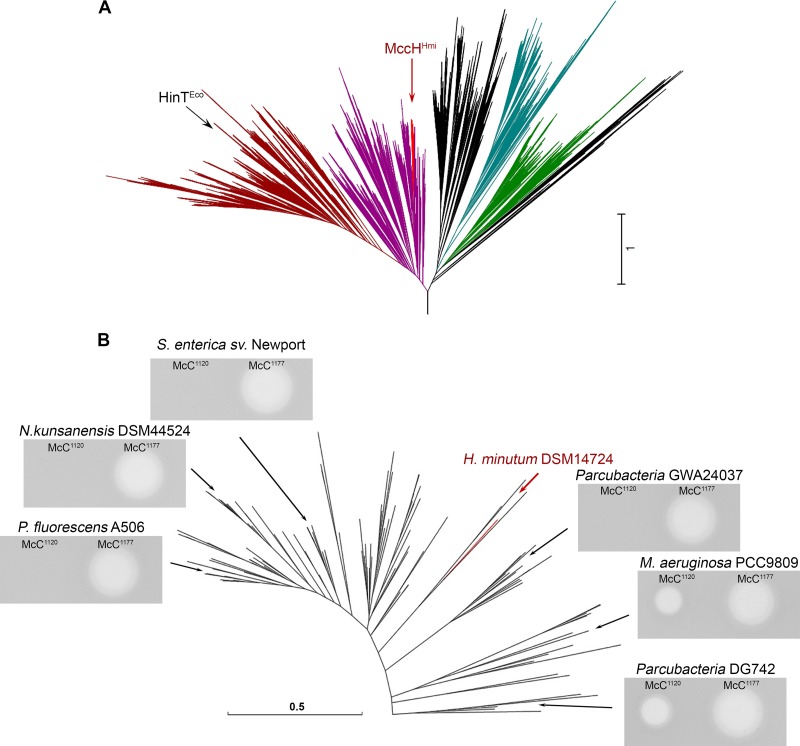
FIG 6.
Diversity of McC-specific phosphoramidases. (A) Approximate maximum likelihood (ML) phylogenetic tree of the HIT domain proteins from completely sequenced genomes. Dark magenta and dark red indicate HinT-like proteins (dark red shows the protein kinase C interacting protein-related subgroup), bright red indicates the MccHHmi clade that is expanded in panel B, cyan indicates GalT, and green indicates FHIT. In black are clades without any clear profile signature. The arrows point to HinTEco (within the PKCI clade) and MccHHmi. (B) Approximate ML phylogenetic tree of proteins within the MccHHmi clade and growth inhibition zones formed by McC1120 and McC1177 on lawns of E. coli B cells transformed with plasmids expressing MccHHmi or homologs from the indicated positions on the tree. The MccHHmi branch is highlighted in red. sv., serovar.