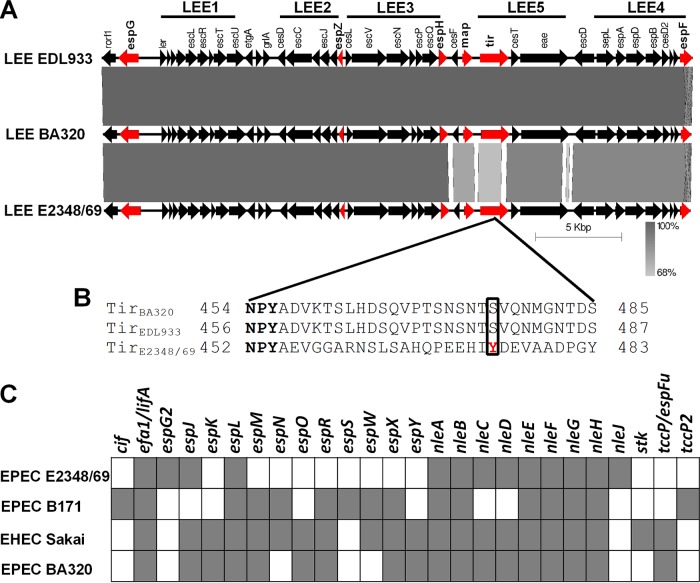
FIG 1.
Effector repertoire of EPEC BA320. (A) Homology comparisons among the LEE regions of EPEC BA320, EHEC EDL933, and EPEC E2348/69. Effector-encoding genes are labeled in red. The size and direction of the arrows indicates the size of each predicted gene and the direction of transcription. The scale of 5 kb is indicated at the bottom. (B) Sequence alignment of the C-terminal region of Tir. The NPY motif is conserved in all three strains (in bold). Phosphorylated tyrosine (Y474) in TirE2348/69 and equivalent residues in TirBA320 and TirEDL933 are boxed with a solid line. (C) In silico detection of the non-LEE-encoded effectors of EPEC E2348/69, EPEC B171, and EHEC Sakai in the EPEC BA320 genome using TBLASTN. Each square indicates the presence (in gray) or absence (in white) of an effector-encoding gene.