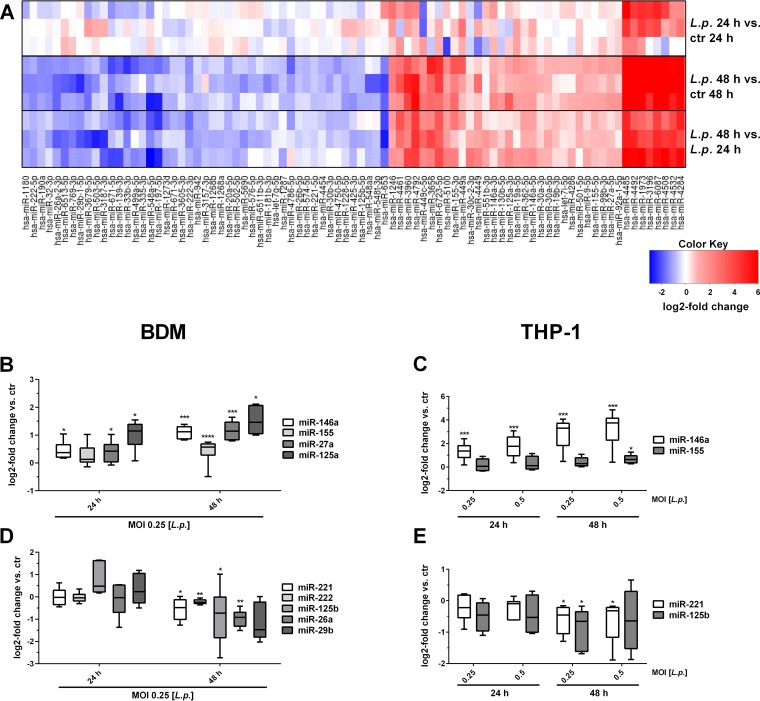
FIG 1.
Differentially expressed miRNAs in blood-derived macrophages after L. pneumophila infection. BDMs from three different donors were infected with L. pneumophila at an MOI of 0.25 or left untreated as a control (ctr). Samples were taken 24 and 48 h postinfection. RNA was isolated and used for library preparation and enriched for small RNAs using a TruSeq small RNA kit. Sequencing was performed in a multiplexed run of 1 × 51 cycle +7 (index). (A) Heatmap of the miRNA log2 fold change over the respective controls. The expression of distinct upregulated (B and C) and downregulated (D and E) miRNAs in BDMs and THP-1 cells after L. pneumophila infection was validated by qPCR using TaqMan assays and are displayed as log2 fold changes. Boxes show the upper and lower quartiles with medians and whiskers indicating minimal and maximal values of five to nine independent biological replicates for dysregulated miRNAs. Paired t tests were performed. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ .001; ****, P ≤ 0.0001 (compared to the corresponding control).