA, coenzyme A; HTH, helix turn helix; FAD, flavin adenine dinucleotide.
Sensor kinase QseC is not involved in mechanosensing. (A) Distributions of fluorescence levels of PfliC-gfp in E. coli Z36 and its qseC::Km knockout strain grown in TBA 0.27% or in TB, measured as described for Fig. 1. (B) Corresponding percentage of GFP-positive cells in each population. Means from a minimum of four independent replicas are shown; error bars represent standard deviations. P values were calculated using the Mann-Whitney test (***, P < 0.0005; **, P < 0.005; *, P < 0.05; ns, not significant). Download FIG S7, EPS file, 0.1 MB (116.8KB, eps) .
Copyright © 2020 Laganenka et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Proteins with MotA-independent changes in expression between growth on TBA 0.5% and that in liquid TB as revealed by whole-proteome analysis of E. coli Z36. Shown are only proteins with a ≥3-fold change between samples. Download Table S2, DOCX file, 0.02 MB (21.2KB, docx) .
Copyright © 2020 Laganenka et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In this study, we demonstrate that the expression of flagellar genes in a number of pathogenic E. coli strains is inhibited in liquid but that their expression is induced when bacteria are grown either in porous medium or on a surface. Because this regulation was not observed in commensal or laboratory strains of E. coli, it likely represents a specific mechanism to avoid the adaptive immune response of the host, akin to genetic or transcriptional variation in the expression of flagellar genes observed in other bacterial pathogens (6–12, 28). However, unlike with the stochastic antigenic variation or transcriptional switching observed in these bacteria, the control of flagellar gene expression in pathogenic E. coli apparently relies on sensing the mechanical load on the flagellar motor.
Although surface sensing-mediated control of gene expression, including control of bacterial flagellation, was reported before, most notably in Vibrio, Caulobacter, and Bacillus species (24, 29–35), this is, to our knowledge, the first example of such regulation in E. coli. It was previously shown that enterohemorrhagic E. coli (EHEC) can regulate the expression of EHEC-specific virulence genes upon sensing host attachment and fluid shear (36), but this surface sensing is not known to rely on flagella or to regulate motility. Moreover, this gene-regulatory mechanosensing controls motility in a different way than was previously described for posttranslational remodeling of the E. coli flagellar motor under a mechanical load (23, 37).
We hypothesize that such motility-dependent mechanosensing allows pathogenic E. coli to express flagella upon contact with epithelial cells or in mucus when motility is beneficial for the initial attachment to the host but that pathogenic E. coli shuts down the expression of flagellar genes upon entry into the host in order to delay detection by the immune system. In line with apparent trade-offs of flagellin expression in pathogenic E. coli, we also observed bimodal flagellar expression in these strains, with a subpopulation of cells remaining in the flagellum-off state even in soft agar. A similar bimodal expression was previously observed in other bacteria, including Salmonella (16) and Bacillus subtilis (38), but whereas in Salmonella bimodal fliC expression is known to depend on RflP activity (16, 17), deletion of rflP in E. coli Z36 did not abolish the bimodality of fliC expression, suggesting a different origin of bimodality.
Though the underlying mechanism of signal transduction from the flagellar motor to transcriptional regulation remains to be established, it apparently requires flagellar rotation under a load. The motility-off state of gene expression was observed not only for the filament-deficient ΔfliC strain of E. coli that cannot efficiently sense the load but also for the ΔmotA strain, which has immobilized flagella. This contrasts with observations of B. subtilis and Vibrio parahaemolyticus; these organisms with deletions of motor proteins mimic strains under high loads (30, 32), but their phenotype is similar to the lock-off mechanosensing phenotype of mot mutants of Caulobacter crescentus and Pseudomonas aeruginosa (24, 39).
Our results demonstrate that mechanosensing occurs in pathogenic E. coli strains due to the activity of the motility repressor RflP, which in these strains inhibits the expression of flagellar genes in liquid. Consistently, artificial upregulation of RflP in E. coli K-12 unmasks its mechanosensing response, demonstrating that other sensory determinants are present in the laboratory strain. While at this stage it cannot be ruled out that RflP-stimulated proteolysis of the FlhDC complex is required only to generally lower the basal expression of flagellar genes, its involvement in mechanosensing might also be direct, as occurred with a previously reported surface contact-dependent relief of proteolysis of a master flagellar regulator in B. subtilis (31).
Bacterial strains and culture conditions.
All strains and plasmids used in this study are listed in Table S1. E. coli strains were grown in liquid tryptone broth (TB) medium (10 g tryptone and 5 g NaCl per liter) or in lysogeny broth (LB) medium (10 g tryptone, 10 g NaCl, and 5 g yeast extract per liter) supplemented with antibiotics where necessary.
Cloning and knockout strain construction.
In order to construct the rflP-gfp fluorescent reporter, the bp –253-to-bp +90 promoter region of the rflP gene was amplified from the E. coli Z36 chromosome and cloned into the pUA66 vector using XhoI and BamHI restriction sites (40). For rflP-FLAG plasmid construction, the rflP gene sequence without a stop codon was amplified from the E. coli Z36 chromosome with a FLAG coding sequence, with the stop codon being added via a primer overhang. The resulting fusion construct was cloned into the pTrc99A vector using SacI and XbaI restriction sites (41).
All the knockout strains described in this work were obtained using the lambda red recombination system as described previously (42). Where necessary, kanamycin cassettes were flipped out using FLP-FLP recombination target (FRT) recombination (43).
Motility assay in soft agar.
Single colonies of different E. coli strains were transferred onto the surfaces of soft agar plates (TB containing 0.27% agar), and the motile behavior of E. coli strains was assessed after 6 to 8 h of incubation at 37°C by analyzing the motility halo diameter. The cells from the edges of such halos were subsequently transferred into 3 ml of fresh TB medium and grown with shaking at 37°C to an optical density at 600 nm (OD600) of 0.5 to 0.6. The motility of the cells was then analyzed using phase-contrast microscopy.
Single-bacterium movement tracking.
E. coli MG1655, Z36, and Z36 ΔrflP overnight cultures were diluted in fresh TB and grown at 37°C with shaking to the OD600 of 0.6. The samples were subsequently diluted to final OD600s of 0.05 to 0.1 in TB and loaded into ibidi channels (μ-Slide Chemotaxis3D; ibidi GmbH, Germany). Movies were recorded at ×10 magnification with a phase-contrast microscope at a rate of 20 frames per second using a complementary metal-oxide semiconductor (CMOS) camera (exposure time, 1 ms), normalized by dividing each frame by an image of the background illumination under the same conditions and inverted in order for the cells to appear bright. A projection of the temporal maximum of each pixel was computed to produce an image of the cell trajectories.
Flow cytometry.
Promoter activities of the flagellar genes flhD, fliA, and fliC and the motility regulator rflP were assayed using plasmid-based reporters containing the respective promoter regions fused to gfp (Table S1). Promoter activities were analyzed in the cells collected from the edges of motility halos formed on 0.27% TBA plates and after subsequent cultivation in liquid TB medium. The samples were diluted in 2 ml tethering buffer (10 mM KH2PO4, 100 μM EDTA, 1 μM l-methionine, and 10 mM sodium lactate, pH 7.0), and fluorescence was measured with a BD LSRFortessa SORP cell analyzer (BD Biosciences, Germany).
In order to compare PfliC-gfp activity in attached and planktonic cells, 300 μl of WT Z36 and ΔmotA mutant day cultures in TB (OD600 = 0.6, diluted in fresh TB to the final OD600 of 0.05) were inoculated into the wells of a 96-well microtiter plate (ibidi, Germany), and the cells were centrifuged for 45 min (37°C, 4,000 rpm). The supernatant with nonattached planktonic cells was transferred into new wells. The wells with attached cells were washed three times with 500 μl tethering buffer, and the presence of attached cells was verified with phase-contrast microscopy. Fresh TB (300 μl) was subsequently added to the wells, and the plates were incubated at 37°C. The measurements of PfliC-gfp activity in attached and planktonic cells were performed with flow cytometry after 60 and 90 min of incubation.
Proteomic analysis.
To compare the proteomes of WT E. coli Z36 and ΔmotA strains grown on the surfaces of soft agar plates and in the liquid medium, the cells were grown on the surface of 0.5% TBA for 4 h at 37°C. Flagellar gene expression was confirmed with flow cytometry using PfliC-gfp promoter fusion activity as a readout. The cells were then washed off the plates with 2 ml cold phosphate-buffered saline (PBS) (8 g NaCl, 0.2 g KCl, 1.42 g Na2HPO4, 0.27 g KH2PO4 per liter), and the final sample size was adjusted to 5 ml with an OD600 of 0.6. The cells were then washed twice in 5 ml cold PBS, and the pellets were stored at –20°C. Before the samples were frozen, 50 μl of the suspension was reinoculated into 5 ml of liquid TB and grown at 37°C with shaking to an OD600 of 0.6. The cells were then washed twice in 5 ml cold PBS, and the pellets were stored at –20°C. The collected samples were subsequently subjected to proteomic analysis using mass spectrometry and subsequent data analysis as described before (44, 45). Although no reference database for E. coli Z36 is available, using the E. coli CFT073 protein database resulted in full coverage of the analyzed proteome.
ACKNOWLEDGMENTS
We thank Timo Glatter and Jörg Kahnt for their help with the proteomics analysis.
This work was supported by the Max Planck Society.
L.L., M.E.L., R.C., and V.S. designed the experiments. L.L., M.E.L., and R.C. performed the experiments and analyzed the data. L.L. and V.S. wrote the manuscript.
Footnotes
Citation Laganenka L, López ME, Colin R, Sourjik V. 2020. Flagellum-mediated mechanosensing and RflP control motility state of pathogenic Escherichia coli. mBio 11:e02269-19. https://doi.org/10.1128/mBio.02269-19.
REFERENCES
- 1.Soutourina OA, Bertin PN. 2003. Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol Rev 27:505–523. doi: 10.1016/S0168-6445(03)00064-0. [DOI] [PubMed] [Google Scholar]
- 2.Chevance FFV, Hughes KT. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol 6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lertsethtakarn P, Ottemann KM, Hendrixson DR. 2011. Motility and chemotaxis in Campylobacter and Helicobacter. Annu Rev Microbiol 65:389–410. doi: 10.1146/annurev-micro-090110-102908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Josenhans C, Suerbaum S. 2002. The role of motility as a virulence factor in bacteria. Int J Med Microbiol 291:605–614. doi: 10.1078/1438-4221-00173. [DOI] [PubMed] [Google Scholar]
- 5.Yoon S, Kurnasov O, Natarajan V, Hong M, Gudkov AV, Osterman AL, Wilson IA. 2012. Structural basis of TLR5-flagellin recognition and signaling. Science 335:859–864. doi: 10.1126/science.1215584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonifield HR, Hughes KT. 2003. Flagellar phase variation in Salmonella enterica is mediated by a posttranscriptional control mechanism. J Bacteriol 185:3567–3574. doi: 10.1128/jb.185.12.3567-3574.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ikeda JS, Schmitt CK, Darnell SC, Watson PR, Bispham J, Wallis TS, Weinstein DL, Metcalf ES, Adams P, O'Connor CD, O'Brien AD. 2001. Flagellar phase variation of Salmonella enterica serovar Typhimurium contributes to virulence in the murine typhoid infection model but does not influence Salmonella-induced enteropathogenesis. Infect Immun 69:3021–3030. doi: 10.1128/IAI.69.5.3021-3030.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park SF, Purdy D, Leach S. 2000. Localized reversible frameshift mutation in the flhA gene confers phase variability to flagellin gene expression in Campylobacter coli. J Bacteriol 182:207–210. doi: 10.1128/jb.182.1.207-210.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Josenhans C, Eaton KA, Thevenot T, Suerbaum S. 2000. Switching of flagellar motility in Helicobacter pylori by reversible length variation of a short homopolymeric sequence repeat in fliP, a gene encoding a basal body protein. Infect Immun 68:4598–4603. doi: 10.1128/iai.68.8.4598-4603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caldwell MB, Guerry P, Lee EC, Burans JP, Walker RI. 1985. Reversible expression of flagella in Campylobacter jejuni. Infect Immun 50:941–943. doi: 10.1128/IAI.50.3.941-943.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stewart MK, Cummings LA, Johnson ML, Berezow AB, Cookson BT. 2011. Regulation of phenotypic heterogeneity permits Salmonella evasion of the host caspase-1 inflammatory response. Proc Natl Acad Sci U S A 108:20742–20747. doi: 10.1073/pnas.1108963108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simm R, Remminghorst U, Ahmad I, Zakikhany K, Römling U. 2009. A role for the EAL-like protein STM1344 in regulation of CsgD expression and motility in Salmonella enterica serovar Typhimurium. J Bacteriol 191:3928–3937. doi: 10.1128/JB.00290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stewart MK, Cookson BT. 2014. Mutually repressing repressor functions and multi-layered cellular heterogeneity regulate the bistable Salmonella fliC census. Mol Microbiol 94:1272–1284. doi: 10.1111/mmi.12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hengge R, Galperin MY, Ghigo J-M, Gomelsky M, Green J, Hughes KT, Jenal U, Landini P. 2016. Systematic nomenclature for GGDEF and EAL domain-containing cyclic di-GMP turnover proteins of Escherichia coli. J Bacteriol 198:7–11. doi: 10.1128/JB.00424-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wada T, Morizane T, Abo T, Tominaga A, Inoue-Tanaka K, Kutsukake K. 2011. EAL domain protein YdiV acts as an anti-FlhD4C2 factor responsible for nutritional control of the flagellar regulon in Salmonella enterica serovar Typhimurium. 193:1600–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koirala S, Mears P, Sim M, Golding I, Chemla YR, Aldridge PD, Rao CV. 2014. A nutrient-tunable bistable switch controls motility in Salmonella enterica serovar Typhimurium. mBio 5:e01611-14. doi: 10.1128/mBio.01611-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spöring I, Felgner S, Preuße M, Eckweiler D, Rohde M, Häussler S, Weiss S, Erhardt M. 2018. Regulation of flagellum biosynthesis in response to cell envelope stress in Salmonella enterica serovar Typhimurium. mBio 9:e00736-17. doi: 10.1128/mBio.00736-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Macnab RM. 1992. Genetics and biogenesis of bacterial flagella. Annu Rev Genet 26:131–158. doi: 10.1146/annurev.ge.26.120192.001023. [DOI] [PubMed] [Google Scholar]
- 19.Feng L, Liu B, Liu Y, Ratiner YA, Hu B, Li D, Zong X, Xiong W, Wang L. 2008. A genomic islet mediates flagellar phase variation in Escherichia coli strains carrying the flagellin-specifying locus flk. J Bacteriol 190:4470–4477. doi: 10.1128/JB.01937-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wada T, Hatamoto Y, Kutsukake K. 2012. Functional and expressional analyses of the anti-FlhD4C2 factor gene ydiV in Escherichia coli. Microbiology 158:1533–1542. doi: 10.1099/mic.0.056036-0. [DOI] [PubMed] [Google Scholar]
- 21.Belas R. 2014. Biofilms, flagella, and mechanosensing of surfaces by bacteria. Trends Microbiol 22:517–527. doi: 10.1016/j.tim.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 22.Gordon VD, Wang L. 2019. Bacterial mechanosensing: the force will be with you, always. J Cell Sci 132:jcs227694. doi: 10.1242/jcs.227694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lele PP, Hosu BG, Berg HC. 2013. Dynamics of mechanosensing in the bacterial flagellar motor. Proc Natl Acad Sci U S A 110:11839–11844. doi: 10.1073/pnas.1305885110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hug I, Deshpande S, Sprecher KS, Pfohl T, Jenal U. 2017. Second messenger-mediated tactile response by a bacterial rotary motor. Science 358:531–534. doi: 10.1126/science.aan5353. [DOI] [PubMed] [Google Scholar]
- 25.Sperandio V, Torres AG, Kaper JB. 2002. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol Microbiol 43:809–821. doi: 10.1046/j.1365-2958.2002.02803.x. [DOI] [PubMed] [Google Scholar]
- 26.Moreira CG, Weinshenker D, Sperandio V. 2010. QseC mediates Salmonella enterica serovar Typhimurium virulence in vitro and in vivo. Infect Immun 78:914–926. doi: 10.1128/IAI.01038-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hughes DT, Clarke MB, Yamamoto K, Rasko DA, Sperandio V. 2009. The QseC adrenergic signaling cascade in enterohemorrhagic E. coli (EHEC). PLoS Pathog 5:e1000553. doi: 10.1371/journal.ppat.1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simm R, Lusch A, Kader A, Andersson M, Römling U. 2007. Role of EAL-containing proteins in multicellular behavior of Salmonella enterica serovar Typhimurium. J Bacteriol 189:3613–3623. doi: 10.1128/JB.01719-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guttenplan SB, Kearns DB. 2013. Regulation of flagellar motility during biofilm formation. FEMS Microbiol Rev 37:849–871. doi: 10.1111/1574-6976.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cairns LS, Marlow VL, Bissett E, Ostrowski A, Stanley-Wall NR. 2013. A mechanical signal transmitted by the flagellum controls signalling in Bacillus subtilis. Mol Microbiol 90:6–21. doi: 10.1111/mmi.12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mukherjee S, Bree AC, Liu J, Patrick JE, Chien P, Kearns DB. 2015. Adaptor-mediated Lon proteolysis restricts Bacillus subtilis hyperflagellation. Proc Natl Acad Sci U S A 112:250–255. doi: 10.1073/pnas.1417419112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCarter L, Hilmen M, Silverman M. 1988. Flagellar dynamometer controls swarmer cell differentiation of V. parahaemolyticus. Cell 54:345–351. doi: 10.1016/0092-8674(88)90197-3. [DOI] [PubMed] [Google Scholar]
- 33.Li G, Brown PJB, Tang JX, Xu J, Quardokus EM, Fuqua C, Brun YV. 2012. Surface contact stimulates the just-in-time deployment of bacterial adhesins. Mol Microbiol 83:41–51. doi: 10.1111/j.1365-2958.2011.07909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diethmaier C, Chawla R, Canzoneri A, Kearns DB, Lele PP, Dubnau D. 2017. Viscous drag on the flagellum activates Bacillus subtilis entry into the K-state. Mol Microbiol 106:367–380. doi: 10.1111/mmi.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hölscher T, Schiklang T, Dragoš A, Dietel AK, Kost C, Kovács ÁT. 2018. Impaired competence in flagellar mutants of Bacillus subtilis is connected to the regulatory network governed by DegU. Environ Microbiol Rep 10:23–32. doi: 10.1111/1758-2229.12601. [DOI] [PubMed] [Google Scholar]
- 36.Alsharif G, Ahmad S, Islam MS, Shah R, Busby SJ, Krachler AM. 2015. Host attachment and fluid shear are integrated into a mechanical signal regulating virulence in Escherichia coli O157:H7. Proc Natl Acad Sci U S A 112:5503–5508. doi: 10.1073/pnas.1422986112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tipping MJ, Delalez NJ, Lim R, Berry RM, Armitage JP. 2013. Load-dependent assembly of the bacterial flagellar motor. mBio 4:e00551-13. doi: 10.1128/mBio.00551-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kearns DB, Losick R. 2005. Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev 19:3083–3094. doi: 10.1101/gad.1373905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laventie BJ, Sangermani M, Estermann F, Manfredi P, Planes R, Hug I, Jaeger T, Meunier E, Broz P, Jenal U. 2019. A surface-induced asymmetric program promotes tissue colonization by Pseudomonas aeruginosa. Cell Host Microbe 25:140–152.e6. doi: 10.1016/j.chom.2018.11.008. [DOI] [PubMed] [Google Scholar]
- 40.Zaslaver A, Bren A, Ronen M, Itzkovitz S, Kikoin I, Shavit S, Liebermeister W, Surette MG, Alon U. 2006. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat Methods 3:623–628. doi: 10.1038/nmeth895. [DOI] [PubMed] [Google Scholar]
- 41.Amann E, Ochs B, Abel K-J. 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 42.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 44.Yuan J, Jin F, Glatter T, Sourjik V. 2017. Osmosensing by the bacterial PhoQ/PhoP two-component system. Proc Natl Acad Sci U S A 114:E10792–E10798. doi: 10.1073/pnas.1717272114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rudenko I, Ni B, Glatter T, Sourjik V. 2019. Inefficient secretion of anti-sigma factor FlgM inhibits bacterial motility at high temperature. iScience 16:145–154. doi: 10.1016/j.isci.2019.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laganenka L, Colin R, Sourjik V. 2016. Chemotaxis towards autoinducer 2 mediates autoaggregation in Escherichia coli. Nat Commun 7:12984. doi: 10.1038/ncomms12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of bacterial strains and plasmids used in this study. Download Table S1, DOCX file, 0.02 MB (25.2KB, docx) .
Copyright © 2020 Laganenka et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of flagellar genes in pathogenic and commensal E. coli isolates in TBA and in liquid TB. Experiments were performed as described for Fig. 1. Percentages of GFP-positive cells in E2808 (A), S13 (B), Z36 (C), T111 (D), M185/1-1 (E), and MG1655 (F) populations are shown. Means from a minimum of four independent replicas are shown; error bars represent standard deviations. P values were calculated using the Mann-Whitney test. *, P < 0.05; ns, not significant. Download FIG S1, EPS file, 0.1 MB (100.2KB, eps) .
Copyright © 2020 Laganenka et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of flagellar genes in wild-type and mutant E. coli Z36. (A to D) The percentage of GFP-positive cells in each population and mean levels of fluorescence expressed in arbitrary units (AU) are shown for the wild-type (A), ΔmotA mutant (B), ΔfliC mutant (C), ΔrflP mutant (D), ΔrflP ΔmotA mutant (E), and ΔrflP ΔfliC mutant (F) strains grown and measured as described for Fig. 2. Means from a minimum of four independent replicas are shown; error bars represent standard deviations. P values were calculated using the Mann-Whitney test (**, P < 0.005; *, P < 0.05; ns, not significant). Download FIG S2, EPS file, 0.2 MB (161.7KB, eps) .
Copyright © 2020 Laganenka et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Complementation of ΔmotA and ΔfliC deletions. (A) Motility halos formed on TBA 0.27% by wild-type E. coli Z36, the ΔmotA mutant, and the ΔfliC mutant expressing motAB and fliC under the control of a leaky trc promoter without IPTG induction. (B) Distributions of fluorescence levels of PfllhD-gfp, PfliA-gfp, and PfliC-gfp reporters in the wild-type, ΔmotA, and ΔfliC strains expressing motAB and fliC under the control of a leaky trc promoter without IPTG induction. Bacterial growth and measurements were as described for Fig. 1C to H. Download FIG S3, PDF file, 0.1 MB (124.2KB, pdf) .
Copyright © 2020 Laganenka et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Class I and III flagellar gene expression on TBA 0.5%. Distributions of fluorescence levels of PflhD-gfp and PfliC-gfp in E. coli Z36 and its ΔmotA knockout strain grown on TBA 0.5%, quantified as described for Fig. 1C to H. Download FIG S4, EPS file, 0.1 MB (121.4KB, eps) .
Copyright © 2020 Laganenka et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Complementation of rflP deletion in E. coli Z36. Distributions of fluorescence levels of PfliC-gfp in E. coli Z36 ΔrflP expressing an rflP-FLAG fusion from an IPTG-inducible promoter, without induction or induced with 10 μM IPTG. Cells were grown in liquid TB as described for Fig. 1. Download FIG S5, EPS file, 0.1 MB (118.9KB, eps) .
Copyright © 2020 Laganenka et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of the PrflP-gfp promoter in E. coli Z36 in TBA and in liquid TB. Mean levels of fluorescence expressed in arbitrary units (AU) are shown for the wild type and the indicated mutant strains grown and measured as described for Fig. 1. Means from a minimum of four independent replicas are shown; error bars represent standard deviations. P values were calculated using the Mann-Whitney test (ns, not significant). Download FIG S6, EPS file, 0.1 MB (99.3KB, eps) .
Copyright © 2020 Laganenka et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sensor kinase QseC is not involved in mechanosensing. (A) Distributions of fluorescence levels of PfliC-gfp in E. coli Z36 and its qseC::Km knockout strain grown in TBA 0.27% or in TB, measured as described for Fig. 1. (B) Corresponding percentage of GFP-positive cells in each population. Means from a minimum of four independent replicas are shown; error bars represent standard deviations. P values were calculated using the Mann-Whitney test (***, P < 0.0005; **, P < 0.005; *, P < 0.05; ns, not significant). Download FIG S7, EPS file, 0.1 MB (116.8KB, eps) .
Copyright © 2020 Laganenka et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Proteins with MotA-independent changes in expression between growth on TBA 0.5% and that in liquid TB as revealed by whole-proteome analysis of E. coli Z36. Shown are only proteins with a ≥3-fold change between samples. Download Table S2, DOCX file, 0.02 MB (21.2KB, docx) .
Copyright © 2020 Laganenka et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.