Abstract
The emergence and spread of drug-resistant Mycobacterium tuberculosis strains (including MDR, XDR, and TDR) force scientists worldwide to search for new anti-tuberculosis drugs. We have previously reported a number of imidazo[1,2-b][1,2,4,5]tetrazines–putative inhibitors of mycobacterial eukaryotic-type serine-threonine protein-kinases, active against M. tuberculosis. Whole genomic sequences of spontaneous drug-resistant M. smegmatis mutants revealed four genes possibly involved in imidazo[1,2-b][1,2,4,5]tetrazines resistance; however, the exact mechanism of resistance remain unknown. We used different approaches (construction of targeted mutants, overexpression of the wild-type (w.t.) and mutant genes, and gene-expression studies) to assess the role of the previously identified mutations. We show that mutations in MSMEG_1380 gene lead to overexpression of the mmpS5-mmpL5 operon in M. smegmatis, thus providing resistance to imidazo[1,2-b][1,2,4,5]tetrazines by increased efflux through the MmpS5-MmpL5 system, similarly to the mechanisms of resistance described for M. tuberculosis and M. abscessus. Mycobacterial MmpS5-MmpL5 transporters should be considered as an MDR-efflux system and they should be taken into account at early stages of anti-tuberculosis drug development.
Keywords: Mycobacterium smegmatis; imidazo[1,2-b][1,2,4,5]tetrazine; MmpS5-MmpL5; efflux; drug discovery; drug resistance; tuberculosis
1. Introduction
Tuberculosis (TB), caused by Mycobacterium tuberculosis, is currently the leading killer among the infectious diseases caused by one infectious agent, responsible for an estimate of 1.2 million deaths in 2018 [1]. According to WHO, 1.7 billion people globally are infected with M. tuberculosis, and thus are at risk of developing the disease [1]. The emergence and spread of multidrug resistant TB (MDR-TB, defined as TB resistant to rifampicin and isoniazid), extensively drug-resistant TB (defined as MDR-TB with resistance to the fluoroquinolones and second-line injectables) and totally drug-resistant TB (TDR-TB) is a global threat to world-wide TB control [2,3,4,5]. Long treatment times (six months for drug-susceptible TB and up to two years for DR-TB) often lead to bad patient compliance, which is one of the causes of drug resistance development and results in worse treatment outcomes. Thus, researchers are forced to search for novel anti-TB drugs and shorter regimens [6].
Eukaryotic-type serine-threonine protein-kinases (ESTPKs) play a key role in M. tuberculosis life cycle regulation, controlling some of its vital aspects such as cell division and survival within host macrophages, and, therefore, they represent attractive targets for drug development [7,8]. We have previously described a number of imidazo[1,2-b][1,2,4,5]tetrazines with a promising antibacterial activity on M. tuberculosis and M. smegmatis [9]. Most of these compounds showed activity as potential ESTPK inhibitors in the original M. smegmatis aphVIII+ test-system [10,11], and two of them were able to bind to the M. tuberculosis PknB adenine-binding pocket according to docking studies [10]. Despite the predicted activity as ESTPK-inhibitors, both the exact mechanism of action and the mechanism of resistance to these compounds are still unknown.
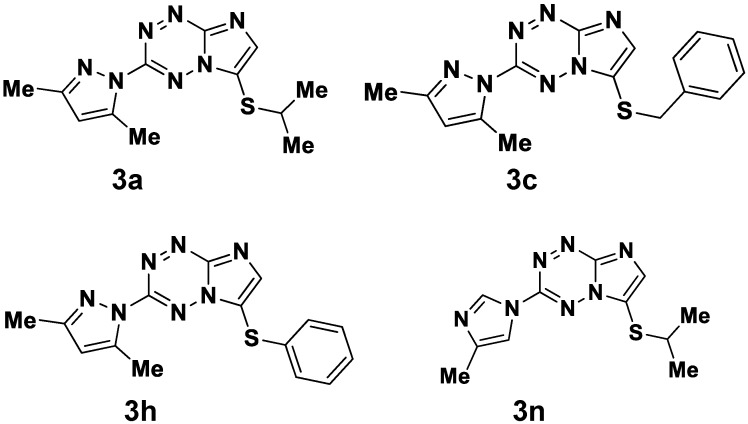
We were able to obtain spontaneous M. smegmatis mutants resistant to four imidazo[1,2-b][1,2,4,5]tetrazines (3a, 3c, 3h and 3n, Figure 1), which had cross-resistance among them, suggesting a common mechanism of drug-resistance [9]. Whole-genomic sequencing and comparative genomic analysis revealed mutations in MSMEG_0641 (binding-protein-dependent transporters inner membrane component) in 1 mutant, in MSMEG_1601 (hypothetical protein) in seven mutants, in MSMEG_2087 (transporter small conductance mechanosensitive ion channel (MscS) family protein) in one mutant [12], while all the mutants carried different mutations in MSMEG_1380 (AcrR/TetR_N transcriptional regulator) – 1 nonsynonymous SNP, 2 insertions leading to a frameshift, 2 duplications (6 and 501 base pairs-long) and one deletion [13].
Figure 1.
Chemical structures of imidazo[1,2-b][1,2,4,5]tetrazines [9].
In this article we describe the investigation of these mutations’ role in mycobacterial drug resistance to imidazo[1,2-b][1,2,4,5]tetrazines by different approaches: construction of targeted mutants, overexpression of the wild-type (w.t.) and mutant genes, and gene-expression studies.
2. Results
2.1. Mutations in MSMEG_1380 Gene Lead to Imidazo[1,2-b][1,2,4,5]tetrazines Resistance in M. smegmatis
The list of nonsynonymous mutations found in spontaneous drug-resistant M. smegmatis mutants used in this study is presented in Table 1. We were able to construct targeted M. smegmatis mutants harboring each mutation in genes MSMEG_0641, MSMEG_1601, and MSMEG_2087, as well as five mutations found in MSMEG_1380 gene using the p2NIL/pGOAL19 suicide system [14] for homologous recombination (Table 1).
Table 1.
Bacterial strains used in the study.
| Bacterial Strains | ||
|---|---|---|
| Name | Comment | Origin |
| M. smegmatis mc2 155 | Wild-type (w.t.) strain | |
| M. smegmatis atR1 | Spontaneous mutant of mc2 155. Mutations: Y52H (TAC>CAC) in MSMEG_1601; del LLA41-43 (del GCTGCTCGC480-488) in MSMEG_1380. | [9] |
| M. smegmatis atR2 | Spontaneous mutant of mc2 155. Mutations: Y52H (TAC>CAC) in MSMEG_1601; ins GC425-426 (frameshift) in MSMEG_1380. | [9] |
| M. smegmatis atR9 | Spontaneous mutant of mc2 155. Mutations: Y188C (TAC>TGC) in MSMEG_2087; ins C8 (frameshift) in MSMEG_1380. | [9] |
| M. smegmatis atR10 | Spontaneous mutant of mc2 155. Mutations: R233S (CGT>AGT) in MSMEG_0641; ins C8 (frameshift) in MSMEG_1380. | [9] |
| M. smegmatis atR14 | Spontaneous mutant of mc2 155. Mutations: Y52H (TAC>CAC) in MSMEG_1601; ins G448 (frameshift) in MSMEG_1380. | [9] |
| M. smegmatis atR19 | Spontaneous mutant of mc2 155. Mutations: Y52H (TAC>CAC) in MSMEG_1601; T52V (ACG>GTG) in MSMEG_1380. | [9] |
| M. smegmatis atR33 | Spontaneous mutant of mc2 155. Mutations: ins VG52-53 (ins GTGGGC154-159) in MSMEG_1380. | [9] |
| M. smegmatis atR37 | Spontaneous mutant of mc2 155. Mutation: del C 662 (frameshift) in MSMEG_1380. | [9] |
| M. smegmatis atR1c | Recombinant strain, mutation: del LLA41-43 (del GCTGCTCGC480-488) in MSMEG_1380. | This study |
| M. smegmatis atR2c | Recombinant strain, mutation: ins GC425-426 (frameshift) in MSMEG_1380. | This study |
| M. smegmatis atR9c | Recombinant strain, mutation: ins C8 (frameshift) in MSMEG_1380. | This study |
| M. smegmatis atR14c | Recombinant strain, mutation: ins G448 (frameshift) in MSMEG_1380. | This study |
| M. smegmatis atR33c | Recombinant strain, mutation: ins VG52-53 (ins GTGGGC154-159) in MSMEG_1380 | This study |
| M. smegmatis 0641c | Recombinant strain, mutation: R233S (CGT>AGT) in MSMEG_0641. | This study |
| M. smegmatis 1601c | Recombinant strain, mutation: Y52H (TAC>CAC) in MSMEG_1601 | This study |
| M. smegmatis 2087c | Recombinant strain, mutation: Y188C (TAC>TGC) in MSMEG_2087 | This study |
We examined the minimal inhibitory concentrations (MICs) of the four compounds on the recombinant M. smegmatis strains and found that mutations in MSMEG_1380 gene lead to elevated MICs as compared to the w.t. strain (4×MIC for the compound 3a, at least 2×MIC for the compound 3c, at least 2×MIC for the compound 3h, and 4×MIC for the compound 3n), while recombinants harboring mutations in genes MSMEG_0641, MSMEG_1601, and MSMEG_2087 had the same MICs as the w.t. strain (Table 2).
Table 2.
Imidazo[1,2-b][1,2,4,5]tetrazines MICs on M. smegmatis strains in liquid medium.
| Compound | M. smegmatis Strains MICs, μg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| mc2 155 | atR1c | atR2c | atR9c | atR14c | atR33c | 0641c | 1601c | 2087c | |
| 3a | 128 | 512 | 512 | 512 | 512 | 512 | 128 | 128 | 128 |
| 3c | 64 | >128 * | >128 * | >128 * | >128 * | >128 * | 64 | 64 | 64 |
| 3h | 128 | >256 * | >256 * | >256 * | >256 * | >256 * | 128 | 128 | 128 |
| 3n | 64 | 256 | 256 | 256 | 256 | 256 | 64 | 64 | 64 |
* The compounds were not soluble at higher concentrations; bacterial growth was observed at the stated concentrations.
Thus we have shown that only the mutations in MSMEG_1380 are responsible for imidazo[1,2-b][1,2,4,5]tetrazines resistance in M. smegmatis.
2.2. W.t. MSMEG_1380 Overexpression Increases M. smegmatis Susceptibility to imidazo[1,2-b][1,2,4,5]tetrazines
In order to investigate further the role of MSMEG_1380 in M. smegmatis resistance to imidazo[1,2-b][1,2,4,5]tetrazines, we cloned the w.t. MSMEG_1380 gene and two of its mutant variants in the tetracycline inducible plasmid pMINDKm- [15].
We used the paper-disc assay to assess the drug susceptibility of M. smegmatis strains to imidazo[1,2-b][1,2,4,5]tetrazines and found that the overexpression of the w.t. MSMEG_1380 gene increases M. smegmatis susceptibility to the tested compounds, while the overexpression of its mutant variants had no effect on the phenotype (Table 3), thus suggesting that the disruption of MSMEG_1380 protein’s function leads to the drug-resistant phenotype.
Table 3.
Growth inhibition halos, produced by imidazo[1,2-b][1,2,4,5]tetrazines on M. smegmatis strains.
| Compound | Concentration, nmole/disc | Growth Inhibition Halo, mm | |||
|---|---|---|---|---|---|
| M. smegmatis Transformants | |||||
| pMINDKm- | pMINDKm-:msmeg_1380 | pMINDKm-:msmeg_1380-19 | pMINDKm-:msmeg_1380-33 | ||
| 3a | 300 | 9.8 ± 1.5 | 17.0 ± 3.6 | 8.8 ± 0.8 | 8.2 ± 0.6 |
| 3c | 300 | 7.0 ± 0.8 | 15.0 ± 2.9 | 6.3 ± 0.5 | 6.3 ± 0.5 |
| 3h | 40 | 6.7 ± 0.5 | 11.5 ± 0.4 | 6.3 ± 0.5 | 6.5 ± 0.4 |
| 3n | 100 | 9.7 ± 2.4 | 16.0 ± 0.8 | 9.2 ± 1.3 | 9.3 ± 1.2 |
2.3. MSMEG_1380 Represses the Expression of the mmpS5-mmpL5 Operon in M. smegmatis
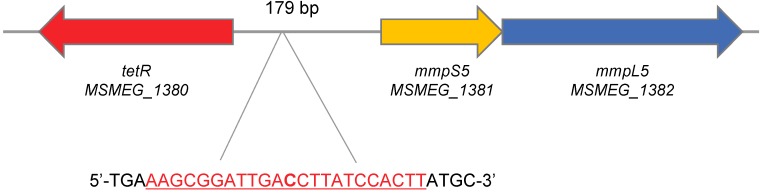
MSMEG_1380 gene lies 179 b.p. upstream the mmpS5-mmpL5 operon (genes MSMEG_1381-MSMEG_1382) in the M. smegmatis genome and is transcribed in the opposite direction (Figure 2). The structure of this operon is conserved in different mycobacterial species: the mmpS5-mmpL5 genes are controlled by a TetR-family transcriptional repressor, encoded by a gene located upstream the operon. Mutations in genes encoding the TetR-repressor, which lead to the upregulation of the mmpS5-mmpL5 genes, are involved in M. abscessus resistance to the derivatives of thiacetazone [16], as well as in cross-resistance of M. tuberculosis to bedaquiline and clofazimine [17]. We have also identified a possible operator sequence in the 5′-untranslated region of MSMEG_1381, similar to the one described in [16]: 5′-AAGCGGATTGACCTTATCCACTT-3′.
Figure 2.
Schematic representation of the mmpS5-mmpL5 operon structure in M. smegmatis genome. The putative operator sequence is shown in red.
To test the hypothesis that resistance to imidazo[1,2-b][1,2,4,5]tetrazines in M. smegmatis has a similar origin to the ones described for M. tuberculosis and M. abscessus [16,17], we analyzed the expression of MSMEG_1380 gene and mmpL5 (MSMEG_1382) genes in different conditions.
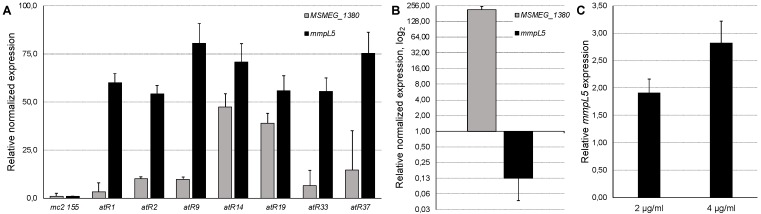
All the spontaneous M. smegmatis mutants had increased mmpL5 expression (54.16–80.45 times) as compared to the w.t. M. smegmatis mc2 155 strain (Figure 3A). The overexpression of the w.t. MSMEG_1380 gene, cloned into the pMINDKm- plasmid led to a 7.90-fold repression of the mmpL5 gene expression (p < 0.001, Figure 3B), confirming that MSMEG_1380 encodes the repressor of the mmpS5-mmpL5 operon, and explaining the drug-susceptible phenotype, observed in the MSMEG_1380 overexpressing strain. On the contrary, the expression of MSMEG_1380 was upregulated in the mutant strains (Figure 3A), indicating that this transcriptional repressor is self-regulatory and that mutations lead to the loss of its function.
Figure 3.
Relative expression levels of mmpS5-mmpL5 operon genes in different conditions: expression levels of MSMEG_1380 and mmpL5 genes in spontaneous M. smegmatis imidazo[1,2-b][1,2,4,5]tetrazine-resistant mutants (A); expression levels of MSMEG_1380 and mmpL5 genes in M. smegmatis pMINDKm-:msmeg_1380 (B); expression levels of the mmpL5 gene after the addition of different concentrations of the compound 3a (shown on the X-axis) (C). Error bars represent standard deviations from triplicates.
We also observed that the addition of subinhibitory concentrations of the compound 3a upregulated the expression of mmpL5 in a dose-dependent manner (Figure 3C).
3. Discussion
Deorphaning phenotypic screening hits, that is determining their mechanism of action and/or resistance, is a key part in the early-stage anti-TB drug development [18]. Here we determine the mechanism of M. smegmatis imidazo[1,2-b][1,2,4,5]tetrazines resistance based on the previously obtained whole-genome sequencing data for 12 spontaneous mutants with cross-resistance to four compounds [9,13].
The construction of targeted mutants showed that only mutations in MSMEG_1380 are responsible for drug resistance. In M. smegmatis, MSMEG_1380 encodes a TetR-family transcriptional repressor, which controls the mmpS5-mmpL5 operon, encoding transmembrane transporters, conserved throughout mycobacterial species. Mutations occurring in MSMEG_1380 led to the upregulation of the mmpS5-mmpL5 operon and increased efflux of the drug-candidates from the cells, similarly to the mechanisms described for M. tuberculosis and M. abscessus [16,17]. Overexpression of the w.t. MSMEG_1380 led to the repression of the mmpS5-mmpL5 operon and expectedly to an increased drug susceptibility phenotype. Interestingly, we observed a dose-dependent upregulation of the mmpS5-mmpL5 operon upon the addition of one of the compounds, which may indicate the ability of this compound to bind to the MSMEG_1380 protein, inhibiting its affinity to the operator sequence; however, this needs to be examined in vitro in future studies.
The tested compounds showed activity as ESTPK inhibitors [9,11]; however, we have not observed any mutations in ESTPK genes. One or more ESTPKs might still be the biotargets of imidazo[1,2-b][1,2,4,5]tetrazines but determining them by spontaneous mutagenesis might be difficult: some of the ESTPKs may fulfill the functions of others in the situation when they might be inhibited [8], and there is a possibility that more than one mutation might be required.
The primary biological role of the MmpS5-MmpL5 system consists in siderophore transport, which is crucial for M. tuberculosis survival under low-iron conditions within macrophages [19]. Yet, this efflux system has also shown itself to be an important factor of drug resistance: besides the mentioned efflux-mediated resistance to thiacetazone derivatives, bedaquiline and clofazimine [16,17], it has also been reported to provide M. tuberculosis resistance to azoles [20]. We can expect that M. tuberculosis strains resistant to bedaquiline and clofazimine might also be resistant to imidazo[1,2-b][1,2,4,5]tetrazines; however, a 36% mismatch in the amino acid sequences of the MmpL5 proteins in M. smegmatis and M. tuberculosis may affect the drug-specificity of the transporter, and this should be additionally examined in future studies. Still, the MmpS5-MmpL5-mediated resistance mechanism needs to be considered during early stages of anti-TB drug development, and convenient in silico and in vitro test-systems for rapid analysis should be developed.
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
M. smegmatis strains described in this study are presented in Table 1. Middlebrook 7H9 medium (Himedia, India) supplemented with OADC (Himedia, India), 0.1% Tween-80 (v/v), and 0.4% glycerol (v/v) was used as liquid medium, while the M290 Soyabean Casein Digest Agar (Himedia, India) was used as solid medium. Escherichia coli DH5α was used for plasmids propagation. Cultures in liquid medium were incubated in the Multitron incubator shaker (Infors HT, Switzerland) at 37 °C and 250 rpm.
4.2. Targeted M. smegmatis mutants’ Construction
Targeted M. smegmatis mc2 155 mutants were constructed by homologous recombination using the p2NIL/pGOAL19 suicide vector system [14]. Briefly, genes MSMEG_0641, MSMEG_1380, MSMEG_1601, and MSMEG_2087 with adjacent 1-1,5 kb fragments were amplified from genomic DNA, isolated from respective mutants by phenol-chloroform/isoamyl alcohol extraction after enzymatic cell lysis [21], with Phusion High-Fidelity DNA Polymerase (Thermo Scientific, USA) using the following primers, picked with primer-BLAST [22]: pN_0641_f 5′- TTTTCTGCAGCCAACAACGATCCAGATGTCCGT-3′ and pN_0641_r 5′- TTTTAAGCTTCAATGGCGGCGTCTTCATTCTG-3′ for MSMEG_0641; pN_1380_f 5′- TTTTAAGCTTGTACTACTCGCTGGTGGCGTC-3′ and pN_1380_r 5′- TTTTGGATCCTGCTGCACGTGTTCGGTGTC-3′ for MSMEG_1380; pN_1601_f 5′- CCCATGACGGGCATCATCAACC-3′ and pN_1601_r 5′- TTTTTTAATTAACGACGATCAGCACGTCCACAC-3′ for MSMEG_1601; pN_2087_f 5′- TTTTAAGCTTCCAGAAGGTCACCAGCGATCTG-3′ and pN_2087_r. The amplified products were digested with respective restriction enzymes (Thermo Scientific, USA) and ligated in the p2NIL plasmid. The cassette from pGOAL19 was subsequently cloned in the obtained plasmids at the PacI restriction site. The plasmids were electroporated in M. smegmatis mc2 155 cells as described in [23] and plated on M290 plates supplemented with kanamycin (50 μg/mL), hygromycin (50 μg/mL), and X-Gal (50 μg/mL); blue single-crossover colonies were selected. Blue colonies were grown overnight in liquid 7H9 medium with ADC, and serial 10-fold dilutions were plated on M290 plates supplemented with X-Gal (50 μg/mL) and sucrose (2% w/v); white double-crossover colonies were selected and tested for Km susceptibility. Target genes were then Sanger-sequenced for a final confirmation of the mutation.
4.3. MIC Determination
MICs of the studied compounds on M. smegmatis were determined in liquid medium. M. smegmatis strains were cultured overnight in 7H9 medium, then diluted in the proportion of 1:200 in fresh medium (to approximately OD600 = 0.05). 196 μl of the diluted culture was poured in sterile nontreated 96-well flat-bottom culture plates (Eppendorf, Germany) and 4 μL of serial two-fold dilutions of the tested compounds in DMSO were added to the wells. The plates were incubated at 37 °C and 250 rpm for 48 h. The MIC was determined as the lowest concentration of the compound with no visible bacterial growth.
4.4. MSMEG_1380 Cloning, Expression and Drug-Susceptibility Testing
MSMEG_1380 genes from respective strains were amplified by Phusion High-Fidelity DNA Polymerase (Thermo Scientific, USA) using primers pM_1380_f 5′-GACACATATGGGAGGAAATGTTGTGAGTGCCCCCGAGACG-3′ and pM_1380_r 5′-TTTTACTAGTTCAGGTGGCGCAGGGCG-3′ picked with primer-BLAST [22] and cloned in the pMINDKm- plasmid [15], a modification of pMIND [24] lacking the kanamycin resistance gene, at the NdeI and SpeI restriction sites, to obtain the following plasmids: pMINDKm-:msmeg_1380, pMINDKm-:msmeg_1380-19, and pMINDKm-:msmeg_1380-33, containing, respectively, the w.t. msmeg_1380 gene as well as its mutant variants from strains atR19 and atR33. The resulting plasmids were electroporated in M. smegmatis mc2 155 cells as described in [23].
M. smegmatis transformants were grown in Middlebrook 7H9 broth supplemented with hygromycin (50 μg/mL) and tetracycline (10 ng/mL) to midexponential phase (OD600 = 1.2). Afterwards the cultures were diluted in the proportion of 1:9:10 (culture:water:M290 medium) and 5 mL were poured as the top layer on Petri dishes with agarized M290 medium. Both top- and bottom-layers were supplemented with hygromycin (50 μg/mL) and tetracycline (10 ng/mL). The plates were allowed to dry for at least 30 min, afterwards sterile paper discs with impregnated imidazo[1,2-b][1,2,4,5]tetrazines were plated. The plates were incubated for 2–3 days at 37 °C, until the bacterial lawn was fully grown. Growth inhibition halos were measured to the nearest 1 mm. The experiments were carried out as triplicates; the average diameter and standard deviation (SD) were calculated.
4.5. Mycobacterial RNA Isolation and Real-Time qPCR
M. smegmatis strains were grown overnight in Middlebrook 7H9 broth to midexponential phase (OD600 = 1.0–1.2); cells from 10 mL culture were harvested by centrifugation for 10 min at 3000× g and washed by 1 mL of RNAprotect Bacteria Reagent (Qiagen, USA). Total RNA was extracted by homogenization in Trizol solution (Invitrogen, USA) [25], followed by phenol (pH = 4.5)-chloroform/isoamyl alcohol (25:24:1) purification and precipitation in high salt solution (0.8 M Na citrate, 1.2 M NaCl) with isopropanol. Remaining genomic DNA was removed by DNAse I, Amplification grade (Invitrogen, USA). 50 ng of total RNA was used for cDNA synthesis by iScript Select cDNA Synthesis Kit (Bio-Rad, USA). 1 ng of cDNA was used for real-time qPCR with the qPCRmix-HS SYBR kit (Evrogen, Russia) on a CFX96 Touch machine (Bio-Rad, USA). CFX Manager V 3.1 (Bio-Rad, USA) was used to analyze the qPCR results: relative normalized expression of three biological replicates was calculated as ΔΔCq and genes sigA and ftsZ were used as reference. The following primers were picked by primer-BLAST [22] for qPCR: q1380-f 5′-CTGCTCGACGAACCATGCGAAAC-3′ and q1380-r 5′-AAGGGTCTTGAGCCGAATCTCAACG-3′ (MSMEG_1380), q1382-f 5′-ACCACGCAGATCATGAACAACGACT-3′ and q1382-r 5′-GAAATCGTCGAAGTCCGCCAGATGA-3′ (MSMEG_1382), qsigAs-sm-f 5′-CGAGCTTGTTGATCACCTCGACCAT-3′ and qsigAs-sm-r 5′-CTCGACCTCATCCAGGAAGGCAAC-3′ (sigA), qftsZs-sm-f 5′-AGCAGCTCCTCGATGTCGTCCTT-3′ and qftsZs-sm-r 5′-GCCTGAAGGGCGTCGAGTTCAT-3′ (ftsZ).
Acknowledgments
We would like to thank Acad. V.N. Charushin and G.L. Rusinov of the Laboratory of Heterocyclic Compounds, Postovsky Institute of Organic Synthesis, Ural Branch of RAS, for generously providing compounds for this study.
Author Contributions
Conceptualization, D.A.M.; formal analysis, D.A.M., K.V.S., A.A.V.; investigation, D.A.M., K.V.S., A.A.V.; resources, D.A.M., K.V.S., V.N.D.; writing—original draft preparation, D.A.M.; writing review & editing, D.A.M., K.V.S., A.A.V., V.N.D.; visualization, D.A.M.; supervision, V.N.D.; project administration, D.A.M.; funding acquisition, D.A.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Russian Science Foundation (RSF), grant number 17-75-20060.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.World Health Organization Global Tuberculosis Report 2019. WHO; Geneva, Switzerland: 2019. pp. 1–297. [Google Scholar]
- 2.Gandhi N.R., Nunn P., Dheda K., Schaaf H.S., Zignol M., Van Soolingen D., Jensen P., Bayona J. Multidrug-resistant and extensively drug-resistant tuberculosis: A threat to global control of tuberculosis. Lancet. 2010;375:1830–1843. doi: 10.1016/S0140-6736(10)60410-2. [DOI] [PubMed] [Google Scholar]
- 3.Caminero J.A., Sotgiu G., Zumla A., Migliori G.B. Best drug treatment for multidrug-resistant and extensively drug-resistant tuberculosis. Lancet Infect Dis. 2010;10:621–629. doi: 10.1016/S1473-3099(10)70139-0. [DOI] [PubMed] [Google Scholar]
- 4.Klopper M., Warren R.M., Hayes C., Gey van Pittius N.C., Streicher E.M., Müller B., Sirgel F.A., Chabula-Nxiweni M., Hoosain E., Coetzee G., et al. Emergence and Spread of Extensively and Totally Drug-Resistant Tuberculosis, South Africa. Emerg. Infect. Dis. 2013;19:449–455. doi: 10.3201/eid1903.120246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Velayati A.A., Farnia P., Farahbod A.M. Overview of drug-resistant tuberculosis worldwide. Int. J. Mycobacteriol. 2016;5:S161. doi: 10.1016/j.ijmyco.2016.09.066. [DOI] [PubMed] [Google Scholar]
- 6.Muñoz-Torrico M., Duarte R., Dalcolmo M., D’Ambrosio L., Migliori G.B. New drugs and perspectives for new anti-tuberculosis regimens. Rev. Port. De Pneumol. 2018;24:86–98. doi: 10.1016/j.rppnen.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 7.Danilenko V.N., Osolodkin D.I., Lakatosh S.A., Preobrazhenskaya M.N., Shtil A.A. Bacterial eukaryotic type serine-threonine protein kinases: From structural biology to targeted anti-infective drug design. Curr. Top. Med. Chem. 2011;11:1352–1369. doi: 10.2174/156802611795589566. [DOI] [PubMed] [Google Scholar]
- 8.Prisic S., Husson R.N. Mycobacterium tuberculosis Serine/Threonine Protein Kinases. Microbiol. Spectr. 2014;2:1–26. doi: 10.1128/microbiolspec.MGM2-0006-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maslov D.A., Korotina A.V., Shur K.V., Vatlin A.A., Bekker O.B., Tolshchina S.G., Ishmetova R.I., Ignatenko N.K., Rusinov G.L., Charushin V.N., et al. Synthesis and antimycobacterial activity of imidazo[1,2-b][1,2,4,5]tetrazines. Eur. J. Med. Chem. 2019;178:39–47. doi: 10.1016/j.ejmech.2019.05.081. [DOI] [PubMed] [Google Scholar]
- 10.Bekker O.B., Danilenko V.N., Ishmetova R.I., Maslov D.A., Rusinov G.L., Tolshchina S.G., Charushin V.N. Substituted azolo[1,2,4,5]tetrazines-inhibitors of antibacterial serine-threonine protein kinases. RU 2462466. Patent. 2012 Sep 27; NPO SRC “BIOAN”, Moscow, Russia.
- 11.Bekker O.B., Danilenko V.N., Maslov D.A. Test system of Mycobacterium smegmatis aphVIII+ for screening of inhibitors of serine-threonine protein kinases of eukaryotic type. RU 2566998. Patent. 2015 Oct 27; NPO SRC “BIOAN”, Moscow, Russia.
- 12.Maslov D.A., Bekker O.B., Shur K.V., Vatlin A.A., Korotina A.V., Danilenko V.N. Whole-genome sequencing and comparative genomic analysis of Mycobacterium smegmatis mutants resistant to imidazo[1,2-b][1,2,4,5]tetrazines, antituberculosis drug candidates. BRSMU. 2018:19–22. doi: 10.24075/brsmu.2018.039. [DOI] [Google Scholar]
- 13.Vatlin A.A., Shur K.V., Danilenko V.N., Maslov D.A. Draft Genome Sequences of 12 Mycolicibacterium smegmatis Strains Resistant to Imidazo[1,2-b][1,2,4,5]Tetrazines. Microbiol. Resour. Announc. 2019;8:e00263-19. doi: 10.1128/MRA.00263-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parish T., Stoker N.G. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology. 2000;146:1969–1975. doi: 10.1099/00221287-146-8-1969. [DOI] [PubMed] [Google Scholar]
- 15.Maslov D.A., Bekker O.B., Alekseeva A.G., Kniazeva L.M., Mavletova D.A., Afanasyev I.I., Vasilevich N.I., Danilenko V.N. Aminopyridine- and aminopyrimidine-based serine/threonine protein kinase inhibitors are drug candidates for treating drug-resistant tuberculosis. BRSMU. 2017:38–43. doi: 10.24075/brsmu.2017-01-04. [DOI] [Google Scholar]
- 16.Richard M., Gutiérrez A.V., Viljoen A.J., Ghigo E., Blaise M., Kremer L. Mechanistic and Structural Insights Into the Unique TetR-Dependent Regulation of a Drug Efflux Pump in Mycobacterium abscessus. Front. Microbio. 2018;9:649. doi: 10.3389/fmicb.2018.00649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andries K., Villellas C., Coeck N., Thys K., Gevers T., Vranckx L., Lounis N., de Jong B.C., Koul A. Acquired resistance of Mycobacterium tuberculosis to bedaquiline. PLoS ONE. 2014;9:e102135. doi: 10.1371/journal.pone.0102135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooper C.B. Development of Mycobacterium tuberculosis Whole Cell Screening Hits as Potential Antituberculosis Agents. J. Med. Chem. 2013;56:7755–7760. doi: 10.1021/jm400381v. [DOI] [PubMed] [Google Scholar]
- 19.Sandhu P., Akhter Y. Siderophore transport by MmpL5-MmpS5 protein complex in Mycobacterium tuberculosis. J. Inorg. Biochem. 2017;170:75–84. doi: 10.1016/j.jinorgbio.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 20.Milano A., Pasca M.R., Provvedi R., Lucarelli A.P., Manina G., de Jesus Lopes Ribeiro A.L., Manganelli R., Riccardi G. Azole resistance in Mycobacterium tuberculosis is mediated by the MmpS5-MmpL5 efflux system. Tuberculosis. 2009;89:84–90. doi: 10.1016/j.tube.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 21.Belisle J.T., Mahaffey S.B., Hill P.J. Mycobacteria Protocols. Volume 465. Humana Press; Totowa, NJ, USA: 2010. Isolation of Mycobacterium Species Genomic DNA; pp. 1–12. Methods in Molecular Biology. [DOI] [PubMed] [Google Scholar]
- 22.Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goude R., Parish T. Mycobacteria Protocols. 2nd ed. Volume 465. Humana Press; Totowa, NJ, USA: 2010. Electroporation of Mycobacteria; pp. 203–215. Methods in Molecular Biology. [DOI] [PubMed] [Google Scholar]
- 24.Blokpoel M.C.J. Tetracycline-inducible gene regulation in mycobacteria. Nucleic Acids Res. 2005;33:e22. doi: 10.1093/nar/gni023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rustad T.R., Roberts D.M., Liao R.P., Sherman D.R. Mycobacteria Protocols. Volume 465. Humana Press; Totowa, NJ, USA: 2010. Isolation of Mycobacterial RNA; pp. 13–22. Methods in Molecular Biology. [DOI] [PubMed] [Google Scholar]