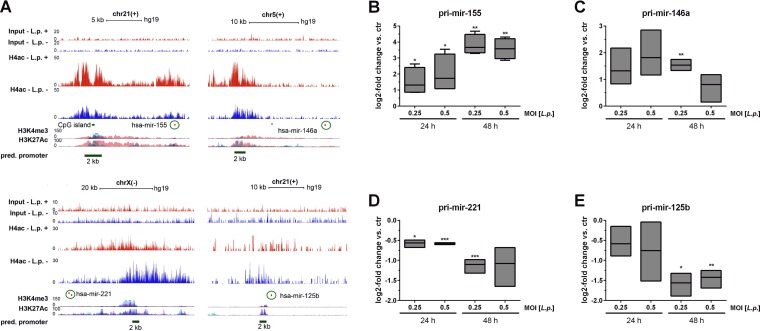
FIG 2.
Infection-related chromatin changes on miRNA promoters. The alterations of the acetylation pattern at the miRNA promoter regions of miR-155, miR-146a, miR-221, and miR-125b after L. pneumophila infection in BDMs were investigated. BDMs were infected with L. pneumophila for 1 h, and ChIP was performed using a pan-ac-H4-antibody (based on data published by Du Bois et al. [32]). (A) After sequencing, the enrichment of H4 acetylation at the miR promoters was analyzed. As confirmation for active sites in the promoter region, IP data for H3K4me3 and H3K27Ac provided by the Encode UCSC browser are shown. Horizontal dark green lines indicate the genomic locations of the promoter regions (2,000 bp) for the four analyzed miRNAs corresponding to given coordinates. Circles highlight the mature transcript of each miRNA. The expression of the pri-miRs (B, C, D, and E) in BDMs after L. pneumophila infection (MOI of 0.25 or 0.5) was examined by qPCR and is displayed as log2 fold changes. Boxes show the upper and lower quartiles, with medians (when n = 3 independent biological replicates) and whiskers indicating minimal and maximal values (when n = 4 independent biological replicates). Paired t tests were performed. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 (compared to the corresponding control).