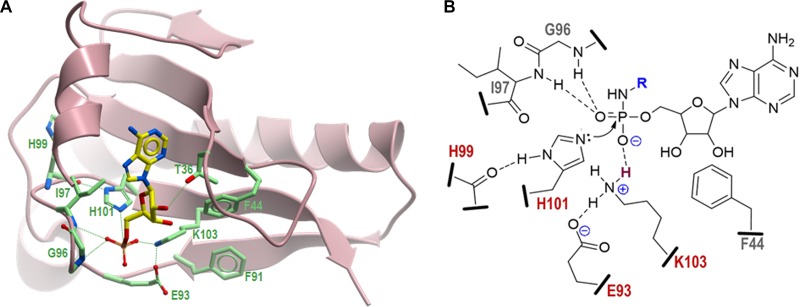
FIG 8.
Three-dimensional structural model of MccHHmi and proposed catalytic mechanism. (A) Model structure of MccHHmi generated by the SWISS-MODEL homology modeling server (23) using as the template the crystal structures of the HIT-like protein from Mycobacterium paratuberculosis (PDB 3P0T) (24) and human HINT1-AMP complex (PDB 3TW2) (28). Positions of catalytic residues in the MccHHmi active site in complex with AMP are shown as atom type-colored sticks: N, blue; O, red; P, orange. The C atoms in AMP are in yellow, and the C atoms in the side chains are in light green. (B) Schematic diagram representing the active site of MccHHmi with the bound substrate, aspartamide-adenylate (the aspartyl moiety is represented by R and shown in blue). Residues of the ExxxxxHxHxKxx motif conserved among all members of the MccH clade that are directly involved in the catalysis are shown in red. Potential hydrogen bonds between peptide side chains, backbone atoms, and phosphate oxygens are indicated by dashed lines. The proton to be transferred from the ε-amino group of K103 to the unbridged oxygen of the phosphoramide is depicted in dark red. Nucleophilic attack of the unprotonated nitrogen of H101 on electrophilic phosphorus atom is shown by a curved arrow.