Aquifer systems may hold up to 40% of the total microbial biomass on Earth. However, little is known about the composition of microbial communities within these critical freshwater ecosystems. Here, we took advantage of Florida’s first-magnitude springs (the highest spring classification based on water discharge), each discharging at least 246 million liters of water each day from the Floridan aquifer system (FAS), to investigate prokaryotic and viral communities from the aquifer. The FAS serves as a major source of potable water in the Southeastern United States, providing water for large cities and citizens in three states. Unfortunately, the health of the FAS and its associated springs has declined in the past few decades due to nutrient loading, increased urbanization and agricultural activity in aquifer recharge zones, and saltwater intrusion. This is the first study to describe the prokaryotic and viral communities in Florida’s first-magnitude springs, providing a baseline against which to compare future ecosystem change.
KEYWORDS: Florida, bacilladnavirus, ecology, freshwater, microbial ecology, phage, prokaryote, springs, ssDNA virus, viral, virome, virus
ABSTRACT
Aquifers, which are essential underground freshwater reservoirs worldwide, are understudied ecosystems that harbor diverse forms of microbial life. This study investigated the abundance and composition of prokaryotic and viral communities in the outflow of five springs across northern Florida, USA, as a proxy of microbial communities found in one of the most productive aquifers in the world, the Floridan aquifer. The average abundances of virus-like particles and prokaryotic cells were slightly lower than those reported from other groundwater systems, ranging from 9.6 × 103 ml−1 to 1.1 × 105 ml−1 and 2.2 × 103 ml−1 to 3.4 × 104 ml−1, respectively. Despite all of the springs being fed by the Floridan aquifer, sequencing of 16S rRNA genes and viral metagenomes (viromes) revealed unique communities in each spring, suggesting that groundwater microbial communities are influenced by land usage in recharge zones. The prokaryotic communities were dominated by Bacteria, and though the most abundant phyla (Proteobacteria, Cyanobacteria, and Bacteroidetes) were found in relatively high abundance across springs, variation was seen at finer taxonomic resolution. The viral sequences were most similar to those described from other aquatic environments. Sequencing resulted in the completion of 58 novel viral genomes representing members of the order Caudovirales as well as prokaryotic and eukaryotic single-stranded DNA (ssDNA) viruses. Sequences similar to those of ssDNA viruses were detected at all spring sites and dominated the identifiable sequences at one spring site, showing that these small viruses merit further investigation in groundwater systems.
INTRODUCTION
Natural freshwater springs form when groundwater from an aquifer meets the land surface. Florida, USA, has one of the highest spring densities in the world, with >700 springs located mainly in the northern half of the state (1, 2). These unique aquatic environments are ecologically and economically important ecosystems, serving as hubs of biodiversity and generating millions of dollars for the state through tourism (3, 4). Thirty-three of Florida’s springs are classified as first-magnitude springs (the highest spring classification based on water discharge), each discharging upwards of 246 million liters of water per day (1, 5, 6). All first-magnitude springs in Florida are fed by the Floridan aquifer system (FAS), which keeps the water clear and thermally insulated at 23°C (1, 2). The FAS is one of the most productive aquifer systems in the world, providing ∼10 million people in Florida and the nearby states of Georgia and Alabama with potable water (7). While the hydrology and residence time (average ∼20 years) of the water discharged through first-magnitude springs vary, the large volume of water discharged by first-magnitude springs makes them a mirror of the aquifer and therefore useful for monitoring the health of the FAS (1, 2, 8).
The health of the FAS and of Florida’s springs is impacted by anthropogenic activities (9). There has been an increase in urbanization and agricultural activity within the drainage basins that supply water to the springs, known as springsheds or spring recharge basins (6). Concentrations of phosphate and nitrate have increased dramatically in springs across the state, resulting in a decline in water quality (6, 8, 10, 11). Saltwater intrusion into the aquifer has become an imminent threat due to rising sea level combined with increased water withdrawals from the aquifer to meet water demands (12, 13). The precarious status of springs throughout Florida, combined with the necessity of these ecosystems as water supplies, emphasizes the need for comprehensive studies of physiochemical and biological parameters within the springs and FAS to provide a baseline against which to compare future conditions.
Aquifer systems host diverse microbial life and harbor a significant fraction (up to 40%) of the total microbial biomass on Earth (14–22). Prokaryotes play vital roles in mediating biogeochemical cycling and ultimately aid in maintenance of aquifers’ water quality via removal of organic carbon, nitrates, and even micropollutants (23, 24). Viruses are also increasingly recognized as important members of aquatic environments, and yet only a small number of studies have looked at viruses in groundwater systems (19–22), which represent a reservoir of unexplored viral diversity. However, despite their significance, little is known about the naturally occurring prokaryotic communities in the FAS or its associated springs. Previous groundwater quality monitoring studies have looked for specific bacterial species in Florida’s springs as indicators of potential contaminants that might have permeated the FAS (25, 26). There are currently no reports providing a comprehensive picture of the microbial community composition, including viruses, within these critical aquatic ecosystems.
Here, we surveyed five first-magnitude springs across the northern half of the state of Florida to describe their prokaryotic and viral community composition. Given that all of the springs are fed by the FAS, we hypothesized that all of the springs would share similar microbial communities. To investigate this, we measured basic physiochemical parameters, determined the abundance of prokaryotic cells and virus-like particles (VLPs) by epifluorescence microscopy, and sequenced 16S rRNA gene amplicons and viral metagenomes (viromes) from each spring site. This report constitutes the first description of the prokaryotic and viral communities of Florida’s first-magnitude springs, providing a glimpse into the microbial communities that inhabit the Floridan aquifer. In contrast to our hypothesis, both chemical and microbial analyses indicated that each spring site is unique, likely due in part to distinct land usage patterns in groundwater recharge zones.
RESULTS
Spring physiochemical characteristics.
The vent outflow waters of five first-magnitude springs (Ichetucknee, Jackson, Manatee, Rainbow, and Volusia Springs) were sampled during May and June of 2017 (Fig. 1 and Table 1). All five springs are fed by the Floridan aquifer and exhibit similar temperatures ranging from 21.2 to 25.4°C. Large variations were measured for several of the physiochemical parameters. Nitrate concentrations ranged from 23.1 μM at Volusia Springs to 230.9 μM at Jackson Springs (Table 1). In contrast, Volusia Springs had the highest phosphate concentration (1.30 μM), an order of magnitude greater than that measured in Ichetucknee Springs, the site with the lowest phosphate concentration (0.12 μM). Rainbow Springs had the lowest conductivity (146.8 μS cm−1) and highest pH (8.33) and dissolved oxygen (DO; 83.3%) levels of the sites, while Manatee Springs represented the opposite end of the spectrum, with the highest conductivity (504.0 μS cm−1) and lowest pH (7.33) and dissolved oxygen (14.0%) levels.
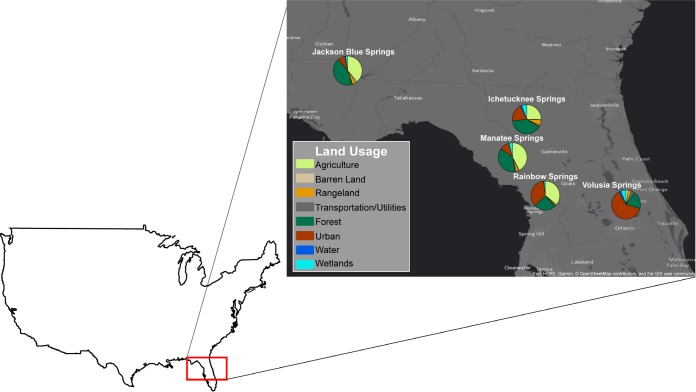
FIG 1.
Map showing the location of investigated springs and the estimated land usage profile around each spring based on the inferred recharge zone. Data are from the Florida Department of Environmental Protection (DEP). This figure was made using ArcGIS (85).
TABLE 1.
Dates of sample collection and available metadata from each spring sitea
| Spring site |
Collection date |
Temp (°C) |
pH | Turbidity (NTU) |
% DO | Conductivity (μS/cm) |
Nitrate (μM) |
Nitrite (μM) |
Ammonium (μM) |
Phosphate (μM) |
|---|---|---|---|---|---|---|---|---|---|---|
| Ichetucknee | 30 May 2017 | 25.4 | 7.67 | 3.8 | 43.6 | 327.9 | 28.5 | 0.06 | 1.10 | 0.12 |
| Jackson | 1 June 2017 | 21.2 | 7.70 | 3.1 | 35.7 | 270.5 | 230.9 | 0.13 | 8.75 | 0.30 |
| Manatee | 5 May 2017 | 22.4 | 7.33 | 3.5 | 14.0 | 504.0 | 153.1 | 0.04 | 0.12 | 0.56 |
| Rainbow | 26 May 2017 | 23.0 | 8.33 | 3.5 | 83.3 | 146.8 | 85.4 | 0.07 | 0.33 | 0.38 |
| Volusia | 22 May 2017 | 22.3 | 7.43 | 3.4 | 39.3 | 183.6 | 23.1 | 0.10 | 2.68 | 1.30 |
NTU, nephelometric turbidity units; DO, dissolved oxygen.
Abundance of prokaryotic cells and virus-like particles (VLPs).
SYBR gold staining and epifluorescence microscopy were used to determine the abundance of prokaryotic cells and VLPs in each of the spring outflow samples. These abundances were fairly consistent between Ichetucknee, Jackson, Manatee, and Rainbow Springs, with cell abundances ranging from 2.2 × 103 to 5.6 × 103 per milliliter of spring water and VLP counts ranging from 9.5 × 103 to 2.5 × 104 per milliliter of spring water (Table 2). Prokaryotic and VLP abundances in Volusia Springs were an order of magnitude higher than those in the other spring sites (3.4 × 104 cells ml−1 and 1.1 × 105 VLP ml−1). The virus-to-prokaryote ratio (VPR) ranged between 2 and 10 in each of the springs, with an average VPR value of 5.
TABLE 2.
Concentrations of prokaryotic cells and virus-like particles in samples from each spring site as determined by SYBR gold staining and epifluorescence microscopy, and calculated virus-to-prokaryote ratiosa
| Spring site | Mean (± SD) no. of cells/ml |
Mean (± SD) no. of VLP/ml |
VPR |
|---|---|---|---|
| Ichetucknee | 2.7 (± 0.65) × 103 | 1.9 (± 0.15) × 104 | 6.9 |
| Jackson | 2.2 (± 0.27) × 103 | 2.1 (± 0.23) × 104 | 9.7 |
| Manatee | 3.6 (± 0.89) × 103 | 9.5 (± 2.5) × 103 | 2.6 |
| Rainbow | 5.6 (± 1.7) × 103 | 2.5 (± 0.37) × 104 | 4.6 |
| Volusia | 3.4 (± 0.99) × 104 | 1.1 (± 0.28) × 105 | 3.2 |
VLP, virus-like particles; VPR, calculated virus-to-prokaryote ratio.
Prokaryotic community composition.
Approximately 700,000 paired-end 16S rRNA gene amplicon reads were obtained from the spring sites, resulting in the identification of 8,230 amplicon sequence variants (ASVs). The majority of these ASVs (7,327) were found in only a single spring site (Fig. 2A). Roughly 10% of the ASVs were found in multiple spring sites, with 567 ASVs found in two spring sites, 164 ASVs found in three, 60 ASVs found in four, and only 24 ASVs found at all five (Fig. 2A). Volusia Springs had the lowest number of ASVs shared with any other spring site, and Ichetucknee, Jackson, and Rainbow Springs shared the most ASVs (Fig. 2A). The shared ASVs comprised only 3% to 6% of the sequences from Ichetucknee and Jackson Springs and 19% to 28% of the sequences from Manatee, Rainbow, and Volusia Springs. A nonmetric multidimensional scaling (NMDS) plot based on the Bray-Curtis dissimilarity of the relative abundances of ASVs in each spring showed clear grouping based on spring site (permutational multivariate analysis of variance [PERMANOVA], P = 0.002), with biological replicates clustering tightly together (Fig. 2B). The statistically significant physiochemical parameters mapped onto the NMDS were phosphate, nitrite, and ammonium (Fig. 2B).
FIG 2.
(A) Venn diagram depicting the distribution of ASVs among springs. (B) NMDS plot showing the similarity of the 16S rRNA gene community structures of each spring site, based on a Bray-Curtis dissimilarity matrix. Biological replicates of each spring are represented by points of identical colors and shapes. The ordinal ellipses represent the 95% confidence interval of the standard error. The arrows represent physiochemical parameters significantly (P < 0.05) correlating with the ordination of the communities. The direction of each arrow indicates direction, and the length denotes magnitude of influence.
Taxonomic assignments of the ASVs based on the SILVA database demonstrated that Bacteria dominated the prokaryotic community of the springs, accounting for >95% of the 16S rRNA gene sequences from each site. The 24 ASVs shared between all five spring sites belonged to members of the phyla Proteobacteria, Cyanobacteria, Verrucomicrobia, and Bacteroidetes. Notably, ASVs representing these four phyla and Planctomycetes composed between 89% and 95% of the total reads from each spring (Fig. 3A). The 25 most abundant phyla composed >99% of the total sequences from each spring (Fig. 3A).
FIG 3.
(A) Heat map representing the percentages of 16S rRNA gene sequences in each spring belonging to the 25 most abundant phyla. Dark orange indicates a higher percentage of sequence abundance, and white represents the absence of the phylum. (B) Stacked bar chart representing the 10 most abundant families in each spring site. The spotted pattern indicates that the family was in the top 10 most abundant families in all five springs, vertical stripes indicate that it was in the 10 most abundant families in 4 springs, and horizontal stripes indicate that it was in the 10 most abundant families in 3 springs.
Although not all ASVs were classified to the family or species level due to database limitations, some trends became apparent in looking at taxonomic resolution finer than phyla. Each spring was dominated by a different family, with Phormidiaceae (26%) being the most abundant family in Ichetucknee Springs, Microcystaceae (10.5%) in Jackson Springs, Pseudomonadaceae (20%) in Manatee Springs, Sphingomonadaceae (9%) in Rainbow Springs, and Methylomonaceae (13%) in Volusia Springs (Fig. 3B). Of the top 10 most abundant families at each spring site, only two families (Sphingomonadaceae and Burkholderiaceae) were common to all sites (Fig. 3B). Six other families (Pseudomonadaceae, Caulobacteraceae, Chitinophagaceae, Hydrogenophilaceae, Rhodobacteraceae, and Moraxellaceae) were among the top 10 most abundant families of three or four spring sites (Fig. 3B). Pseudomonadaceae, Moraxellaceae, and Caulobacteraceae were the Proteobacteria families that appeared most often in the shared ASVs among all sites, accounting for 1.5% to 7.5% of the 24 shared ASV sequences.
Archaea represented a small portion of the springs’ prokaryotic communities, at only 0.2% to 4.7% of the 16S rRNA gene sequences. Manatee and Volusia Springs had the highest proportions of archaeal sequences, accounting for 4.7% and 1.6% of the total 16S rRNA gene sequences from each spring, respectively. Thaumarchaeota was the dominant archaeal phylum in both Manatee and Volusia Springs, representing 3.6% and 1.6% of the 16S rRNA gene sequences, respectively (Fig. 3A). In contrast, archaeal sequences comprised less than 0.5% of the 16S rRNA gene sequences recovered from the other three spring sites.
Viral community composition.
Assembled contigs from our viral metagenomes were curated by size (removal of contigs <1 kb) and through a series of BLAST analyses to increase our confidence of their viral origin (see Materials and Methods). Viral community structure was assessed by comparing the coverage of the raw reads from each spring to the curated viral contigs. Of 55,254 contigs, only 53 were identified in all five spring sites (based on the presence of reads covering 75% of the contig) (Fig. 4A). The majority of the shared contigs represented double-stranded DNA (dsDNA) tailed phage (n = 40) belonging to the order Caudovirales. The remainder of the contigs found at all five spring sites were most similar to those associated with single-stranded DNA (ssDNA) viruses, specifically, eukaryotic circular Rep-encoding single-stranded DNA (CRESS DNA) viruses (n = 11), and phage belonging to the family Microviridae (n = 2) (27). The viral communities were compared between springs using an NMDS plot with Bray-Curtis dissimilarity based on contig coverage (i.e., the normalized number of raw reads mapping back to the curated viral contigs). A PERMANOVA confirmed that there were significant differences (P = 0.002) among the viral assemblages from the different spring sites based on the curated contigs, though there is overlap of the 95% confidence intervals of Ichetucknee, Jackson and Rainbow Springs (Fig. 4B). It should be noted that Ichetucknee Springs was represented by only two replicates, as one sample was lost during processing.
FIG 4.
(A) Venn diagram depicting the distribution of curated viral contigs among springs. (B) NMDS plot showing the similarity of viral community structures of each spring site based on a Bray-Curtis dissimilarity matrix of the relative abundances of viral contigs measured by read coverage normalized by contig length and library size. Biological replicates of each spring are represented by points of identical shapes and colors. The ordinal ellipses represent the 95% confidence interval of the standard error. The arrows represent physiochemical parameters significantly (P < 0.05) correlating with the ordination of the communities. The direction of each arrow indicates direction, and length denotes magnitude of influence.
The vast majority (>90%) of the contigs were more similar to sequences found in the Integrated Microbial Genomes/Virus (IMG/VR) database (containing ∼700,000 metagenomic viral contigs) than to sequences in the NCBI RefSeq viral database (containing ∼10,000 curated representative complete viral genomes). In addition, BLASTx comparison of the curated viral contigs against the IMG/VR database revealed that roughly half of the contigs (42% to 62%) from each spring were similar to sequences previously found in aquatic environments (Fig. 5). Between 15% and 30% of the sequences of the contigs from each spring site were similar to sequences originating from freshwater environments (Fig. 5).
FIG 5.

Stacked bar plot showing the ecosystem types represented by the sequences in the IMG/VR database that most closely matched the curated viral contig sequences from the springs. Shades of blue represent aquatic ecosystems.
Since environmental virome sequences in the IMG/VR database are uncharacterized, the nucleic acid type (ssDNA versus dsDNA) and host type (prokaryotic or eukaryotic) could be assigned only for curated viral contigs that had significant (E value ≤ 10−10) BLASTx similarities to contigs in the NCBI RefSeq viral database. Between 28% and 73% of the contigs from each site had significant matches (E value ≤ 10−10) to those in the NCBI Refseq viral database. With the exception of Ichetucknee Springs, all of the springs were dominated by dsDNA viruses (>82% of the sequences that mapped to viral contigs), with prokaryotic dsDNA viruses belonging to the order Caudovirales comprising >57% of the sequences that mapped to viral contigs (Fig. 6). In contrast, Ichetucknee Springs was dominated by CRESS DNA viruses (27).
FIG 6.

Stacked bar plot of the relative abundances of reads most similar to ssDNA (green) versus dsDNA (blue) eukaryotic viruses (light colors) and prokaryotic viruses (dark colors) in each spring based on best BLASTx hit to the NCBI RefSeq virus database. Relative abundance was estimated through read coverage normalized by contig length and library size. Unassigned viruses were manually curated, and any contigs that could not be assigned to one of the four categories were excluded from analysis. Excluded contigs comprised <1% of the total contigs analyzed for each spring.
A total of 58 complete circular viral genomes and 3 satellites (circular sequences encoding a Rep similar to those encoded by ssDNA viral genomes but with no discernible capsid protein) were identified and annotated using a combination of the Cenote-taker2 Pipeline and manual screening of the resulting circular contigs (28). The viral genomes ranged in size from a 1.6-kb eukaryotic CRESS DNA virus found in Manatee Springs to a 90-kb prokaryotic dsDNA myovirus recovered from Jackson Springs (see Table S1 in the supplemental material). The pipeline identified 20 members of the dsDNA phage order Caudovirales. However, the majority of the complete genomes belonged to ssDNA viruses (n = 38). Fifteen complete genomes of ssDNA phage were identified, including 14 genomes representing members of the family Microviridae and one genome from the Inoviridae (Table S1). The remainder of the complete ssDNA genomes represented CRESS DNA viruses. No single complete viral genome was shared among all five springs (Fig. 7). Despite the fact that the springs’ viral communities were largely unique, two genomes (a podovirus and CRESS DNA virus) and one satellite were shared between four of the five spring sites (Fig. 8). Interestingly, the CRESS DNA virus (CRESS DNA virus sp. Ctin 15) is most similar to members of the Bacilladnaviridae family and shares key features with members of this family, including four major open reading frames (ORFs) arranged on both virion and complementary genome strands (29). Based on BLASTx matches to the Rep, CRESS DNA virus sp. Ctin 15 shares 54.64% amino acid identity with Bacilladnavirus sp. isolate ctcc592 (GenBank accession number MH617605.1).
FIG 7.
Heat maps showing the relative abundance of each complete circular viral genome (calculated as read coverage normalized by genome length and library size). Each row represents one viral genome with 20 dsDNA circular virus genomes on the bottom half of the map and 38 ssDNA circular genomes (including the 3 satellites) on the top. White represents absence of the genome.
FIG 8.
Genome maps of the two viruses (A and B) and one satellite (C) found in four of five spring sites. The graph in the center of each map represents normalized read coverage from each spring site. nts, nucleotides.
List of complete viral genome names, Genbank accession numbers, taxonomic assignments, and genome lengths. Download Table S1, DOCX file, 0.02 MB (19.1KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
DISCUSSION
Prokaryotic cell and viral abundances relative to spring physiochemical characteristics.
Averages of 9.6 × 103 prokaryotic cells and 3.4 × 104 VLPs per milliliter of water were present in the samples from the Ichetucknee, Jackson, Manatee, and Rainbow Springs. However, the abundance of both groups was approximately an order of magnitude higher in the samples from Volusia Springs. Prior studies in groundwater systems have revealed cellular abundances ranging from 103 to 105 ml−1 and viral abundances of 104 to 107 ml−1 (21, 30–34). Both the bacterial and viral counts from the springs fall in the lower range of reported abundance values from other groundwater systems. The average VPR of the springs sites (value = 5) was consistent with the mean VPR of other groundwater studies (5.9) and lower than the mean VPR of the more commonly studied marine pelagic and freshwater pelagic environments (26.5 and 17.2, respectively) (35). Due to the oligotrophic nature of aquifers, the low VPR in the springs may indicate low viral production and/or the prevalence of the lysogenic infection cycle, though further research is needed to examine this (36–39).
Physiochemical parameters were examined for each spring to provide potential explanations for the variation in cell and viral abundances. Notably, Volusia Springs had a much higher phosphate concentration (more than double) than any other spring, which possibly contributed to the higher prokaryotic and VLP counts in this spring site (Tables 1 and 2). Nitrate concentrations varied by spring, with Volusia Springs having the lowest and Jackson Springs having the highest (23.1 μM and 230.9 μM, respectively). To put this into context, groundwater nitrate concentrations are usually around 30 μM and shallow groundwater with agricultural influence has been reported to have concentrations as high as 161 μM in the United States (40). Previous measurements of nitrate concentrations in Florida’s springs specifically have ranged between 0.81 and 67.74 μM (8). Therefore, Jackson Springs is affected by unusually high nitrate loading. Variations in nutrient concentrations might be attributable to differences in land usage around the springs. Jackson Springs, with the highest concentrations of nitrate, nitrite, and ammonium, has a springshed largely comprised of forests and agricultural land, while Volusia Springs, with the highest phosphate concentration, has the most urbanized springshed (Fig. 1). However, note that although Jackson and Manatee Springs had seemingly similar land usage profiles (Fig. 1), the physicochemical parameters (Table 1) and prokaryotic communities (Fig. 2B) were distinct between these two sites. Delving into the subcategories within the broader agricultural use category, we found that row crop growth contributed to over 60% of the agricultural land usage around Jackson and only 37% of that around Manatee. The growth of row crops requires more fertilizer use than other agricultural activities (e.g., maintenance of livestock and tree farms), which may explain the differences in nutrient levels between the two springs.
Prokaryotic community composition.
Differences in nutrient concentrations could contribute to or result from differences in the prokaryotic communities of each spring. The low number of shared ASVs and the results of the ASV NMDS analysis suggest that each spring harbors a unique prokaryotic community (Fig. 2). The nitrite and ammonium levels of Jackson and Volusia Springs were elevated in comparison to those of the other springs, seemingly driving the divergence of their prokaryotic communities from the communities seen at other three sites (Fig. 2B). However, of the physiochemical parameters mapping to the NMDS, phosphate was the only one elevated in only Volusia Springs, and the direction of the arrow on the plot (Fig. 2B) suggests that it is driving the divergence of Volusia’s prokaryotic community. On the other hand, the nitrate concentration, which was elevated in Jackson Springs, did not significantly influence the clustering of prokaryotic communities represented on the NMDS plot.
The relative abundances of bacterial and archaeal 16S rRNA gene sequences in the springs were similar to the abundances found in other aquifer studies, and some similar taxa have also been reported previously (41–44). Proteobacteria and Cyanobacteria composed a large portion of the 16S rRNA gene sequences from all of the Florida springs investigated here. While the phylum Proteobacteria encompasses a wide variety of bacteria capable of successfully inhabiting an extremely oligotrophic and sunless environment, such as the aquifer, Cyanobacteria are most commonly associated with a photosynthetic lifestyle. Although other studies have previously reported the presence of nonphotosynthesizing cyanobacteria in groundwater (45), it is possible that the sequences detected here were derived from cyanobacterial mats located in and around the springs’ vents (46). However, this was an unexpected finding given that we observed mats near the vents of only two of the springs (Ichetucknee and Manatee) during sample collection.
The most abundant phyla were present in all of the springs, though there was variation regarding the relative abundances at each site. Along with the variations seen at finer taxonomic resolution, this suggests that the nutrient disparities between the springs are selecting for different taxa at each site, though parameters unaccounted for in this study, such as micronutrients, may also be playing a role. The few bacterial families shared among the springs does suggest that there might be a core microbiome being delivered through the aquifer water and that various selective pressures (i.e., nutrient inputs) shape the prokaryotic communities before they even reach the surface. Once prokaryotes from the aquifer reach the springs, it is likely that the communities continue to change as they enter the oxygen- and sunlight-rich springs, resulting in prokaryotic communities that are distinct at each site with respect to both their relative abundances and their taxonomic compositions.
Viral community composition.
Here, we investigated the DNA viral community as a whole, including the relative abundances of dsDNA and ssDNA viruses, by using a next-generation sequencing library construction kit that directly incorporates both types of genome templates into the workflow (47). However, various steps used in the viral purification process may impose biases on the types of viruses recovered. For example, samples were filtered through a 0.2-μm-pore-size filter, which may have selectively removed large viruses, and the treatment of viral samples with chloroform may have excluded the identification of lipid-containing viruses in this study (48, 49). Additionally, during our processing of the viral contigs, CD-HIT-EST was used to remove redundant contigs. This program does not account for different start points in assemblies, and the contigs used in the analyses may therefore have included redundant sequences. Despite these potential limitations, the approach applied here revealed that the Florida springs investigated in this study harbor viral communities that range in composition from being almost exclusively dsDNA (Rainbow and Volusia Springs) to >54% of raw reads mapping to ssDNA viral contigs (Fig. 6). Ichetucknee Springs was particularly interesting as it was the only spring that had a greater abundance of ssDNA viruses than dsDNA viruses. This was unexpected given that the only other study investigating aquatic viral communities using the same approach concluded that ssDNA viruses represented only a small portion (<5%) of DNA virus communities in the marine and freshwater environments examined (46). However, the same study also suggested that additional ssDNA viral sequences could be overlooked due to database limitations and, after reanalyzing data using contig sequences that likely represented ssDNA viruses, the relative abundance of ssDNA viruses increased to more than 50% of the total viral community in samples from two freshwater lakes (Lake Michigan and Lake Superior) (47). Considering that only four complete ssDNA viral genomes were identified in Ichetucknee Springs, our study supports the idea that in some aquatic environments, a few ssDNA viruses can be among the most abundant viruses contributing to the community composition (47). These findings are also consistent with a previous study that demonstrated elevated levels of ssDNA viruses in aquifer systems (22). However, the 2013 study by Smith et al. used multiple-displacement amplification, which has a known bias toward small circular genomes such as those of ssDNA viruses, making it difficult to derive quantitative information from their sequence data (22, 50). Ichetucknee Springs is one of few environments reported where certain ssDNA viruses are more abundant than dsDNA viruses (47). Now that library construction methods allow quantitative recovery of ssDNA and dsDNA viruses, we anticipate that future work will identify other “hot spots” of ssDNA viruses, leading to prediction of their hosts and elucidation of the factors that contribute to their success.
The viral contigs from the springs were most closely related to sequences from environmental viromes found in the IMG/VR database, and the majority of these sequences were similar to those previously identified in aquatic environments. This finding highlights the proportion of viral sequences (27% to 72%) from each spring that would be overlooked if researchers compared the sequences only against the NCBI virus databases and emphasizes the value of custom databases composed of environmental virus sequences. However, since the environmental virome sequences in the IMG/VR database represent uncharacterized sequences, taxonomic associations could be attributed based only on similarities to the NCBI RefSeq virus database.
Almost half of the complete circular genomes were ssDNA viruses, mainly CRESS DNA viruses. Though it has been suggested previously that their small capsid size (15 to 38 nm in diameter) contributes to their elevated presence in aquifers (22), the abundance of these complete ssDNA genomes is also influenced by the fact that their small genome size increases the likelihood of complete genome assembly. Tailed phage with dsDNA genomes belonging to the order Caudovirales are ubiquitous in all aquatic environments examined to date, and their presence in our study is consistent with findings of other aquatic virome studies. Genomes of ssDNA phage are of interest since less is known about the geographical distribution and ecology of these phage (51). Among the ssDNA viruses detected in publicly available viromes from a variety of ecosystems, sequences similar to those of members of the Microviridae have been the most frequently reported, though a recent study showed that lysogenic Inoviridae sequences are also prevalent (52–54). While there is growing acknowledgment that ssDNA viruses may play an important role in aquatic environments, we still know little about their diversity and host range. Ichetucknee Springs, which was dominated by ssDNA viruses, may be an ideal ecosystem for further investigation (51).
As seen with the prokaryotic data, it is possible that some of the identified viruses originated from areas surrounding the spring vent, rather than directly from the aquifer. Notably, a CRESS DNA virus found in four of the five springs sites was found to be most similar to members of the family Bacilladnaviridae. Bacilladnaviruses are believed to infect diatoms and have been recovered from marine and estuarine environments (29). To our knowledge, the CRESS Ctin 15 virus species identified here represents the first bacilladnavirus identified from freshwater. Although we can only speculate with the available data, it is possible that this virus originated from benthic diatoms attached to substrate near the spring vents. Nevertheless, the detection of this novel bacilladnavirus expands the known diversity and environmental range for these CRESS DNA viruses.
Several levels of resolution were used to demonstrate distinct viral community structures among the springs. Each spring had a different composition of viral types, and the NMDS analysis of read coverage of the curated viral contigs showed statistically different viral communities in Manatee and Volusia Springs. Analysis of read coverage of the complete circular viral genomes demonstrated distinct viral community structures in all five of the springs. Additionally, only 53 contigs (partial genomes) were shared among all five springs and not a single complete genome had adequate read coverage from all five springs. The variation that we see in the viral communities may reflect the variations in their host availability, as revealed by the distinct prokaryotic communities identified in the same samples. This suggests that changes in land usage and nutrient inputs might be impacting even the smallest members of these essential ecosystems.
Concluding remarks.
The health of the Floridan aquifer and its associated springs has declined in the past few decades due to anthropogenic impacts. This study demonstrated that despite the magnitude of water coming from the FAS, each spring site has a unique prokaryotic and viral community. The differences can be attributed in part to the differing nutrient inputs, originating from differences in land usage in each springshed. This report serves to develop a baseline of the microbiology and virology of Florida’s springs against which future changes, resulting from natural and/or anthropogenic factors, can be compared.
MATERIALS AND METHODS
Sample collection.
Samples were collected from five first-magnitude springs (Ichetucknee Springs [29.98°N 82.76°W], Jackson Springs [30.79°N 85.14°W], Manatee Springs [29.50°N 82.98°W], Rainbow Springs [29.08°N 82.42°W], and Volusia Springs [28.95°N 81.31°W]) within the FAS (Fig. 1). Sampling sites were selected based on each spring’s inferred recharge zone, where each site should be independent of other spring sites sampled, and to represent a range of land use profiles (Fig. 1). Land usage data and springshed areas were obtained from the Florida Department of Environmental Protection (DEP) (https://geodata.dep.state.fl.us/datasets/statewide-land-use-land-cover/; https://geodata.dep.state.fl.us/datasets/1eea00d9f3794a12bc658e77de29574f_1).
At each site, triplicate 50-liter samples were collected for microbial analyses at locations as close to the spring vent outflow as possible using a 5-liter horizontal polyvinyl chloride (PVC) water sampler (Forestry Suppliers) and an inflatable raft in accordance with research permit 08301620 from the Florida DEP. Conductivity, temperature, pH, turbidity, and dissolved oxygen levels were measured alongside collection of water samples at each site using a YSI Pro DSS handheld multiparameter water quality meter. For chemical analyses, samples were filtered through a 0.2-μm-pore-size Anotop filter (Millipore) and the filtrates were collected into acid-washed Falcon tubes. The samples were stored at –20°C until analysis was performed with a Lachat QuickChem 8500 Series 2 system for determination of nitrate, nitrite, ammonium, and phosphate concentrations. Additionally, triplicate 100-ml water samples were collected for quantifying prokaryotic and viral abundance. All field sampling occurred between May and June 2017, and samples were taken to the laboratory within 8 h of collection for processing.
Prokaryotic and viral abundances.
Bacterial and viral abundances were determined via SYBR gold staining and epifluorescence microscopy (55). For this purpose, triplicate samples from each spring were fixed with paraformaldehyde (2% final concentration) and 100-ml volumes were filtered onto 0.02-μm-pore-size Anodisc filters (Whatman). Filters were stained using 100 μl of 100× Invitrogen SYBR gold solution (Thermo Fisher) for 12 min and set on 100 μl of MilliQ water (filtered with a 0.02-μm-pore-size filter) to remove excess stain. After blotting on a Kimwipe, filters were mounted on slides with an antifade solution of 50:50 phosphate-buffered saline (PBS)/glycerin and 10% p-phenylenediamine. Bacteria and viruses were then enumerated (10 fields per replicate) using a Zeiss Axio Scope.A1 epifluorescence microscope and Media Cybernetics Image-Pro software (56).
Processing of water samples for sequencing.
Prokaryotic cells and virus particles were concentrated from the 50-liter water samples using a 30-kDa tangential flow filter (TFF) (GE Healthcare), resulting in a final volume of 150 to 200 ml (57, 58). The concentrates were then filtered through a 0.22-μm-pore-size Sterivex filter (Millipore) to capture prokaryotic cells, and these filters were stored at –80°C until DNA extraction. Viral particles were further concentrated from filtrate obtained from the 0.22-μm-pore-size filter by incubation overnight at 4°C with 10% (wt/vol) polyethylene glycol (PEG) 8000. The PEG samples were then centrifuged at 11,000 × g for 45 min at 4°C to pellet the viral particles, which were subsequently resuspended in 1-ml volumes of water (filtered with a 0.02-μm-pore-size filter) from their respective sampling sites. The resuspended viral particles were further purified by treating the samples with 20% chloroform to lyse any remaining cells or vesicles (59) and were then incubated for 30 min at 37°C with 10 U DNase I (Thermo Fisher) per milliliter of sample to degrade free DNA. Purified virus particles were stored at 4°C until further processing was performed.
16S rRNA gene amplicon sequencing and bioinformatics.
DNA was extracted from 0.22-μm-pore-size Sterivex filters by opening the cartridge using crab leg crackers and removing the membrane filter with tweezers and sterile razor blades. DNA extractions were performed using a PowerSoil DNA isolation kit (MoBio) following the manufacturer’s protocol. The extracted DNA was submitted to the Michigan State University Research Technology Support Genomics Core Facility for amplicon library preparation and Illumina sequencing of the V4 hypervariable region of the 16S rRNA gene using primers 16S-V4 515f (60) and 806r (61). These primers amplify both Bacteria and Archaea, though recent studies have shown that universal prokaryotic primers do miss some Archaea and members of Candidate Phyla Radiation (62, 63). The 16S-V4 PCR products from each sample were batch normalized using SequalPrep DNA normalization plates (Invitrogen), and samples in a dually indexed pool were paired-end sequenced (2 by 250 bp) on an Illumina MiSeq Nano v2 system (see Fig. S1 in the supplemental material for rarefaction curves). Sequences are available at the NCBI Sequence Read Archive database (study number PRJNA541276, accession numbers SAMN11581363 to SAMN11581377).
Rarefaction curve for the 16S rRNA gene sequences. The first letter denotes the spring site (I, Ichetucknee; J, Jackson; M, Manatee; R, Rainbow; V, Volusia). The second letter (a to c) represents the replicate. Replicates of each spring are the same color. Download FIG S1, PDF file, 0.7 MB (684.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Approximately 700,000 paired-end reads from 16S rRNA gene amplicons were processed using bioinformatic applications available in CyVerse cyberinfrastructure and R with RStudio version 1.0.153 (64–66). Initially, each replicate sample was processed individually, although triplicates from each spring were subsequently combined for taxonomic assignments. Primers and adapters were removed from sequences, and reads were filtered based on quality scores using Trimmomatic v 0.36.0 (67). FastQC v 0.11.5 was used to verify that sequences were trimmed successfully (68). Trimmed sequences were then processed using the Divisive Amplicon Denoising Algorithm (DADA2) package v 1.6.0 in RStudio (69), which uses a modeling approach to correct Illumina sequencing amplicon errors. After controlling for sequencing errors was performed, the DADA2 package merged paired-end reads and removed chimeric sequences, outputting amplicon sequence variants (ASVs). The ASVs were then compared against the SILVA v 132 rRNA database for taxonomic assignments (default bootstrapping value of 50) (70), and the data were normalized based on library size using the DESeq2 v 1.18.1 package in RStudio (71). Non-prokaryotic and plastid signatures identified by the SILVA database were manually removed from the data set before analysis.
Statistical analyses were carried out in RStudio to compare the compositions of the prokaryotic communities from all spring sites. The metaMDS and envfit functions in vegan v 2.3-1 were used to create a nonmetric, multidimensional scaling (NMDS) plot based on a Bray-Curtis dissimilarity matrix comparing the community structures of all springs and to map the significantly correlated environmental parameters, respectively (72). The adonis function was then used to calculate a Bray-Curtis distance matrix and apply permutational multivariate analysis of variance (PERMANOVA) to determine whether the biological replicates clustered significantly according to the spring of origin. The replicates were then merged using the merge.samples function within the phyloseq v 1.22.3 package, the most abundant 25 phyla across all five sites were ranked, and a heat map was constructed using superheat package v 1.0.0 (73, 74).
Virome sequencing and bioinformatics.
Viral DNA was extracted from 200 μl of concentrated and purified virus particles using a QIAamp MinElute virus spin kit (Qiagen). The extracted viral DNA was fragmented to 300 bp at the H. Lee Moffitt Cancer Center & Research Institute Molecular Genomics Core Facility using a Covaris M220 instrument. Fragmented DNA samples were processed with an Accel-NGS 1S Plus DNA library kit for Illumina platforms (Swift Biosciences) following the manufacturer’s protocol and using 20 cycles of indexing PCR. This kit was chosen based on its ability to recover both single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA) viruses without the need for preamplification, enabling comparison of their relative abundances in environmental samples (47). The dually indexed sequencing libraries were sent to the University of Colorado BioFrontiers Next-Gen Sequencing Core Facility for pooling and paired-end sequencing using a high output V2 kit (300 cycles, 2 by 150 bp) on a NextSeq 500 platform. Raw virome sequences are available at the NCBI Sequence Read Archive database (study number PRJNA540674; accession numbers SAMN11552880 to SAMN11552894).
Virome paired-end reads were processed using bioinformatic applications available through the CyVerse cyberinfrastructure (64). The adapters were trimmed from sequences, and low-quality reads were removed using Trimmomatic v 0.36.0 (67). FastQC v 0.11.5 was used to verify successful trimming of sequences (68). Sequences from each replicate were assembled separately using metaSPAdes v 3.11.1 with default parameters (75) through the University of South Florida high-performance computing cluster. Assembled contigs were then filtered by size on the Galaxy Web-based platform (76), and contigs from each replicate were merged according to spring site. The contigs sharing >95% identity in each merged file were clustered using CD-HIT-EST (77–79). Contigs were then compared (BLASTx v 2.7.1; E value of <10−10) against a custom viral database (80). The custom viral database was built using the Create BLAST database application on CyVerse (64, 81) with sequences from the NCBI RefSeq viral database (release 87, March 2018) and IMG/VR metagenomic viral contigs (mVCs) (released January 2018). Note that IMG/VR includes predicted viral sequences from metagenomic studies that are not found in RefSeq (77). BLAST results were viewed using MEGAN v 6.13.1 (82), and only contigs with significant matches to sequences in the custom viral database were retained. These contigs were then subjected to a secondary curation procedure to identify and remove sequences with significant hits to cellular organisms via megaBLASTn searches (E value of <10−7) against the GenBank nt database. Before elimination of contigs with matches to cellular organisms, these sequences were screened through the VirSorter v 1.0.3 application on CyVerse, which is designed to predict integrated viral sequences from cellular genomes (83). Contig sequences that were matched to cellular organisms based on megaBLASTn results but that were predicted to be viral by VirSorter (categories 1 to 6) were added back to the list of curated viral contigs obtained from each of the spring sites (Fig. S2). Curated viral contigs were compared (BLASTx v 2.7.1; E value of <10−10) against the NCBI RefSeq viral database and the IMG/VR database to identify the viruses and ecosystem types, respectively, to which the spring sequences were most similar. For taxonomic assignment, BLASTx results from the NCBI RefSeq viral database were viewed in MEGAN v 6.13.1 (82) and the contigs for which an assignment was possible were manually divided into four categories (ssDNA eukaryotic viruses, ssDNA prokaryotic viruses, dsDNA eukaryotic viruses, and dsDNA prokaryotic viruses). Curated viral contigs are available at https://figshare.com/articles/Supplemental_Files_for_Malki_et_al_mBio/11900055.
Flow chart depicting the processing of viral contigs. The percentages of initial (≥200-bp) contigs remaining after each step are listed. (Step 1) The (≥200-bp) contigs are filtered by size (≥1 kb) and put through CD-HIT-EST for (95%) dereplication. (Step 2) Contigs are compared by BLASTx against the RefSeq/IMG/VR database. (Step 3) After BLASTn, the contigs are compared to entries in the nr database. (Step 4a) Contigs with no cellular hits in the nr database are moved to a final contigs file. (Step 4b) Contigs with cellular hits to the nr database are put through VirSorter. (Step 5) Contigs classified as viral by VirSorter (categories 1 to 6) are moved into the final contigs file. Curated viral contigs are available at https://figshare.com/articles/Supplemental_Files_for_Malki_et_al_mBio/11900055. Download FIG S2, PDF file, 0.5 MB (509.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Viral communities from each spring were compared based on the abundance of trimmed forward reads mapping to each of the curated viral contigs using the Bowtiebatch v 1.0.1 and Read2RefMapper v 1.0.1 applications as suggested in the iVirus pipeline (84). Reads were mapped with a minimum of 90% identity, and contigs were considered present only if reads mapped to at least 75% of the contig length. The number of reads mapping to a given contig was normalized by contig length and library size. Normalized values were used to create an NMDS plot based on a Bray-Curtis dissimilarity matrix for each spring site using the metaMDS function in vegan v 2.3-1 (72). The adonis function was then used to calculate the PERMANOVA data using the biological replicates from each spring. It should be noted that Ichetucknee Springs had only two replicates because one sample was lost during processing. To assess the abundance of each viral type in each spring, read mapping was also performed in the same manner for contigs that were divided into the four viral categories (ssDNA eukaryotic, ssDNA prokaryotic, dsDNA eukaryotic, and dsDNA prokaryotic contigs). Values were used to create a stacked bar plot in R using the ggplot2 package representing the percentage of raw reads within each spring assigned to each virus type, normalizing by contig length and dividing the normalized numbers of reads mapping to dsDNA viruses by 2 to account for the fact that dsDNA templates were present at twice the concentration of ssDNA templates (47).
Identification and annotation of complete circular viral genomes.
The Cenote-taker-2 Pipeline (28) (still in alpha testing) (https://github.com/mtisza1/Cenote-Taker2) was used to identify circular sequences within the curated viral contigs that likely represent complete viral genomes. Circular contigs were considered complete genomes if they fell within the size range of the viral families as defined by the International Committee on the Taxonomy of Viruses (ICTV) and were screened manually to ensure that they contained the hallmark genes of their respective families. Following the methods outlined above, read mapping was used to compare the distributions and abundances of these genomes in the springs. The complete genomes were separated by genome type, namely, ssDNA or dsDNA, and after averaging of the values from replicate samples from each spring was performed, a heat map was created using the superheat v 1.0.0 package (73, 74). The complete viral genomes have been submitted to GenBank under accession numbers MN582052 to MN582112.
Data availability.
Prokaryotic 16S rRNA gene sequences are available at the NCBI Sequence Read Archive database (study number PRJNA541276, accession numbers SAMN11581363 to SAMN11581377). Raw virome sequences are available at the NCBI Sequence Read Archive database (study number PRJNA540674, accession numbers SAMN11552880 to SAMN11552894). The complete viral genomes have been submitted to GenBank under accession numbers MN582052 to MN582112. Curated viral contigs are available at https://figshare.com/articles/Supplemental_Files_for_Malki_et_al_mBio/11900055. The complete list of complete viral genome names, Genbank accession numbers, taxonomic assignments, and genome lengths is available in Table S1.
ACKNOWLEDGMENTS
We are grateful to Katherine Bruder, Noemi Van Bogaert, Anni Djurhuus, Grace Koziol, William E. Landry, and Natalia López-Figueroa for assistance during sample collections; the Florida Department of Environmental Protection Division of Recreation and Parks Florida Park Service for their assistance in obtaining the proper sampling permits; Salvatore Caprara, William Abbott, and Kristen Buck for performing the nutrient analysis; and Anthony Green and Benjamin Bolduc for technical and bioinformatic support.
This project was funded by grant DEB-1555854 from the National Science Foundation to M.B. K.M. was supported by a National Science Foundation Graduate Research Fellowship (award no. 3900101301). A.J.S. was supported by grants from the Swedish Research Council Formas and a Marie Curie International Outgoing Fellowship within the 7th European Community Framework Program.
Footnotes
This article is a direct contribution from Mya Breitbart, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Steven Wilhelm, University of Tennessee at Knoxville, and Karin Holmfeldt, Linnaeus University.
Citation Malki K, Rosario K, Sawaya NA, Székely AJ, Tisza MJ, Breitbart M. 2020. Prokaryotic and viral community composition of freshwater springs in Florida, USA. mBio 11:e00436-20. https://doi.org/10.1128/mBio.00436-20.
REFERENCES
- 1.Berndt MP, Oaksford ET, Mahon GL, Schmidt W. 1998. Water resources atlas of Florida. Florida State University, Tallahassee, Florida, USA. [Google Scholar]
- 2.Scott TM, Means GH, Meegan RP, Means RC, Upchurch S, Copeland R, Jones J, Roberts T, Willet A. 2004. Springs of Florida. US Geological Survey, Tallahassee, FL. [Google Scholar]
- 3.Bonn M, Bell F. 2003. Economic impact of selected Florida springs on the surrounding local areas. Florida Department of Environmental Protection, Tallahassee, FL. [Google Scholar]
- 4.Borisova T, Hodges AW, Stevens TJ. 2014. Economic contributions and ecosystem services of springs in the Lower Suwannee and Santa Fe River basins of North-Central Florida. University of Florida, Food and Resource Economics Department, Gainesville, FL. [Google Scholar]
- 5.Miller JA. 1990. Ground water atlas of the United States Alabama, Florida, Georgia, and South Carolina (HA 730-G). US Geological Survey, Tallahassee, FL. [Google Scholar]
- 6.Copeland RE. 2003. Florida spring classification system and spring glossary. Florida Geological Survey, Tallahassee, Florida. [Google Scholar]
- 7.Marella RL, Berndt MP. 2005. Water withdrawals and trends from the Floridan aquifer system in the southeastern United States, 1950–2000. US Department of the Interior, US Geological Survey, Reston, Virginia. [Google Scholar]
- 8.Katz BG. 2004. Sources of nitrate contamination and age of water in large karstic springs of Florida. Env Geol 46:689–706. doi: 10.1007/s00254-004-1061-9. [DOI] [Google Scholar]
- 9.Heffernan JB, Liebowitz DM, Frazer TK, Evans JM, Cohen MJ. 2010. Algal blooms and the nitrogen-enrichment hypothesis in Florida springs: evidence, alternatives, and adaptive management. Ecol Appl 20:816–829. doi: 10.1890/08-1362.1. [DOI] [PubMed] [Google Scholar]
- 10.Albertin AR, Sickman JO, Pinowska A, Stevenson RJ. 2012. Identification of nitrogen sources and transformations within karst springs using isotope tracers of nitrogen. Biogeochemistry 108:219–232. doi: 10.1007/s10533-011-9592-0. [DOI] [Google Scholar]
- 11.Bacchus ST, Barile PJ. 2005. Discriminating sources and flowpaths of anthropogenic nitrogen discharges to Florida springs, streams and lakes. Environ Eng Geosci 11:347–369. doi: 10.2113/11.4.347. [DOI] [Google Scholar]
- 12.Ardón M, Morse JL, Colman BP, Bernhardt ES. 2013. Drought-induced saltwater incursion leads to increased wetland nitrogen export. Glob Chang Biol 19:2976–2985. doi: 10.1111/gcb.12287. [DOI] [PubMed] [Google Scholar]
- 13.White E, Kaplan D. 2017. Restore or retreat? Saltwater intrusion and water management in coastal wetlands. Ecosyst Health Sustainability 3:e01258. doi: 10.1002/ehs2.1258. [DOI] [Google Scholar]
- 14.Whitman WB, Coleman DC, Wiebe WJ. 1998. Prokaryotes: the unseen majority. Proc Natl Acad Sci U S A 95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kallmeyer J, Pockalny R, Adhikari RR, Smith DC, D'Hondt S. 2012. Global distribution of microbial abundance and biomass in subseafloor sediment. Proc Natl Acad Sci U S A 109:16213–16216. doi: 10.1073/pnas.1203849109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McMahon S, Parnell J. 2014. Weighing the deep continental biosphere. FEMS Microbiol Ecol 87:113–120. doi: 10.1111/1574-6941.12196. [DOI] [PubMed] [Google Scholar]
- 17.Griebler C, Lueders T. 2009. Microbial biodiversity in groundwater ecosystems. Freshwater Biol 54:649–677. doi: 10.1111/j.1365-2427.2008.02013.x. [DOI] [Google Scholar]
- 18.Goldscheider N, Hunkeler D, Rossi P. 2006. Microbial biocenoses in pristine aquifers and an assessment of investigative methods. Hydrogeol J 14:926–941. doi: 10.1007/s10040-005-0009-9. [DOI] [Google Scholar]
- 19.Kallies R, Hölzer M, Brizola Toscan R, Nunes da Rocha U, Anders J, Marz M, Chatzinotas A. 2019. Evaluation of sequencing library preparation protocols for viral metagenomic analysis from pristine aquifer groundwaters. Viruses 11:484. doi: 10.3390/v11060484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daly RA, Roux S, Borton MA, Morgan DM, Johnston MD, Booker AE, Hoyt DW, Meulia T, Wolfe RA, Hanson AJ, Mouser PJ, Moore JD, Wunch K, Sullivan MB, Wrighton KC, Wilkins MJ. 2019. Viruses control dominant bacteria colonizing the terrestrial deep biosphere after hydraulic fracturing. Nat Microbiol 4:352–361. doi: 10.1038/s41564-018-0312-6. [DOI] [PubMed] [Google Scholar]
- 21.Kyle JE, Eydal HS, Ferris FG, Pedersen K. 2008. Viruses in granitic groundwater from 69 to 450 m depth of the Äspö hard rock laboratory, Sweden. ISME J 2:571–574. doi: 10.1038/ismej.2008.18. [DOI] [PubMed] [Google Scholar]
- 22.Smith RJ, Jeffries TC, Roudnew B, Seymour JR, Fitch AJ, Simons KL, Speck PG, Newton K, Brown MH, Mitchell JG. 2013. Confined aquifers as viral reservoirs. Environ Microbiol Rep 5:725–730. doi: 10.1111/1758-2229.12072. [DOI] [PubMed] [Google Scholar]
- 23.Maeng SK, Ameda E, Sharma SK, Grützmacher G, Amy GL. 2010. Organic micropollutant removal from wastewater effluent-impacted drinking water sources during bank filtration and artificial recharge. Water Res 44:4003–4014. doi: 10.1016/j.watres.2010.03.035. [DOI] [PubMed] [Google Scholar]
- 24.Mermillod-Blondin F, Simon L, Maazouzi C, Foulquier A, Delolme C, Marmonier P. 2015. Dynamics of dissolved organic carbon (DOC) through stormwater basins designed for groundwater recharge in urban area: assessment of retention efficiency. Water Res 81:27–37. doi: 10.1016/j.watres.2015.05.031. [DOI] [PubMed] [Google Scholar]
- 25.Griffin DW, Stokes R, Rose J, Paul J. III, 2000. Bacterial indicator occurrence and the use of an F+ specific RNA coliphage assay to identify fecal sources in Homosassa Springs, Florida. Microb Ecol 39:56–64. doi: 10.1007/s002489900193. [DOI] [PubMed] [Google Scholar]
- 26.Katz BG, Griffin DW. 2008. Using chemical and microbiological indicators to track the impacts from the land application of treated municipal wastewater and other sources on groundwater quality in a karstic springs basin. Environ Geol 55:801–821. doi: 10.1007/s00254-007-1033-y. [DOI] [PubMed] [Google Scholar]
- 27.Zhao L, Rosario K, Breitbart M, Duffy S. Eukaryotic circular rep-encoding single-stranded DNA (CRESS DNA) viruses: ubiquitous viruses with small genomes and a diverse host range. Adv Virus Res 103:71. doi: 10.1016/bs.aivir.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 28.Tisza MJ, Pastrana DV, Welch NL, Stewart B, Peretti A, Starrett GJ, Pang Y-YS, Krishnamurthy SR, Pesavento PA, McDermott DH, Murphy PM, Whited JL, Miller B, Brenchley J, Rosshart SP, Rehermann B, Doorbar J, Ta’ala BA, Pletnikova O, Troncoso JC, Resnik SM, Bolduc B, Sullivan MB, Varsani A, Segall AM, Buck CB. 2020. Discovery of several thousand highly diverse circular DNA viruses. eLife 9:e51971. doi: 10.7554/eLife.51971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kazlauskas D, Dayaram A, Kraberger S, Goldstien S, Varsani A, Krupovic M. 2017. Evolutionary history of ssDNA bacilladnaviruses features horizontal acquisition of the capsid gene from ssRNA nodaviruses. Virology 504:114–121. doi: 10.1016/j.virol.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 30.Roudnew B, Seymour JR, Jeffries TC, Lavery TJ, Smith RJ, Mitchell JG. 2012. Bacterial and virus-like particle abundances in purged and unpurged groundwater depth profiles. Groundwater Monit R 32:72–77. doi: 10.1111/j.1745-6592.2011.01393.x. [DOI] [Google Scholar]
- 31.Pedersen K. 1993. The deep subterranean biosphere. Earth Sci Rev 34:243–260. doi: 10.1016/0012-8252(93)90058-F. [DOI] [Google Scholar]
- 32.Nilsen RK, Beeder J, Thorstenson T, Torsvik T. 1996. Distribution of thermophilic marine sulfate reducers in North Sea oil field waters and oil reservoirs. Appl Environ Microbiol 62:1793–1798. doi: 10.1128/AEM.62.5.1793-1798.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan D, Nolan J, Williams KH, Robbins MJ, Weber KA. 2017. Abundance and distribution of microbial cells and viruses in an alluvial aquifer. Front Microbiol 8:1199. doi: 10.3389/fmicb.2017.01199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roudnew B, Lavery TJ, Seymour JR, Smith RJ, Mitchell JG. 2013. Spatially varying complexity of bacterial and virus-like particle communities within an aquifer system. Aquat Microb Ecol 68:259–266. doi: 10.3354/ame01615. [DOI] [Google Scholar]
- 35.Parikka KJ, Le Romancer M, Wauters N, Jacquet S. 2017. Deciphering the virus-to-prokaryote ratio (VPR): insights into virus–host relationships in a variety of ecosystems. Biol Rev Camb Philos Soc 92:1081–1100. doi: 10.1111/brv.12271. [DOI] [PubMed] [Google Scholar]
- 36.Maurice CF, Bouvier T, Comte J, Guillemette F, Del Giorgio P. 2010. Seasonal variations of phage life strategies and bacterial physiological states in three northern temperate lakes. Environ Microbiol 12:628–641. doi: 10.1111/j.1462-2920.2009.02103.x. [DOI] [PubMed] [Google Scholar]
- 37.Evans C, Brussaard CP. 2012. Regional variation in lytic and lysogenic viral infection in the Southern Ocean and its contribution to biogeochemical cycling. Appl Environ Microbiol 78:6741–6748. doi: 10.1128/AEM.01388-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Payet JP, Suttle CA. 2013. To kill or not to kill: the balance between lytic and lysogenic viral infection is driven by trophic status. Limnol Oceanogr 58:465–474. doi: 10.4319/lo.2013.58.2.0465. [DOI] [Google Scholar]
- 39.Brum JR, Hurwitz BL, Schofield O, Ducklow HW, Sullivan MB. 2017. Seasonal time bombs: dominant temperate viruses affect Southern Ocean microbial dynamics. ISME J 11:588. doi: 10.1038/ismej.2016.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hallberg GR. 1989. Nitrate in ground water in the United States, p 35–74. In Developments in agricultural and managed forest ecology, vol 21 Elsevier, Philadelphia, PA. [Google Scholar]
- 41.Smith RJ, Jeffries TC, Roudnew B, Fitch AJ, Seymour JR, Delpin MW, Newton K, Brown MH, Mitchell JG. 2012. Metagenomic comparison of microbial communities inhabiting confined and unconfined aquifer ecosystems. Environ Microbiol 14:240–253. doi: 10.1111/j.1462-2920.2011.02614.x. [DOI] [PubMed] [Google Scholar]
- 42.Hershey OS, Kallmeyer J, Wallace A, Barton MD, Barton HA. 2018. High microbial diversity despite extremely low biomass in a deep karst aquifer. Front Microbiol 9:2823. doi: 10.3389/fmicb.2018.02823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bell E, Lamminmäki T, Alneberg J, Andersson AF, Qian C, Xiong W, Hettich RL, Balmer L, Frutschi M, Sommer G, Bernier-Latmani R. 2018. Biogeochemical cycling by a low-diversity microbial community in deep groundwater. Front Microbiol 9:2129. doi: 10.3389/fmicb.2018.02129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bruno A, Sandionigi A, Bernasconi M, Panio A, Labra M, Casiraghi M. 2018. Changes in the drinking water microbiome: effects of water treatments along the flow of two drinking water treatment plants in a urbanized area, Milan (Italy). Front Microbiol 9:2557. doi: 10.3389/fmicb.2018.02557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luef B, Frischkorn KR, Wrighton KC, Holman H-YN, Birarda G, Thomas BC, Singh A, Williams KH, Siegerist CE, Tringe SG, Downing KH, Comolli LR, Banfield JF. 2015. Diverse uncultivated ultra-small bacterial cells in groundwater. Nat Commun 6:6372. doi: 10.1038/ncomms7372. [DOI] [PubMed] [Google Scholar]
- 46.Stevenson RJ, Pinowska A, Albertin A, Sickman JO. 2007. Ecological condition of algae and nutrients in Florida springs: the synthesis report. Florida Department of Environmental Protection, Tallahassee, FL. [Google Scholar]
- 47.Roux S, Solonenko NE, Dang VT, Poulos BT, Schwenck SM, Goldsmith DB, Coleman ML, Breitbart M, Sullivan MB. 2016. Towards quantitative viromics for both double-stranded and single-stranded DNA viruses. PeerJ 4:e2777. doi: 10.7717/peerj.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laanto E, Mäntynen S, De Colibus L, Marjakangas J, Gillum A, Stuart DI, Ravantti JJ, Huiskonen JT, Sundberg L-R. 2017. Virus found in a boreal lake links ssDNA and dsDNA viruses. Proc Natl Acad Sci U S A 114:8378–8383. doi: 10.1073/pnas.1703834114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hingamp P, Grimsley N, Acinas SG, Clerissi C, Subirana L, Poulain J, Ferrera I, Sarmento H, Villar E, Lima-Mendez G, Faust K, Sunagawa S, Claverie J-M, Moreau H, Desdevises Y, Bork P, Raes J, de Vargas C, Karsenti E, Kandels-Lewis S, Jaillon O, Not F, Pesant S, Wincker P, Ogata H. 2013. Exploring nucleo-cytoplasmic large DNA viruses in Tara Oceans microbial metagenomes. ISME J 7:1678–1695. doi: 10.1038/ismej.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim K-H, Bae J-W. 2011. Amplification methods bias metagenomic libraries of uncultured single-stranded and double-stranded DNA viruses. Appl Environ Microbiol 77:7663–7668. doi: 10.1128/AEM.00289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Székely AJ, Breitbart M. 2016. Single-stranded DNA phages: from early molecular biology tools to recent revolutions in environmental microbiology. FEMS Microbiol Lett 363:fnw027. doi: 10.1093/femsle/fnw027. [DOI] [PubMed] [Google Scholar]
- 52.Creasy A, Rosario K, Leigh BA, Dishaw LJ, Breitbart M. 2018. Unprecedented diversity of ssDNA phages from the family Microviridae detected within the gut of a protochordate model organism (Ciona robusta). Viruses 10:404. doi: 10.3390/v10080404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roux S, Krupovic M, Daly RA, Borges AL, Nayfach S, Schulz F, Sharrar A, Carnevali PBM, Cheng J-F, Ivanova NN. 2019. Cryptic inoviruses revealed as pervasive in bacteria and archaea across Earth’s biomes. Nat Microbiol 4:1895–1906. doi: 10.1038/s41564-019-0510-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roux S, Krupovic M, Poulet A, Debroas D, Enault F. 2012. Evolution and diversity of the Microviridae viral family through a collection of 81 new complete genomes assembled from virome reads. PLoS One 7:e40418. doi: 10.1371/journal.pone.0040418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Noble RT. 2001. Enumeration of viruses. Methods Microbiol 30:43–51. doi: 10.1016/S0580-9517(01)30038-7. [DOI] [Google Scholar]
- 56.Suttle CA, Fuhrman JA. 2010. Enumeration of virus particles in aquatic or sediment samples by epifluorescence microscopy, p 145–153. In Wilhelm SW, Weinbauer MG, Suttle CA (ed), Manual of aquatic viral ecology. American Society of Limnology and Oceanography, Waco, TX. [Google Scholar]
- 57.Wommack KE, Sime-Ngando T, Winget DM, Jamindar S, Helton RR. 2010. Filtration-based methods for the collection of viral concentrates from large water samples, p 110–117. In Wilhelm SW, Weinbauer MG, Suttle CA (ed), Manual of aquatic viral ecology. American Society of Limnology and Oceanography, Waco, TX. [Google Scholar]
- 58.Colombet J, Robin A, Lavie L, Bettarel Y, Cauchie H, Sime-Ngando T. 2007. Virioplankton ‘pegylation’: use of PEG (polyethylene glycol) to concentrate and purify viruses in pelagic ecosystems. J Microbiol Methods 71:212–219. doi: 10.1016/j.mimet.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 59.Soler N, Krupovic M, Marguet E, Forterre P. 2015. Membrane vesicles in natural environments: a major challenge in viral ecology. ISME J 9:793–796. doi: 10.1038/ismej.2014.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parada AE, Needham DM, Fuhrman JA. 2016. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol 18:1403–1414. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- 61.Apprill A, McNally S, Parsons R, Weber L. 2015. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol 75:129–137. doi: 10.3354/ame01753. [DOI] [Google Scholar]
- 62.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goff SA, Vaughn M, McKay S, Lyons E, Stapleton AE, Gessler D, Matasci N, Wang L, Hanlon M, Lenards A, Muir A, Merchant N, Lowry S, Mock S, Helmke M, Kubach A, Narro M, Hopkins N, Micklos D, Hilgert U, Gonzales M, Jordan C, Skidmore E, Dooley R, Cazes J, McLay R, Lu Z, Pasternak S, Koesterke L, Piel WH, Grene R, Noutsos C, Gendler K, Feng X, Tang C, Lent M, Kim S-J, Kvilekval K, Manjunath BS, Tannen V, Stamatakis A, Sanderson M, Welch SM, Cranston KA, Soltis P, Soltis D, O'Meara B, Ane C, Brutnell T, Kleibenstein DJ, et al. 2011. The iPlant collaborative: cyberinfrastructure for plant biology. Front Plant Sci 2:34. doi: 10.3389/fpls.2011.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.RStudio Team. 2016. RStudio: integrated development environment for R (Version 1.0. 153). RStudio Inc, Boston, MA: https://www.rstudio.com/. [Google Scholar]
- 66.R Core Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/. [Google Scholar]
- 67.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Andrews S. 2016. FastQC: a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 69.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Quast C, Pruesse E, Gerken J, Peplies J, Yarza P, Yilmaz P, Schweer T, Glöckner FO. 2012. The SILVA ribosomal RNA gene database project: improved data processing and Web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’hara R, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H. 2019. Vegan: community ecology package. R package version 2.5-6 https://cran.r-project.org/web/packages/vegan/index.html.
- 73.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Barter RL, Yu B. 2018. Superheat: an R package for creating beautiful and extendable heatmaps for visualizing complex data. J Comput Graph Stat 27:910–922. doi: 10.1080/10618600.2018.1473780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res 27:824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, Miller W, Kent WJ, Nekrutenko A. 2005. Galaxy: a platform for interactive large-scale genome analysis. Genome Res 15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Paez-Espino D, Chen I-MA, Palaniappan K, Ratner A, Chu K, Szeto E, Pillay M, Huang J, Markowitz VM, Nielsen T, Huntemann M, K Reddy TB, Pavlopoulos GA, Sullivan MB, Campbell BJ, Chen F, McMahon K, Hallam SJ, Denef V, Cavicchioli R, Caffrey SM, Streit WR, Webster J, Handley KM, Salekdeh GH, Tsesmetzis N, Setubal JC, Pope PB, Liu W-T, Rivers AR, Ivanova NN, Kyrpides NC. 2017. IMG/VR: a database of cultured and uncultured DNA viruses and retroviruses. Nucleic Acids Res 45:D457–D465. doi: 10.1093/nar/gkw1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li W, Jaroszewski L, Godzik A. 2001. Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics 17:282–283. doi: 10.1093/bioinformatics/17.3.282. [DOI] [PubMed] [Google Scholar]
- 79.Godzik A, Li W. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 80.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 81.Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huson DH, Beier S, Flade I, Górska A, El-Hadidi M, Mitra S, Ruscheweyh H-J, Tappu R. 2016. MEGAN community edition-interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput Biol 12:e1004957. doi: 10.1371/journal.pcbi.1004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Roux S, Enault F, Hurwitz BL, Sullivan MB. 2015. VirSorter: mining viral signal from microbial genomic data. PeerJ 3:e985. doi: 10.7717/peerj.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bolduc B, Youens-Clark K, Roux S, Hurwitz BL, Sullivan MB. 2017. iVirus: facilitating new insights in viral ecology with software and community data sets imbedded in a cyberinfrastructure. ISME J 11:7–14. doi: 10.1038/ismej.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.ESRI. 2011. ArcGIS Desktop: Release 10. Environmental Systems Research Institute, Redlands, CA, USA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of complete viral genome names, Genbank accession numbers, taxonomic assignments, and genome lengths. Download Table S1, DOCX file, 0.02 MB (19.1KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Rarefaction curve for the 16S rRNA gene sequences. The first letter denotes the spring site (I, Ichetucknee; J, Jackson; M, Manatee; R, Rainbow; V, Volusia). The second letter (a to c) represents the replicate. Replicates of each spring are the same color. Download FIG S1, PDF file, 0.7 MB (684.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Flow chart depicting the processing of viral contigs. The percentages of initial (≥200-bp) contigs remaining after each step are listed. (Step 1) The (≥200-bp) contigs are filtered by size (≥1 kb) and put through CD-HIT-EST for (95%) dereplication. (Step 2) Contigs are compared by BLASTx against the RefSeq/IMG/VR database. (Step 3) After BLASTn, the contigs are compared to entries in the nr database. (Step 4a) Contigs with no cellular hits in the nr database are moved to a final contigs file. (Step 4b) Contigs with cellular hits to the nr database are put through VirSorter. (Step 5) Contigs classified as viral by VirSorter (categories 1 to 6) are moved into the final contigs file. Curated viral contigs are available at https://figshare.com/articles/Supplemental_Files_for_Malki_et_al_mBio/11900055. Download FIG S2, PDF file, 0.5 MB (509.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
Prokaryotic 16S rRNA gene sequences are available at the NCBI Sequence Read Archive database (study number PRJNA541276, accession numbers SAMN11581363 to SAMN11581377). Raw virome sequences are available at the NCBI Sequence Read Archive database (study number PRJNA540674, accession numbers SAMN11552880 to SAMN11552894). The complete viral genomes have been submitted to GenBank under accession numbers MN582052 to MN582112. Curated viral contigs are available at https://figshare.com/articles/Supplemental_Files_for_Malki_et_al_mBio/11900055. The complete list of complete viral genome names, Genbank accession numbers, taxonomic assignments, and genome lengths is available in Table S1.