Abstract
Vibrio parahaemolyticus (Vp) is the etiological agent of the acute hepatopancreatic necrosis disease (AHPND) in Penaeus vannamei shrimp. Vp possesses a 63–70 kb conjugative plasmid that encodes the binary toxin PirAvp/PirBvp. The 250 kDa PirABvp complex was purified by affinity chromatography with galactose-sepharose 4B and on a stroma from glutaraldehyde-fixed rat erythrocytes column, as a heterotetramer of PirAvp and PirBvp subunits. In addition, recombinant pirB (rPirBvp) and pirA (rPirAvp) were obtained. The homogeneity of the purified protein was determined by SDS-PAGE analysis, and the yield of protein was 488 ng/100 μg of total protein of extracellular products. The PirABvp complex and the rPirBvp showed hemagglutinating activity toward rat erythrocytes. The rPirAvp showed no hemagglutinating capacity toward the animal red cells tested. Among different mono and disaccharides tested, only GalNH2 and GlcNH2 were able to inhibit hemagglutination of the PirABvp complex and the rPirBvp. Glycoproteins showed inhibitory specificity, and fetuin was the glycoprotein that showed the highest inhibition. Other glycoproteins, such as mucin, and glycosaminoglycans, such as heparin, also inhibited the activity. Desialylation of erythrocytes enhanced the hemagglutinating activity. This confirms that Gal or Gal (β1,4) GlcNAc are the main ligands for PirABvp. The agglutinating activity of the PirABvp complex and the rPirBvp is not dependent on cations, because addition of Mg2+ or Ca2+ showed no effect on the protein capacity. Our results strongly suggest that the PirBvp subunit is a lectin, which is part of the PirA/PirBvp complex, and it seems to participate in bacterial pathogenicity.
Keywords: PirABvp toxin, lectin, binary toxin, AHPND, Vibrio parahaemolyticus, amino-sugars, glycosaminoglycans
1. Introduction
The worldwide production of Penaeus vannamei shrimp has suffered significant losses in Asia [1,2] and Latin America [3,4]. This has been due to the acute hepatopancreatic necrosis disease (AHPND), which is considered an emerging shrimp disease. Specific strains of Vibrio parahaemolyticus (Vp) that were first reported in Mexico in 2013 are the etiological agents of the disease. The Vp strains harbor a 63–70 kb conjugative plasmid [5]. The plasmid called pVA1 encodes a binary PirA/PirB toxin homologous to the Photorhabdus luminiscens insect-related (Pir) binary toxin [5,6,7,8]. Recombinant PirAvp and PirBvp were reported to be the primary virulence factor that causes massive sloughing of the tubule epithelial cells of the shrimp hepatopancreas (Hp), causing 100% cumulative mortality in 24 h post-infection [8].
The PirABvp binary toxin has been identified in other Vibrio species belonging to the Harveyi clade, such as V. harveyi [9], V. campbellii, and V. owensii [5,10,11]. The presence of genes encoding PirA and PirB was reported in the Gram-positive bacterial species Micrococcus luteus [12]. This bacterium was isolated in 2006 from the Hp of farmed P. vannamei shrimp collected in Nayarit, Mexico; it is possible that the toxin was acquired by horizontal gene transfer from AHPND isolates [13,14]. The genome of Vibrio punensis, belonging to the Orientalis clade, also contains the toxigenic genes that cause AHPND in shrimp [15]. The presence of these genes in different bacterial species is a potential risk for the spread of emerging diseases. The clinical signs of shrimp affected with AHPND are a pale Hp, empty gut, anorexia, and lethargy accompanied by pathognomonic lesions: massive sloughing of tubule epithelial cells of the shrimp Hp [6,16]. Most authors suggest that the PirABvp toxin acts as a pore-forming toxin that kills the hepatopancreatic cells of shrimp, although this has not been experimentally demonstrated. Moreover, the specific mechanisms used to recognize specific cellular receptors for the PirABvp toxin have not yet been elucidated [6,16,17].
AB toxins are synthesized by a variety of bacteria, pathogens, and plants. The most well-known AB toxins include the cholera toxin, shiga toxin, pertussis toxin, anthrax, and ricin [18]. Many of these AB toxins contain two functional regions: an enzymatically active cytotoxic subunit and a region that recognizes cell surface receptors [19]. Ricin is a type of AB toxin with lectin activity. It is isolated from the castor tree (Ricinus cummunis), one of the most poisonous plants in the world. Lectins are a group of proteins that share the ability to recognize and bind to specific carbohydrate structures. They are ubiquitous in nature. They are present in various types of organisms, and their particular interaction with carbohydrate structures plays a role in different biological processes, including the control of pathogens and predators [20,21].
Currently, there are no reports of AB toxins with lectin activity in marine bacteria, so the isolation and determination of the specificity of both subunits for sugars are necessary to identify adhesion molecules or receptors on the hepatopancreatic epithelial cells. This work aims to identify the specificity of the toxin from sugars of the subunits of the PirABvp toxin secreted by Vibrio parahaemolyticus, particularly PirBvp.
2. Materials and Methods
2.1. Growth Conditions of E. coli Strains and Vibrio Parahaemolyticus
Escherichia coli BL21 CodonPlus-RIL (Agilent Technologies, Inc., Santa Clara, CA, USA) and E. coli Top10 (Invitrogen, Carlsbad, CA, USA) were used for the transformation and replication, respectively, of the genes coding the toxins PirAvp and PirBvp and cloned into the pET system plasmid. The E. coli culture was maintained on Luria–Bertani (LB) agar plates supplemented with the appropriate antibiotics. For rPirAvp, kanamycin (50 μg/mL) and chloramphenicol (34 μg/mL) were used; for rPirBvp, ampicillin (100 μg/mL) and chloramphenicol (34 μg/mL) were used. For protein expression, cultures were grown in LB at 37 °C and induced at 30 °C with shaking (200 rpm).
The highly virulent M0904 strain of Vibrio parahaemolyticus (Vp), which causes AHPND in shrimp, was used. It was isolated from shrimp hepatopancreas [16]. A pure culture of VpM0904 was obtained by inoculation in tryptic soy broth (TSB, Bioxon, Mexico), according to Soto-Rodriguez et al. [16].
2.2. Cloning and Expression of Recombinant PirAvp and PirBvp
The recombinant proteins rPirAvp and rPirBvp were cloned separately, transformed using the E. coli BL21 CodonPlus-RIL cell line, and induced with isopropyl-β-D-1-thiogalactopyranoside (IPTG). Briefly, the PirAvp sequence was purified using the resulting amplicon for a PCR with primers containing NdeI and HindIII (Thermo-Fisher Co., San José, CA, USA) restriction sites: PirA-NdeI-Forward: 5′-AAT CAT ATG AGT AAC AAT ATA AAA CAT G-3′ and PirA-HindIII-Reverse: 5′-AAT AAG CTT AGT GGT AAT AGA TTG TAC AG-3′. The PCR products were purified and double digested with NdeI and HindIII enzymes before ligation into the vector pET-28a (Promega, Co., Madison, WI, USA) for the transformation of E. coli BL21 CodonPlus-RIL. Protein expression was induced with 0.5 mM of IPTG at 30 °C for 16 h; later, the cells were collected by centrifugation and stored at −20 °C until the purification process. A similar protocol was followed for the rPirBvp expression. The PCR product was obtained using primers containing NcoI and XhoI restriction sites. The primers were PirB-NcoI (Forward-AAT CCA TGG GTA CTA ACG AAT ACG TTG TAA C-3′) and PirB-XhoI (Reverse-TAA CTC GAG TTA CTA CTT TTC TGT ACC AAA TTC AT-3′). The PCR product was purified, double digested with NcoI and XhoI (Thermo-Fisher), and ligated into the plasmid pET32c (Promega) to transform E. coli BL21 CodonPlus-RIL for the expression of a thioredoxin (TRX) fusion protein and His6 tag. Recombinant protein expression was induced by 0.5 mM IPTG at 30 °C for four h; then, it was separated by centrifugation and stored at −20 °C until the purification process.
2.3. Purification of rPirAvp and rPirBvp Toxins
Purification of the recombinant proteins was carried out by one-step Ni-affinity chromatography on a Ni-Sepharose Fast Flow resin (GE Healthcare, Chicago, IL, USA). Briefly, bacterial cells were washed using buffer A (50 mM Tris-HCl, 300 mM NaCl, 10 mM imidazole, 5% glycerol, 0.25 mM DDT, pH 8.0) and the cell suspension was sonicated at 42 W on ice for 5 min (5 s on, 15 s off). Cell debris was removed by centrifugation at 24,000× g for 45 min at 4 °C. After sonication, the supernatant solution was loaded on a Ni-affinity column, which had already been equilibrated with buffer A. To remove any nonspecifically bound proteins, the column was extensively washed with buffer B (50 mM Tris-HCl, 300 mM NaCl, 20 mM imidazole, 5% glycerol, 0.25 mM DDT, pH 8.0). For rPirAvp, the protein was eluted with buffer C (50 mM Tris–HCl, 300 mM NaCl, 5% glycerol, 0.25 mM DTT, pH 8.0) with different concentrations of imidazole (20, 50, 100, 300, and 500 mM) and the TEV protease protocol (Sigma, St. Louis, MO, USA) was used to separate the His6 tag anchored to the purified rPirAvp. For rPirBvp, the protein was eluted using the same buffer with different concentrations of imidazole, and TRX was removed from the purified protein by incubating with enterokinase (ThermoFisher) at 37 °C overnight. The molecular weights of both purified proteins were verified by SDS-PAGE analysis under reducing conditions using Coomassie blue staining.
2.4. Hemagglutinating Activity
The hemagglutinating activity (HA) of rPirAvp and rPirBvp toxins was tested using red blood cells (RBCs) from several animal species and human RBCs from healthy donors. Blood was collected in sterile Alsever’s solution (100 mM glucose, 20 mM NaCl, 30 mM sodium citrate, pH 7.2) and washed three times with phosphate-buffered saline (PBS, 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4) by centrifugation (800× g for 10 min). The HA was performed in 96-well microtiter U plates (NUNC, Denmark) by a two-fold serial dilution according to the protocol of Fragkiadakis [22]. Briefly, after the two-fold dilution in PBS, 25 μL of each protein’s dilution was mixed with 25 μL of 2% (w/v) erythrocytes previously suspended in PBS. The suspension was incubated at room temperature for 120 minutes. The hemagglutinating titer was reported as the inverse of the last dilution exhibiting HA. HA was also assessed in the presence of RBCs treated with sialidase (0.1 U of Vibrio cholerae sialidase (Sigma) for every 0.5 mL of packed RBCs at 37 °C for 45 minutes). To determine whether the subunits have protease activity, remazol brilliant blue R (Sigma) was used according to the manufacturer’s instructions.
2.5. Purification of Wild-Type PirABvp Toxin Complex
The wild-type PirABvp toxin complex was purified from the extracellular products (ECPs) of VpM0904 by two affinity chromatography systems. ECPs were obtained by removing the bacterial cells by centrifugation at 4500× g for 10 min at 4 °C and filtration at 0.22 µm. The supernatant culture broth was separated and the purification process continued.
Affinity chromatography was done on glutaraldehyde-fixed stroma from Wistar rat’s erythrocytes fixed with glutaraldehyde and immobilized on G-25 Sephadex (Sigma, USA). The rat erythrocytes were fixed following the methodology described by Vazquez et al. [23]. The fixed erythrocytes were packed in a column (1.5 × 25 cm) containing Sephadex G-25 (Pharmacia Fine Chem, Uppsala, Sweden). The supernatant culture broth was dialyzed in potassium/sodium phosphate buffer (KNa PBS, 50 mM NaH2PO4, 50 mM K2HPO4, 150 mM NaCl, pH 7.4) and 195.4 mg was loaded onto a column that had been previously equilibrated with the same buffer at room temperature at a flow rate of 15 mL/h. The unbound material was washed from the column with KNa PBS until the absorbance of the eluent was <0.01 at 280 nm. The PirABvp toxin was eluted with 3% acetic acid, 1.5 mL fractions were collected, and the pH was immediately neutralized with 1.0 M NaOH. Fractions were pooled and dialyzed with KNa PBS; then, the protein was quantified by the Bradford method [24] and stored at −20 °C until it was used to determine the hemagglutinating activity according to the methodology previously described.
Affinity chromatography was done on an α-galactose-Sepharose (Invitrogen) column. The α-galactose-sepharose was packed into a column (1 × 5 cm) and equilibrated with sodium/sodium phosphate buffer (NaNa PBS, 50 mM NaH2PO4, 50 mM Na2HPO4, 150 mM NaCl, pH 7.4). The remaining supernatant (249 mg) was dialyzed against NaNa PBS and loaded onto the column. The unbound material was washed from the column with NaNa PBS until the absorbance of the eluent was <0.01 at 280 nm. To obtain a fraction containing the PirABvp toxin, the column was washed with sodium/sodium phosphate buffer supplemented with 200 mM galactose, collected, dialyzed against KNa PBS, and quantified using the Bradford method [24] before being immediately stored at −20 °C until use to determine the hemagglutinating activity according to the methodology previously described.
HA of wild-type PirAvp and PirBvp subunits was not tested because it was not possible to obtain purified subunits with the methodology previously described.
2.6. Sugar Specificity and the Requirement of Divalent Cations
The lectin’s sugar specificity was tested by inhibiting the HA of Wistar rat’s erythrocytes using monosaccharides, oligosaccharides, amino acids, glycosaminoglycans, and glycoproteins. rPirBvp was diluted in PBS to 2560 HA units (HAU, titer = 64); 25 µL of this dilution was placed on microtiter-U plates and incubated for 30 min at room temperature with 25 µL of each sugar, amino acid, glycosaminoglycan, or glycoprotein at several concentrations in triplicate. Subsequently, 25 µL of 2.0% rat erythrocytes suspension in PBS was added. The following reagents were tested: D-lactose, D-maltose, galactosamine (GalNH2), glucosamine (GlcNH2), D-galactose, D-glucose, N-acetyl glucosamine (GlcNAc), N-acetyl-galactosamine (GalNAc), D-fucose (D-Fuc), D-mannose (Man), mannitol, xylose (Xyl), saccharose, sorbitol, methyl-glucose (CH3-Glc), methyl-mannose (CH3-Man), melibiose, N-Ac-diacetyl-chitobiose, galacturonic acid, L-arginine (Arg), L-lysine (Lys), L-serine (Ser), cystine, hyaluronic acid, heparin, fetuin (FET), albumin from chicken egg white (OVO), and mucin from the porcine stomach (PSM) (Sigma). The results were expressed as the minimum concentration of carbohydrate, amino acid, glycosaminoglycan, or glycoprotein able to inhibit 2560 HAU. The effect of divalent cations on the HA was assessed by adding several concentrations (from 0.01 to 5 mM) of CaCl2 and MgCl2. Wild-type and recombinant rPirAvp did not show hemagglutinating activity against rat erythrocytes, so they were not used in this experiment.
2.7. Production of Polyclonal Antibodies (Pabs) in Rabbit: Anti-rPirA and Anti-rPirB
Two male New Zealand white rabbits (1800 g) were used for this study. Before starting the immunization protocol, each rabbit was dewormed by an oral route with sulfamide-trimethoprim (30 mg/kg/12 h for 10 days), unique doses of ivermectin (0.4 mg/kg), toltrazuril (20 mg/kg), and praziquantel (5 mg/kg). The rabbits were subcutaneously injected along the dorsum at the region of the scapula with a 1.0-mL suspension of recombinant protein (in 50 mM Tris-HCl, pH 8.0) in Freund’s complete adjuvant (Sigma), corresponding to 500 μg/mL of protein, confirmed by the Bradford technique. Four subsequent inoculations of rPirAvp and rPirBvp proteins in the same buffer emulsified in Freund’s incomplete adjuvant (FIA) (Sigma) were administered. Rabbits were also subcutaneously injected with 1.0 mL of a suspension of recombinant protein in FIA at the same concentration (500 μg/mL). Blood (3 to 5 mL) was collected from the auricular marginal vein from each rabbit 7 days before the first injection, and then at days 7, 14, 21, and 28 for the measurement of antibody titers. Deep anesthesia was induced with chloroform on day 35, and then blood was withdrawn by cardiac puncture. Blood was incubated at 37 °C for 30 min and was later incubated at 4 °C for 1 h. After the incubation, blood was centrifuged at 6000× g for 30 min and serum samples were collected. The polyclonal antibodies (Pabs) were purified by three ammonium sulfate precipitations according to Harlow and Lane [25]; then, they were dialyzed and resuspended in PBS at pH 7.2 and quantified according to Bradford [24]. Samples were stored at −20 °C until use.
2.8. Polyclonal Antibodies Specificity
Antibody (anti-rPirA and anti-rPirB) titration was determined by Western blot, according to Harlow and Lane [25]. Briefly, several concentrations of rPirAvp and rPirBvp were separated in 12% SDS-PAGE according to the method described by Laemmli under non-reducing conditions [26]. Samples were electrophoresed for 2 h at 100 V and the gels were electro-blotted onto nitrocellulose membranes using a Transblot apparatus (BioRad Laboratories, Hercules, CA, USA) for 1 h with the transfer buffer (50 mM Tris-HCl, 380 mM glycine, 0.1% SDS, 20% methanol, pH 8.3), according to Towbin [27]. The nitrocellulose membrane was blocked, washed, treated with double serial dilutions of the antibody (1:50 to 1:51,200), and incubated for 1 h at room temperature. After extensive washing in PBS-Tween 20, the membrane was incubated with anti-rabbit-HRP (Invitrogen) at 1:3000 for 1 h. To reveal activity, the membrane was extensively washed and incubated for 5 min in a substrate solution containing 0.02% diaminobenzidine (DAB), 0.03% hydrogen peroxide and 0.015% cobalt chloride in PBS-Tween 20. The antibody titer was the value of the highest dilution in which the proteins were observed in the membrane.
2.9. Immunodetection of PirAvp and PirBvp
To detect the presence of PirAvp and PirBvp proteins of VpM0904 in both ECPs and the fractions of purification, the anti-rPirA’s and anti-rPirB’s Pabs were used in the Western-blot assay. After 12% SDS-PAGE under non-reducing conditions, the gels were blotted on a nitrocellulose membrane, incubated in blocking buffer, and probed with the appropriate primary antibody for 1 h at room temperature (1:12,000 for PirAvp and 1:300 for PirBvp). After washing 3 times with PBS-Tween 20, the membrane was incubated with the secondary antibody, horseradish peroxidase-conjugated F(ab’)2 fragment of anti-rabbit IgG (Sigma), for 1 h at room temperature. A spot was revealed after washing with PBS-Tween 20 using DAB and hydrogen peroxide in the same buffer. For the identification of wild-type PirAvp, it was necessary to perform the complete procedure in the presence of 200 mM galactose. Some samples were electrophorezed for two h at 100 V and the proteins were visualized in the gel using the silver staining protocol [28].
3. Results
3.1. Purification of Recombinant Proteins
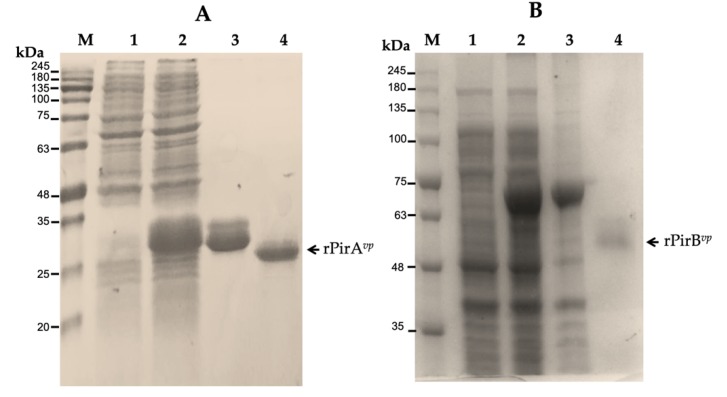
Both recombinant proteins were obtained in E. coli lysates at different incubation times and purified by Ni-affinity. The PirAvp recombinant that was eluted using 500 mM imidazole appeared on the SDS-PAGE as a band with an apparent molecular weight of about 25 kDa, probably the dimeric nature of this protein (Figure 1A). The PirBvp recombinant protein was eluted with 300 mM imidazole, and it was identified as a single band with an apparent molecular weight of about 66 kDa because it was co-expressed with a thioredoxin tag to promote solubility. Treatment with enterokinase produced a band with an apparent molecular weight of 50 kDa (Figure 1B). The protein profile of lysates of E. coli showed differences between induced and non-induced cells. The molecular weights for both proteins were consistent with those calculated as reported previously: 50.1 kDa for PirBvp and 12.7 for PirAvp.
Figure 1.
SDS-PAGE analysis of rPirAvp and rPirBvp toxins of V. parahaemolyticus. (A) M, molecular maker; lane 1, lysate of E. coli non-induced; lane 2, lysate of induced culture after 16 h; lane 3, rPirAvp purified with 500 mM imidazole; lane 4, rPirAvp after TEV protease treatment. (B) M, molecular maker; lane 1, lysate of E. coli non-induced; lane 2, lysate of culture after 4 h; lane 3, rPirBvp purified with 300 mM imidazole; lane 4, rPirBvp after enterokinase treatment.
3.2. HA of the rPirBvp and Wild-Type PirABvp Toxin
The rPirBvp purified by Ni-affinity was capable of agglutinating erythrocytes from rats (Wistar strain) but failed to agglutinate red cells from rabbits; A, B, and O human types; ovine species; and mouse (BalbC strain). rPirAvp did not show HA against any of the erythrocytes tested. Likewise, the wild-type PirABvp, purified from ECPs with stroma and α-galactose column, agglutinated only rat erythrocytes. When the rat erythrocytes were desialylated, there was a 4.0-fold increase in the agglutinating activity of rPirBvp (Table 1).
Table 1.
Hemagglutination activity assay of the recombinant and wild-type toxins. The tests were performed in the presence of 2% erythrocyte suspension in PBS, pH 7.4.
| Red Blood Cells | Hemagglutinating Activity (HA) a | ||
|---|---|---|---|
| rPirAvp | rPirBvp | PirABvp* | |
| Human A | - | - | - |
| Human B | - | - | - |
| Human O | - | - | - |
| Rabbit | - | - | - |
| Mouse (BalbC) | - | - | - |
| Rat (Wistar) | - | 81,476 | 114,285 |
| Desialylated Rat b | - | 325,970 | ND |
| Ovine | - | - | - |
rPirAvp was 1.12 μg/25 μL; rPirBvp was 0.8 μg/25 μL; PirABvp was 0.035 μg/25 μL. a HA: hemagglutinating activity is reported as the inverse of the last dilution showing visible agglutinating activity. b Erythrocytes of rat were incubated in presence of Vibrio cholera sialidase 45 min at 37 °C using rPirBvp for the activity assay. PirABvp* purified using α-galactose column. -: No hemagglutinating activity. ND: not determined hemagglutinating activity.
Native PirAvp and PirBvp subunits could not be purified. In addition, rPirAvp and rPirBvp did not have proteolytic activity, confirmed by the assay with remazol brilliant blue R.
3.3. Purification of Wild-Type PirABvp Toxin Complex
The PirABvp from ECPs of VpM0904 was identified and purified in a single step by affinity chromatography on glutaraldehyde-fixed stroma from rat erythrocytes (Figure 2A) or α-galactose-Sepharose (Figure 2B). The purified complex toxin with stroma had an apparent molecular weight of 70 kDa, which was not observed in the α-galactose affinity purification. The procedure using α-galactose-sepharose showed a 250 kDa band, which apparently corresponded to the tetrameric complex formed of four subunits of PirAvp (12 kDa for each one) and four subunits of PirBvp(50 kDa for each one).
Figure 2.
SDS-PAGE of the PirABvp toxin purified by affinity chromatography. M, molecular marker. (A) Lane 1, fraction obtained from a column with rat stroma of extracellular products (ECPs) eluted with 3% acetic acid. (B) Lane 1, fraction purified from ECPs onto α-galactose column eluted with 200 mM galactose.
In the purification of the PirABvp using a rat stroma column, a 199-fold increase in the specific activity of the toxin complex was observed with this procedure compared with the ECPs (Table 2). The concentration of the toxin eluted from the column represented 0.50% of the total protein present in ECPs.
Table 2.
Purification process of the PirABvp toxin from ECPs of Vibrio parahaemolyticus in rat stroma.
| Fraction | Total Protein (mg) | HAU a | Specific Activity b |
|---|---|---|---|
| ECP | 195.4 | 160.0 | 0.82 |
| Unretained fraction | 194.42 | 0.0 | 0.0 |
| Retained fraction (PirABvp) | 0.980 | 160.0 | 163.26 |
a HAU = hemagglutinating activity units tested in presence of rat erythrocytes. b Specific activity = HAU/mg protein.
In the purification of the PirABvp toxin in the galactose column, a 174-fold increase in the specific activity of the PirABvp toxin was observed with this procedure compared with the ECPs (Table 3). The concentration of PirABvp toxin eluted from the column represented 0.57% of the total protein present in ECPs. Also, there were differences in hemagglutinating activity units of the PirABvp purified by the two affinity chromatographic methods, 160 (titer = 4) for the eluted rat stroma complex and 640 (titer = 16) for the galactose purified complex.
Table 3.
Purification process of the PirABvp toxin of ECPs from Vibrio parahaemolyticus in α-galactose.
| Fraction | Total Protein (mg) | HAU a | Specific Activity b |
|---|---|---|---|
| ECP | 249.24 | 640.0 | 2.56 |
| Unretained fraction | 247.80 | 0.0 | 0.0 |
| Retained fraction (PirABvp) | 1.43 | 640.0 | 447.55 |
a HAU = hemagglutinating activity units tested in presence of rat erythrocytes. b Specific activity = HAU/mg protein.
3.4. Sugar Specificity and the Effect of Divalent Cations on the HA of rPirBvp
GalNH2 and GlcNH2 were better sugar inhibitors for rPirBvp HA; 25 mM was the minimal concentration to inhibit 2560 HAU of rPirBvp, whereas, 4-fold higher concentrations of the disaccharides, Mal and Lac, were necessary to inhibit a similar activity of the rPirBvp. The amino acid, Arg, and the cysteine dimer, cystine, present the same activity as the amino-sugars (Table 4). Other carbohydrates, such as Gal, Glc, Man, GlcNAc, GalNAc, Fuc, mannitol, xylose, sorbitol, Met-glucopyranoside, Met-mannose, melibiose, N,N’-Ac-diacetyl-chitobiose, sialic acid, sucrose, and the amino acids, Lys and Ser, had no inhibitory activity at 200 mM. Heparin showed a relative inhibitory potency, 93-fold higher compared with the amino-sugars tested, whereas hyaluronic acid had no inhibitory activity. The FET glycoprotein showed the strongest inhibition for rPirBvp, whereas OVO showed a lower inhibition, and PSM showed no effect on the rPirBvp HA. No effect of divalent cations, such as Ca2+ and Mg2+, was observed on PirBvp lectin hemagglutinating activity at a concentration of 5 mM (data not shown).
Table 4.
Specificity of PirBvp subunit for carbohydrates and glycoproteins.
| Inhibitor | Concentration (mM) a | Relative Inhibitory Potency b |
|---|---|---|
| Lac | 100 | 1 |
| Mal | 100 | 1 |
| GalNH2 | 25 | 4 |
| GlcNH2 | 25 | 4 |
| Arginine | 25 | 4 |
| Cystine | 25 | 4 |
| Heparin | 0.27 | 370.4 |
| OVO | 0.015 | 6666.7 |
| FET | 0.00031 | 322,580.6 |
a Minimum concentration to inhibit 2560 hemagglutinating activity units (HAU) of the rPirBvp lectin in presence of 2% rat erythrocytes suspension in PBS, pH 7.4. b Relative inhibition capacity compared with lactose or maltose. Other carbohydrates without inhibitory capacity, even at 200 mM, were GalNAc, GlcNAc, Glc, Gal, Man, GlcNAc, GalNAc, Fuc, mannitol, xyl, sorbitol, CH3-glucopyranoside, CH3-mannose, melibiose, N,N’-Ac-diacetyl-chitobiose, sialic acid, sucrose, and amino acids (Lys and Ser), hyaluronic acid; glycoproteins without inhibitory capacity were mucin from porcine stomach and bovine submaxillary mucin.
3.5. Specificity of Polyclonal Antibodies
Each protein was used for the production of polyclonal antibodies in rabbits, and they were used to confirm the presence of proteins during the purification procedure. To determine the minimum dilution for the detection of rPirAvp or rPirBvp, 1 μg of total protein was run on a single-well gel, electro-blotted, and detected by serial dilutions of 1:50 to 1:51, 200 (0.093 to 96 μg of antibody); 0.9 and 16 μg were the concentrations of anti-rPirA and anti-rPirB antibody that were necessary to detect each protein (1:12,000 and 1:300, respectively).
Both recombinant proteins were identified by SDS-PAGE using reducing conditions of each fraction obtained with different concentrations of imidazole (Figure 1). rPirAvp was identified in the dimeric (24 kDa) and tetrameric (48 kDa) forms, the dimeric form was the most abundant (Figure 3A) in non-reducing conditions. rPirBvp was identified as a 50 kDa monomer and probably as a 200–250 kDa tetra- or pentameric form under the same conditions (Figure 3B).
Figure 3.
Specificity and sensitivity of polyclonal antibodies. Minimum detectable concentration of rPirAvp (A) and rPirBvp (B) using anti-rPirA (1:12,000) and anti-PirB (1:300) antibodies. (A) M, molecular marker; lanes 1–7: 1260, 632, 316, 158, 79, 39, and 19.5 ng of rPirAvp; (•) dimeric form; (••) tetrameric form. (B) lanes 1–7: 2940, 1470, 735, 368, 184, 92, and 46 ng of rPirBvp; (*) monomer; (**) tetrameric or pentameric form. (C) antibodies specificity against ECPs from VpM0904 tested with anti-rPirA. Lane 1 and 2, free and plus 200 mM galactose, respectively; lane 3, anti-rPirB antibody.
Sensitivity of the anti-rPirB Pabs versus rPirBvp was approximately 46 ng of the recombinant protein per band (Figure 3B, lane 7); the anti-rPirA Pabs specific for rPirAvp had a sensitivity that corresponded to approximately 19.5 ng of protein per spot (Figure 3A, lane 7).
Similarly, each subunit of the native protein was recognized by the antibodies obtained in this work. Anti-rPirA Pabs recognized a 24 kDa fraction and anti-rPirB Pabs detected a 50 kDa band (Figure 3C, lane 1). Anti-rPirA showed cross-reaction with a 50 kDa protein, which was eliminated using 200 mM galactose (Figure 3C, lane 2), and anti-rPirBvp only detected a 50 kDa band in the ECPs of VpM0904 (Figure 3C, lane 3).
4. Discussion
Bacterial lectins play a key role in eliminating phylogenetically-related antagonists for the adhesion or colonization of specific niches, such as tissues of hosts or for biofilm formation. This implies the need for the secretion of toxins with lectin activity that allows oligo- and polysaccharides to be recognized in the targets. In addition, lectins can act as recognition molecules in cell–molecule and cell–cell interactions in a variety of biological systems [29,30]. The first reports of bacterial lectins were from Pseudomonas; their toxins have been associated with virulence factors [31,32,33,34]. P. aeruginosa produces a variety of toxins with lectin-like activity that are known as bacteriocins and pyocins that agglutinate bacteria to inhibit the growth of competitors [31,32]. This kind of lectin has been characterized in P. putida, P. fluorescens, P. syringae, and Xanthomonas citri [34,35,36,37,38,39]. The Tor biotype of V. cholerae expresses the lectin-like toxin called cell-associated mannose-sensitive hemagglutinin (MSHA), which is a putative attachment factor [40,41,42].
The results from the present study suggest that the PirBvp subunit of the PirABvp binary toxin secreted by the marine bacterium, V. parahaemolyticus, the causal agent of AHPND in shrimp, has lectin activity. The PirBvp subunit binds to a molecule containing amino sugars, such as glycosaminoglycans linked to proteoglycans that could have a receptor function on the membrane of target cells in shrimp tissues.
Although the wild-type PirABvp protein can be purified using two procedures, the amounts obtained of the total protein secreted by VpM0904 was very low in both cases: 0.50% using a stroma column and 0.57% with a galactose column.
In the present study, E. coli cells were successfully transformed. Cells produced 6000-times more recombinant PirAvp and PirBvp than VpM0904 cells can produce. This quantity was enough to determine the hemagglutinating activity and specificity for sugars of both proteins, as reported in previous studies on characterizations of other Vp proteins [8,43,44]. The PirBvp subunit recognized only rat erythrocytes, which contain large amounts of 9-O-acetyl sialic acid (Neu5,9Ac), above 25–30% of sugars in their cell membranes and other O-glycoconjugates with galactose or galactosamine residues [45,46]. The former sugar could bind to the PirBvp subunit and recognize the amino group; however, elimination of the sialic acid residues (Neu5,9Ac) from rat erythrocytes showed an increase in the hemagglutination activity. This result demonstrated that the potential negative charge of the sialic acid was not as decisive for the PirABvp toxin–ligand interactions, such as already been reported for other lectins [47]. Likewise, this indicates that the carbohydrate determinants on these red blood cells are partially accessible, and after partial removal of the sialic acid from the erythrocytes’ surface, they became more accessible. We propose that the lectin activity is specific for glycoconjugates containing N-acetylglucosamine or N-acetylgalactosamine in a terminal non-reducing position [48]. Therefore, the results suggest that the recognition of the receptor is not sialic acid-dependent.
During the purification process of proteins using rat stroma, a complex containing PirAvp and PirBvp subunits was formed. The complex was purified as an oligomer of four PirAvp subunits and four PirBvp subunits when using galactose. Probably, these differences between the structures obtained by each purification process were due to the recognition of the complex of different molecules. Thus, whereas the stroma column has several sugar sequences, the galactose column has only one, that could facilitate the purification of a tetrameric complex by the oligomerization, induced by recognition of the same sugar, as previously reported for the lectin purified from Mytilus californianus [49]. Also, the use of acetic acid in a stroma column to dialysis the protein did not allow a reversion to the same quaternary structure that was previously eluted. This did happen in the column of galactose, which was eluted with the same sugar. For this reason, a band of approximately 70 kDa molecular weight was observed, probably formed by one PirAvp plus one PirBvp subunit. Evidently, the PirABvp toxin belongs to AB binary toxins composed of one A subunit of 12 kDa and one B subunit of 50 kDa.
We found that the hemagglutinating activity of the PirBvp subunit is higher if it is attached to PirAvp. Furthermore, PirAvp alone did not show activity against any type of erythrocytes tested. Possibly the A subunit participates in stabilizing the complex for a better binding to the possible receptor molecule on the shrimp hepatopancreatic epithelial cells [50]. These results highlight the importance of the interaction between the two subunits of the complex, which could act in molecular synergy to recognize the target molecule, as has been reported for the A and B subunits of the R. communis toxin [51]. In agreement with the above, Lin et al. [52] suggested that the PirAvp subunit probably acts to give structural stability to the PirABvp toxin. Our results contribute to the understanding of the structural characterization of the PirABvp toxin and, in turn, the type and chemical structure of the saccharides present in possible receptor molecules. Most authors suggest that the PirABvp binary toxin acts as a pore-forming toxin, killing epithelial cells that induce AHPND in shrimp. In silico analysis based on the structural similarity between PirABvp and Bacillus thuringiensis Cry toxins support this theory [50,53]. However, this has not been experimentally demonstrated yet. The data presented here demonstrate that the PirBvp toxin has lectin activity, which is possibly involved in the recognition of a receptor molecule.
The inhibition with amino sugars suggests that the possible receptor or adhesion molecule can be a proteoglycan or glycolipid of the epithelial cells of the shrimp hepatopancreas [54]. The PirBvp lectin activity was inhibited by GlcNH2, GalNH2, lactose (Galβ1,4Glc), maltose (Glcα1,4Glc), arginine, cystine, heparin, OVO, and FET. The fact that lactose (Galβ1,4Glc) and maltose (Glcα1,4Glc) inhibit the hemagglutinating activity of PirBvp at the same concentration reveals that the recognition can be based on the glycosidic bond coupled with one of the monosaccharides that form the sugar. Probably, the PirBvp subunit may recognize the α glycosidic bond between two sugar units, suggesting that the sequence of the Gal(β1-3 or 4)GlcNAc(α 1-2)Man sugar is essential for the PirBvp subunit interaction with the host structures. The existence of heparan sulfate (HS) chains was reported in shrimp [55]. These are sulfated glycosaminoglycans (GAGs) in syndecans involved in several steps of infection caused by the white spot syndrome virus, including initial attachment and entry of viral proteins in shrimp. Besides, the inhibition in the presence of the amino acids, arginine and cysteine, suggest that a negative charge in the membrane of the erythrocyte can mediate interactions with the positive charge of the arginine residue [56]. Positive residues, like arginine, lysine, and histidine, usually participate in hydrogen bonding with some receptors and the positive charge can enhance the complex stability in protein–protein interactions [57,58].
Heparin is another inhibitor of the hemagglutination activity, and its inhibition was 93-times higher than that of amino acids or amino-sugars. The GAGs, such as heparin/HS, are synthesized and serine-linked to the core proteins of proteoglycans, and they are located on the plasma membrane and in the extracellular matrix [59]. These data reveal a PirBvp–GAGs interaction mediated by a sequence of sugars of heparin or HS binds to a proteoglycan of the epithelial cells. The effects of these GAGs are exerted through their capacity to engage protein ligands [60]. Many pathogens produced proteins that interact with heparin/HS as part of their molecular adaptation to infection of mammals [61]. Probably, the B subunit may recognize a sequence with O-amino sugars, stabilized by the negative charge of the sulfate groups possessed by these GAGs. Also, the presence of numerous negatively charged sulfate groups in GAGs provides an ideal multi-landing pad for proteins and macromolecules through electrostatic interactions [62].
The most powerful glycoprotein inhibitor is fetuin, which contains O-glycosidically-linked oligosaccharide chains [63]. However, ovalbumin, which was also inhibitory, contains a single N-glycosdically-linked glycan chain at Asn-392 [64]. Interestingly, the glycoproteins with inhibitory capacity possess as predominant oligosaccharides either the sequence NeuAc(α2,6)Gal(β1,3)GalNAc(αl,O)Ser/Thr or NeuAC(α2,3)GalNAc(α1,O)Ser/Thr [65], which may explain why both proteins have an inhibitory capacity. Mucin from porcine stomach, a glycoprotein containing O-glycosdically-linked glycan chains (Galβ1,3GalNAc) [66], did not inhibit the hemagglutinating activity of PirBvp. Possibly, this is due to PirBvp recognizing the difference between the α1-O bond of Ser/Thr and the β1-O bond present in mucin. The predominant component of bovine submaxillary mucin is 9-O-acetyl- and 8,9-di-O-acetyl-N-acetyl-NeuAC [47], which explains why this sequence is not recognized by the B subunit. The above suggests that the PirBvp subunit contains a binding site for glycans in the binding site of a possible receptor on the epithelial cells of the shrimp hepatopancreas. Inhibition of the lectin activity of the PirBvp subunit was observed in the presence of Ca2+ and Mg2+ ions; possibly, the ions caused structural changes in the subunit, which resulted in the loss of activity [67]. Data of this study contribute to the knowledge of the interactions of the PirBvp subunit with the receptor. Although the principal interaction can be with glycosaminoglycans in the receptor structure, a positive charge is necessary for protein–protein interactions to have more stability and produce damage in the hepatopancreas of shrimp.
5. Conclusions
The PirBvp subunit integrated into a tetrameric PirABvp complex can recognize glycosaminoglycans molecules (amino sugars) and probably binds to the receptor molecules on the membrane of the hepatopancreatic epithelial cells of shrimp to trigger the massive sloughing of these cells. However, the specific role of the PirBvp subunit as a lectin, as well as the function of the PirAvp subunit, in the pathogenesis of AHPND have not yet been determined.
Acknowledgments
We thank Rodolfo Lozano-Olvera, Karla Aguilar-Redon, Samara García-Flores and Luisa Contreras-López, for providing technical assistance.
Author Contributions
Conceptualization, S.S.-R. and N.V.-P.; data curation, N.V.-P.; formal analysis, A.P.; funding acquisition, S.S.-R. and N.V.-P.; investigation, M.V.-D.L.S. and E.Z.; methodology, M.V.-D.L.S., A.P. and E.Z.; resources, N.V.-P.; software, M.V.-D.L.S., A.P. and E.Z.; validation, M.V.-D.L.S., A.P., and E.Z.; visualization, P.C.-S.; writing—original draft, S.S.-R., N.V.-P. and P.C.-S.; writing—review and editing, S.S.-R. and P.C.-S. All authors have read and agreed to the published version of the manuscript.
Funding
This work received partial support from the PFCE program with agreement number P/PFCE-2019-18MSU0019M-04” and from the CIAD Mazatlan-Unit.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.De la Pena L.D., Cabillon N.A., Catedral D.D., Amar E.C., Usero R.C., Monotilla W.D., Calpe A.T., Fernandez D.D.G., Saloma C.P. Acute hepatopancreatic necrosis disease (AHPND) outbreaks in Penaeus vannamei and P. monodon cultured in the Philippines. Dis. Aquat. Org. 2015;116:251–254. doi: 10.3354/dao02919. [DOI] [PubMed] [Google Scholar]
- 2.Joshi J., Srisala J., Truong V.H., Chend T.I., Nuangsaenge B., Suthienkul O., Lo C.F., Flegel T.W., Sritunyalucksana K., Thitamadee S. Variation in Vibrio parahaemolyticus isolates from a single Thai shrimp farm experiencing an outbreak of acute hepatopancreatic necrosis disease (AHPND) Aquaculture. 2014;428–429:297–302. doi: 10.1016/j.aquaculture.2014.03.030. [DOI] [Google Scholar]
- 3.Nunan L., Lightner D., Pantoja C., Gomez-Jimenez S. Detection of Acute Hepatopancreatic Necrosis disease (AHPND) in Mexico. Dis. Aquat. Org. 2014;111:81–86. doi: 10.3354/dao02776. [DOI] [PubMed] [Google Scholar]
- 4.Restrepo L., Bayot B., Betancourt I., Pinzón A. Draft genome sequence of pathogenic bacteria Vibrio parahaemolyticus strain Ba94C2, associated with acute hepatopancreatic necrosis disease isolate from South America. Genome Data. 2016;11:143–144. doi: 10.1016/j.gdata.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han J.E., Tang K.F.J., Tran L.H., Lightner D.V. Photorhabdus insect-related (Pir) toxin- like genes in a plasmid of Vibrio parahaemolyticus, the causative agent of acute hepatopancreatic necrosis disease (AHPND) of shrimp. Dis. Aquat. Org. 2015;113:33–40. doi: 10.3354/dao02830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nunan L., Redman R.M., Mohney L.L., Pantoja C.R., Fitzsimmons K., Lightner D.V. Determination of the infectious nature of the agent of acute hepatopancreatic necrosis syndrome affecting penaeid shrimp. Dis. Aquat. Org. 2013;9:45–55. doi: 10.3354/dao02621. [DOI] [PubMed] [Google Scholar]
- 7.Yang Y.T., Chen I.T., Lee C.T., Chen C.Y., Lin S.S., Hor L.I., Tseng T.C., Huang Y.T., Sritunyalucksana K., Thitamadee S., et al. Draft genome sequences of four strains of Vibrio parahaemolyticus, three of which cause early mortality syndrome/acute hepatopancreatic necrosis disease in shrimp in China and Thailand. Genome Announc. 2014;2:e00816-14. doi: 10.1128/genomeA.00816-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sirikharin R., Taengchaiyaphum S., Sanguanrut P., Chi D.T., Mavichak R., Proespraiwong P., Nuangsaeng B., Thitamadee S., Flegel T.W., Sritunyalucksana K. Characterization and PCR detection of binary, Pir-like toxins from Vibrio parahaemolyticus isolates that cause acute hepatopancreatic necrosis disease (AHPND) in shrimp. PLoS ONE. 2015;10:e0126987. doi: 10.1371/journal.pone.0126987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kondo H., Van P.T., Dang L.T., Hirono I. Draft genome sequence of non-Vibrio parahaemolyticus acute hepatopancreatic necrosis disease strain KC13.17.5, isolated from diseased shrimp in Vietnam. Genome Announc. 2015;3:e00978-00915. doi: 10.1128/genomeA.00978-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao J., Liu L., Ke Y., Li X., Liu Y., Pan Y., Yan S., Wang Y. Shrimp AHPND-causing plasmids encoding the PirAB toxins as mediated by pirAB-Tn903 are prevalent in various Vibrio species. Sci. Rep. 2017;7:42177. doi: 10.1038/srep42177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong X., Bi D., Wang H., Zou P., Xie G., Wan X., Huang J., Yang Q., Zhu Y., Chen M., et al. PirABvp-bearing Vibrio parahaemolyticus and Vibrio campbellii pathogens isolated from the same AHPND-affected pond possess highly similar pathogenic plasmids. Front. Microbiol. 2017;8:1859. doi: 10.3389/fmicb.2017.01859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durán-Avelar M.D.J., Vázquez-Reyes A., González-Mercado A.L., Zambrano-Zaragoza J.F., Ayón-Pérez M.F., Agraz-Cibrián J.M., Gutiérrez-Franco J., Vibanco-Pérez N. pirA- and pirB-like genes identification in Micrococcus luteus strains in Mexico. J. Fish Dis. 2018;41:1667–1673. doi: 10.1111/jfd.12874. [DOI] [PubMed] [Google Scholar]
- 13.Chonsin K., Matsuda S., Theethakaew C., Kodama T., Junjhon J., Suzuki Y., Suthienkul O., Iida T. Genetic diversity of Vibrio parahaemolyticus strains isolated from farmed Pacific white shrimp and ambient pond water affected by acute hepatopancreatic necrosis disease outbreak in Thailand. FEMS Microbiol. Lett. 2016;363:fnv222. doi: 10.1093/femsle/fnv222. [DOI] [PubMed] [Google Scholar]
- 14.Phiwsaiya K., Charoensapsri W., Taengphu S., Dong H.T., Sangsuriya P., Nguyen G.T.T., Pham H.Q., Amparyup P., Sritunyalucksana K., Taengchaiyaphum S., et al. A natural Vibrio parahaemolyticus Pirvp B+ mutant kills shrimp but produces no Pirvp toxins or AHPND lesions. Appl. Environ. Microbiol. 2017;83:e00680-17. doi: 10.1128/AEM.00680-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Restrepo L., Bayot B., Arciniegas S., Bajaña L., Betancourt I., Panchana F., Reyes Muñoz A. PirVP genes causing AHPND identified in a new Vibrio species (Vibrio punensis) within the commensal Orientalis clade. Sci. Rep. 2018;8:13080. doi: 10.1038/s41598-018-30903-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soto-Rodríguez S., Gomez-Gil B., Lozano-Olvera R., Betancourt-Lozano M., Morales-Covarrubias M.S. Field and experimental evidence of Vibrio parahaemolyticus as the causative agent of acute hepatopancreatic necrosis disease (AHPND) of cultured shrimp (Litopenaeus vannamei) in northwestern Mexico. Appl. Environ. Microbiol. 2015;8:1689–1699. doi: 10.1128/AEM.03610-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lightner D.V., Redman R.M., Pantoja C.R., Noble B.L., Tran L. Early mortality syndrome affects shrimp in Asia. Glob. Aquac. Advocate. 2012;15:40. [Google Scholar]
- 18.Odumosu O., Nicholas D., Yano H., Langridge W. AB toxins: A paradigm switch from deadly to desirable. Toxins. 2010;2:1612–1645. doi: 10.3390/toxins2071612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olsnes S., Pihl A. Toxins lectins and related proteins. In: Cohen P., editor. Molecular Action of Toxins and Viruses. Elsevier; Amsterdam, The Netherlands: 1982. pp. 51–105. [DOI] [Google Scholar]
- 20.Van Damme E.J.M. History of plant lectin research. In: Hirabayashi J., editor. Lectins: Methods and Protocols. Springer; New York, NY, USA: 2014. pp. 3–13. [DOI] [PubMed] [Google Scholar]
- 21.Shang C., Dang L., Van Damme E.J.M. Plant AB toxin with lectin domains. In: Gopalakrishnakone P., editor. Plant Toxins. Springer; Dordrecht, The Netherlands: 2017. pp. 183–198. [DOI] [Google Scholar]
- 22.Fragkiadakis G.A. Isolation of lectins from hemolymph of decapod crustaceans by adsorption on formalinized erythrocytes. J. Biochem. Biophys. Methods. 2000;44:109–114. doi: 10.1016/S0165-022X(00)00089-0. [DOI] [PubMed] [Google Scholar]
- 23.Vazquez L., Masso F., Rosas P., Montano L.F., Zenteno E. Purification and characterization of a lectin from Macrobachium rosenbergii (Crustacea: Decapoda) hemolymph. Comp. Biochem. Physiol. 1993;105:617e23. doi: 10.1016/0305-0491(93)90097-O. [DOI] [Google Scholar]
- 24.Bradford M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 25.Harlow E., Lane D. A Laboratory Manual. 1st ed. Cold Spring Harbor Laboratory; New York, NY, USA: 1988. Antibodies; pp. 139–175. [Google Scholar]
- 26.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Towbin H., Staehelin J., Gordon J. Electrophoresis transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merril C.R., Goldman D., Sedman S., Ebert M. Ultrasensitive stain for proteins in polyacrylamide gels shows regional variation in cerebrospinal fluid proteins. Science. 1981;211:1437–1438. doi: 10.1126/science.6162199. [DOI] [PubMed] [Google Scholar]
- 29.Sharon N., Lis H. History of lectins: From hemagglutinins to biological recognition molecules. Glycobiology. 2004;14:53–62. doi: 10.1093/glycob/cwh122. [DOI] [PubMed] [Google Scholar]
- 30.Gilboa-Garber N., Katco D.J., Garber N.C. Identification and characterization of Pseudomonas aeruginosa PA-IIL lectin gene and protein compared to PA-IL. FEMS Immunol. Med. Mic. 2000;29:53–57. doi: 10.1111/j.1574-695X.2000.tb01505.x. [DOI] [PubMed] [Google Scholar]
- 31.Parret A.H.A., Schoofs G., Proost P., De Mot R. Plant lectin-like bacteriocin from arhizosphere-colonizing Pseudomonas isolate. J. Bacteriol. 2003;185:897–908. doi: 10.1128/JB.185.3.897-908.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parret A.H.A., Temmerman K., De Mot R. Novel lectin-like bacteriocins of biocontrol strain Pseudomonas fluorescens Pf-5. Appl. Environ. Microbiol. 2005;71:5197–5207. doi: 10.1128/AEM.71.9.5197-5207.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghequire M.G.K., Loris R., De Mot R. MMBL proteins: From lectin to bacteriocin. Biochem. Soc. Trans. 2012;40:1553–1559. doi: 10.1042/BST20120170. [DOI] [PubMed] [Google Scholar]
- 34.Ghequire M.K.G., Garcia-Pino A., Lebbe L.K.M., Spaepen S., Loris R., De Mot R. Structural Determinants for Activity and Specificity of the Bacterial Toxin LlpA. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jamet A., Nassif X. New players in the toxin field: Polymorphic toxin systems in bacteria. mBio. 2015;6:e00285-15. doi: 10.1128/mBio.00285-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghequire M.G.K., de Mot R. The tailocin tale: Peeling off phage tails. Trends Microbiol. 2015;23:587–590. doi: 10.1016/j.tim.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 37.Michel-Briand Y., Baysse C. The pyocins of Pseudomonas aeruginosa. Biochimie. 2002;84:499–510. doi: 10.1016/S0300-9084(02)01422-0. [DOI] [PubMed] [Google Scholar]
- 38.Ghequire M.G.K., de Mot R. Ribosomally encoded antibacterial proteins and peptides from Pseudomonas. FEMS Microbiol. Rev. 2014;38:523–568. doi: 10.1111/1574-6976.12079. [DOI] [PubMed] [Google Scholar]
- 39.Grishin A.V., Krivozubov M.S., Karyagina A.S., Gintsburg A.L. Pseudomonas aeruginosa lectins as targets for novel antibacterials. Acta Nat. 2015;7:29–41. doi: 10.32607/20758251-2015-7-2-29-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Häse C., Bauer M., Finkelstein R. Genetic characterization of mannose-sensitive hemagglutinin (MSHA)- negative mutants of Vibrio cholerae derived by Tn.5 mutagenesis. Gene. 1994;150:17–25. doi: 10.1016/0378-1119(94)90852-4. [DOI] [PubMed] [Google Scholar]
- 41.Finkelstein R.A., Mukerjee S. Hemagglutination: A rapid method for differentiating Vibrio cholerae and El Tor vibrios. Proc. Soc. Exp. Biol. Med. 1963;112:355–359. doi: 10.3181/00379727-112-28043. [DOI] [Google Scholar]
- 42.Hanne L.F., Finkelstein R.A. Characterization and distribution of the hemagglutinins produced by Vibrio cholerae. Infect. Immun. 1982;36:209–214. doi: 10.1128/IAI.36.1.209-214.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kadokura K., Sakamoto Y., Saito K., Ikegami T., Hirano T., Hakamata W., Oku T., Nishio T. Production of a recombinant chitin oligosaccharide deacetylase from Vibrio parahaemolyticus in the culture medium of Escherichia coli cells. Biotechnol. Lett. 2007;29:1209–1215. doi: 10.1007/s10529-007-9386-6. [DOI] [PubMed] [Google Scholar]
- 44.Kadokura K., Sakamoto Y., Saito k Ikegami T., Hirano T., Hakamata W., Oku T., Nishio T. Production and Secretion of a Recombinant Vibrio parahaemolyticus Chitinase by Escherichia coli and Its Purification from the Culture Medium. Biosci. Biotechnol. Biochem. 2007;71:2848–2851. doi: 10.1271/bbb.70389. [DOI] [PubMed] [Google Scholar]
- 45.Schauer R. Sialic acids: Fascinating sugars in higher animals and man. Zoology. 2004;107:49–64. doi: 10.1016/j.zool.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 46.Gumus A., Balcan E. Determination of Glycoconjugate Residues of Erythrocytes at Different Age Groups of Rats. Int. J. Hematol. Oncol. 2010;20:6–13. doi: 10.4999/uhod.09070. [DOI] [Google Scholar]
- 47.Zenteno R., Vazquez L., Sierra C., Pereyra A., Slomianny M.C., Bouquelet S., Zenteno E. Chemical. Characterization from lectin from Macrobrachium Rosenbergii (De Man) by MALDI-TOF. Comp. Biochem. Physiol. 2000;127:243–250. doi: 10.1016/S0305-0491(00)00260-1. [DOI] [PubMed] [Google Scholar]
- 48.Schlepper-Schafer J., Kolb-Bachofen V., Kolb H. Analysis of Lectin Dependent Recognition of Desialylated Erythrocytes by Kupffer Cells. Biochem. J. 1980;186:827–831. doi: 10.1042/bj1860827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garcia-Maldonado E., Cano-Sanchez P., Hernandez-Santoyo A. Molecular and functional characterization of a glycosylated Galactose-Binding lectin from Mytilus californianus. Fish Sellfish Immunol. 2017;66:564–574. doi: 10.1016/j.fsi.2017.05.057. [DOI] [PubMed] [Google Scholar]
- 50.Lin S.H., Hsu K., Wang H. Structural insights into the cytotoxic mechanism of Vibrio parahaemolyticus PirAvp and PirBvp toxins. Mar. Drugs. 2017;15:373. doi: 10.3390/md15120373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frigerio L., Roberts L.M. The Synthesis of ricinus communis Lectins. In: Lord J.M., Hartley M.R., editors. Toxic Plant Proteins. Springer; Berlin/Heidelberg, Germany: 2010. Plant Cell Monographs. [DOI] [Google Scholar]
- 52.Lin S.H., Chen Y.F., Hsu K.C., Chen Y., Ko T.P., Lo C.F., Wang H.C. Structural Insights to the Heterotetrameric Interaction between the Vibrio parahaemolyticus PirAvp and PirB Toxins and Activation of the Cry-Like Pore-Forming Domain. Toxins. 2019;1:233. doi: 10.3390/toxins11040233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee C.T., Chen I.T., Yang Y.T., Ko T.P., Huang J.Y., Huang M.F., Lin S.J., Chen C.Y., Lin S.S., Lightner D.V., et al. The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin. Proc. Natl. Acad. Sci. USA. 2015;112:10798–10803. doi: 10.1073/pnas.1503129112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hempel C., Wang C.W., Kurtzhals J.A.L., Staalsø T. Binding of Plasmodium falciparum to CD36 can be shielded by the glycocalyx. Malar. J. 2017;16:193. doi: 10.1186/s12936-017-1844-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang H., Li S., Li F., Wen R., Xiang J. Analysis on the expression and function of syndecan in the Pacific white shrimp Litopenaeus vannamei. Dev. Comp. Immunol. 2015;51:278–286. doi: 10.1016/j.dci.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 56.Xu D., Escko J.D. Demystifying heparan sulfate-protein interactions. Annu. Rev. Biochem. 2014;83:129–157. doi: 10.1146/annurev-biochem-060713-035314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mascotti D.P., Lohman T.M. Thermodynamics of charged oligopeptide–heparin interactions. Biochem. 1995;34:2908–2915. doi: 10.1021/bi00009a022. [DOI] [PubMed] [Google Scholar]
- 58.Sheinerman F.B., Norel R., Honig B. Electrostatic aspects of protein–protein interactions. Curr. Opin. Struct. Biol. 2000;10:153–159. doi: 10.1016/S0959-440X(00)00065-8. [DOI] [PubMed] [Google Scholar]
- 59.Ori A., Wilkinson M.C., Fernig D.G. A Systems Biology Approach for the Investigation of the Heparin/Heparan Sulfate Interactome. J. Biol. Chem. 2011;286:19892–19904. doi: 10.1074/jbc.M111.228114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ori A., Wilkinson M.C., Fernig D.G. The heparanome and regulation of cell function: Structures, functions and challenges. Front. Biosci. 2008;13:4309–4338. doi: 10.2741/3007. [DOI] [PubMed] [Google Scholar]
- 61.Chen Y., Götte M., Liu J., Park P.W. Microbial subversion of heparan sulfate proteoglycans. Mol. Cells. 2008;26:415–426. [PubMed] [Google Scholar]
- 62.Tao L., Tian S., Zhang J., Liu Z., Robinson-McCarthy L., Miyashita S.I., Breault D.T., Gerhard R., Ottamasathien S., Whelan S.P.J., et al. Sulfated glycosaminoglycans and low-density lipoprotein receptor contribute to Clostridium difficile toxin A entry into cells. Nat. Microbiol. 2019;4:1760–1769. doi: 10.1038/s41564-019-0464-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bergh M.L., Hooghwinkel G.J., van den E. Biosynthesis of the O-Glycosidically Linked Oligosaccharide Chains of Fetuin. J. Biol. Chem. 1982;258:7430–7436. [PubMed] [Google Scholar]
- 64.Harvey D.J., Wing D.R.B., Küster B., Wilson I.B.H. Composition of N-Linked Carbohydrates from Ovalbumin and Co-purified Glycoproteins. J. Am. Soc. Mass Spectrom. 2000;11:564–571. doi: 10.1016/S1044-0305(00)00122-7. [DOI] [PubMed] [Google Scholar]
- 65.Schauer R. Chemistry, metabolism and biological function of sialic acid. Adv. Carbohydr. Chem. Biochem. 1982;40:131–235. doi: 10.1016/s0065-2318(08)60109-2. [DOI] [PubMed] [Google Scholar]
- 66.Zenteno E., Vázquez L., Chávez R., Córdoba F., Wieruszeski J., Montreuil J., Debray H. Specificity of the isolectins from the cactus plant Machaerocereus eruca for oligosaccharides from mucin. Glycoconj. J. 1995;12:699–706. doi: 10.1007/BF00731267. [DOI] [PubMed] [Google Scholar]
- 67.Abhilash J., Dileep K.V., Palanimuthu M., Geethanandan K., Haridas M. Metal ions in sugar binding, sugar specificity and structural stability of Spatholobus parviflorus seed lectin. J. Mol. Model. 2013;19:3271–3327. doi: 10.1007/s00894-013-1854-4. [DOI] [PubMed] [Google Scholar]