Abstract
Expanding the number of nucleotides in DNA increases the information density of functional DNA molecules, creating nanoassemblies that cannot be invaded by natural DNA/RNA in complex biological systems. Here, we show how 6-letter GACTZP DNA contributes this property in two parts of a nanoassembly: (1) in an aptamer evolved from a 6-letter DNA library to selectively bind liver cancer cells; and (2) in a 6-letter self-assembling GACTZP nanotrain that carries the drug doxorubicin. The aptamer-nanotrain assembly, charged with doxorubicin, selectively kills liver cancer cells in culture, as the selectivity of the aptamer binding directs doxorubicin into the aptamer-targeted cells. The assembly does not kill untransformed cells that the aptamer does not bind. This architecture, built with an expanded genetic alphabet, is reminiscent of antibodies conjugated to drugs, which presumably act by this mechanism as well, but with the antibody replaced by an aptamer.
Keywords: 6-Nucleotide DNA, nanotrain, aptamer, targeted drug delivery
Graphical Abstract:
Folded DNA molecules were evolved or designed from a 6-letter alphabet to perform two non-genetic functions: (a) bind selectively to liver cancer cells and (b) bind the toxic drug doxorubicin. Combined into an aptamer-nanotrain conjugate, these selectively kill liver cancer cells. This selective drug delivery is reminiscent of antibody-drug conjugates, but without proteins.
Introduction
One branch of DNA nanotechnology uses Watson-Crick pairing to assemble DNA and/or RNA (collectively xNA) molecules to give folded structures more complex than linear double helices. These folded xNA molecules often exhibit functional behavior beyond simple information storage, such as signaling dendrimers,[1] beacons,[2] and geometric shapes[3] that are imaged by atomic force microscopy.[4] Suggested applications of DNA as a programmable material include binding molecules (“aptamers”),[5–6] catalytic molecules (“aptazymes”),[7] and drug-carrying assemblies (such as nanotrains).[8–12] Aptamers, for example, have been seen as possible replacements for antibodies.[13]
However, the limited number (just four) of building blocks in standard xNA limits its functional versatility. The low information density, in turn, can causes ambiguous folding, even in natural xNA.[14] This ambiguity also limits the functional performance of designed DNA, including catalytic DNA evolved in a laboratory.[15]
Further, complex biological systems also contain many xNA molecules that are also built from G, A, C, and T/U). This means that in human blood and other medically relevant environments, the A:T and G:C pairing that gives programmed DNA molecules their functional folds can be invaded by background xNA, destroying both the programmed fold and ist function[16].
Recently, we showed that as many as 8 nucleotides from an Artificially Expanded Genetic Information System (AEGIS) can be added to the nucleic acid “alphabet” to give a “hachimoji” DNA and RNA (from Japanese, hachi = eight and moji = letter).[17] The additional 4 nucleotides form two pairs (the P:Z and B:S pairs) with the same size and shape as the natural A:T and G:C pairs. Subsets of hachimoji DNA have been built into signaling beacons that avoid invasion by natural DNA,[18] translation systems to encode additional amino acids,[19] diagnostics products,[20] and laboratory in vitro evolution (LIVE) platforms[21] that create aptamers with additional nucleotides that bind to cells[22] and proteins.[23]
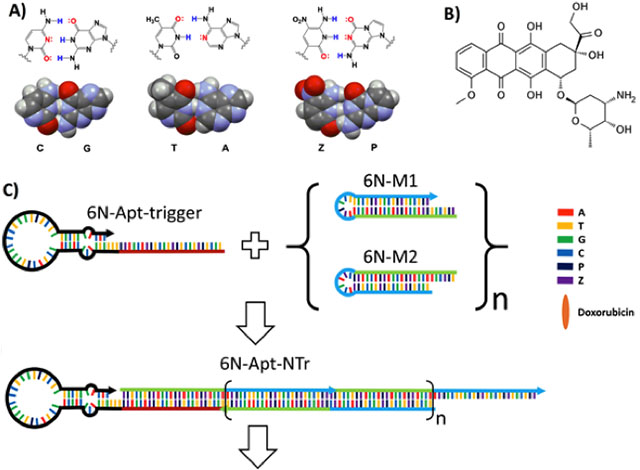
Here, we use components of one of the additional pairs (the P:Z pair, Fig. 1A) to serve two different functional roles in one nanoassembly. In one role, the P:Z pair is exploited to assemble a “nanotrain” that carries the drug doxorubicin (Dox, Fig. 1B). Nanotrains are long, linear duplexes built from two components that individually form stable hairpins with loops; single-stranded overhangs are designed as toeholds at their ends (Fig. 1C, 1D and Fig. S1). When a triggering DNA sequence is added that can hybridize to the toehold of one component, strand exchange opens the hairpin and forces its complementary strand to become single-stranded. The resulting single strand has a region that, in turn, hybridizes to the toehold of the second component and invades its hairpin. The nanoassembly that results is an alternating concatamer of both components, a series of “boxcars” of repeating elements (Fig 1C). In nanotrain assembly, P and Z increase the information density of the sequences, and also ensure that the base-paired regions cannot be invaded by any standard nucleic acids that might be present in any sample.
Figure 1.
Key components of AEGIS aptamer-nanotrain assembly. (A) Structures of three nucleobase pairs (G:C, A:T, and P:Z) that support nanoassembly in GACTZP 6-nucleotide DNA (PDB ID: 4RHD) (B) Doxorubicin. (C) Self-assembling nanostructure triggered by an aptamer (6N-Apt-trigger) that hybridizes to the toehold of 6N-M1, invades its hairpin to generate a single strand that can invade the hairpin of 6N-M2. This yields a 6-nucleotide DNA nanotrain (6N-Apt-NTrs) formed from short 6-nucleotide DNA fragments (6N-M1 and 6N-M2) tethered to a 6-nucleotide aptamer specific for liver cancer cells. The nanotrain may be loaded with the doxorubicin drug. (D) Sequences of 6-nucleotide DNA fragments. Underlined region highlighted with red color shows the sequence of the trigger; Black color underlined region shows the sequence of the aptamer; Green and blue color underlined regions in 6N-M1 and 6N-M2 show the complementary regions when forming nanotrains.
A second role for P and Z is performed in an aptamer that had been selected by 6-letter (GACTZP) laboratory in vitro evolution (LIVE) experiments to bind to liver cancer HepG2 cells.[24] Immortalized non-cancerous liver cells were used in the counter-selection to remove aptamers bound non-cancer related targets. The structure of one of several aptamers that emerged (the LZH5 aptamer) was optimized. Here, the modified version (LZH5B) was modeled to have a long binding loop with a stem containing a bulge (Fig. S2). A trigger sequence was then added to the 5’-end of the LZH5B stem to allow the aptamer itself to initiate formation of the nanotrain.
We hypothesized that doxorubicin, intercalated into the assembled nanotrain, could be guided to an unknown feature on the liver cancer cell (but not on the normal cell) by the tethered aptamer. The feature-aptamer-nanotrain-drug complex would then be internalized into a lysosome, where it would be digested to release doxorubicin. By this mechanism, we hoped to achieve specific cytotoxicity of doxorubicin against the target liver cancer cell.
Further, both the nanotrain and aptamer contain components that do not bind to G, A, C, or T/U. Thus, neither structure could be disrupted via the invasion of any natural DNA or RNA. Thus, the extra Z and P building blocks would prevent the release of doxorubicin prematurely to solution, and thus prevent non-targeted toxicity.
Results
We first asked whether GACTZP repeats placed in a nanotrain could bind doxorubicin. Here, a series of GACTZP oligonucleotides were designed from a report that a single unit of duplex GCA/CGT or CGA/GCT can intercalate one molecule of doxorubicin.[22] The designed sequences were self-complementary, forming 8 consecutive GCA/CGT, GCZ/CGP, or PZA/ZPT paired units. These were connected by TTT linkers (Fig. S3A). The three sequences (DOXCAR1, DOXCAR2 and DOXCAR3) were synthesized by solid-phase synthesis from the corresponding phosphoramidites (firebirdbio.com) and annealed (snap-cooling) to form self-complementary duplexes. The concentration-dependence of doxorubicin binding was studied by mixing different concentrations (0–400 nM) of DOXCARs with doxorubicin (1 μM, 2 h, 5 mM MgCl2, PBS, r.t.) and examining the quenching of doxorubicin fluorescence upon intercalation.
Binding of doxorubicin to PZ-containing DNA was confirmed by the loss of its fluorescence. This loss, studied at various concentrations of different DOXCARs, suggested that replacement of GC/CG by PZ/ZP, or A/T by Z/P, neither impacted the intercalation of doxorubicin nor the quenching of its fluorescence (Fig. S3B). Fitting the binding curve showed that fluorescence decreases as the concentration of DOXCARs increases, with apparent disassociation constants (Kd) of ~20 nM (Fig. S3C); these were similar for all DOXCARs.
We then designed the nanotrain and an interacting modified aptamer, starting with aptamer LZH5 reported previously as a product of GACTZP LIVE targeting liver cancer HepG2 cells. In that LIVE, an immortalized untransformed non-cancerous cell line (Hu1545V) was used in a counter selection. Aptamer LZH5 was chosen from about a dozen other aptamers because of its superior affinity (41 nM) and its ready internalization into HepG2 cells.
The structure of LZH5 was then truncated to give LZH5B, which had shorter sequence and improved affinity (12 nM) (Fig. S2). Internalization of LZH5B was confirmed by a binding assay and confocal microscopy (Fig. S4).
For the nanotrain itself, two hairpin monomers (6N-M1, 6N-M2, Table S1) were designed from a 6-nucleotide GACTZP DNA so that the energy stored in the loops would be protected by the corresponding stems. A DNA trigger probe was appended to the 5’-end of LZH5B to initiate the assembly of the 6-letter DNA nanotrain.
Introduction of the LZH5B-trigger to a mixture of 6N-M1 and 6N-M2 initiated the polymerization of these building blocks through mutual hybridization, and was expected to result in the self-assembly of the DNA nanotrains tethered to the LZH5B aptamer (LZH5B-NTrs, Fig. 1). The molar ratio of the LZH5B-trigger to monomers was optimized.
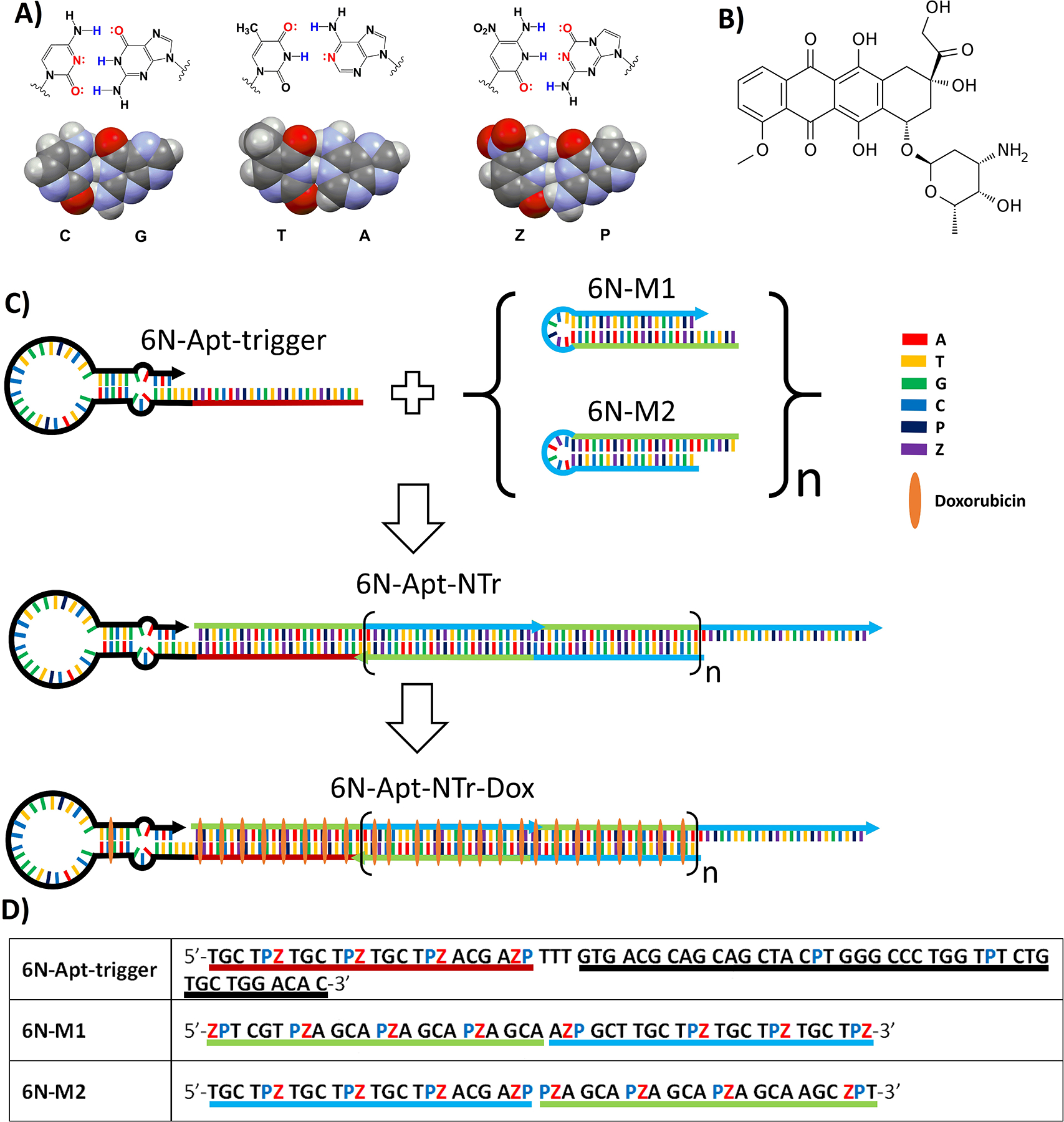
Doxorubicin was then loaded by incubating the assembly with doxorubicin (50:1 doxorubicin:assembly, PBS, 5 mM Mg2+, r.t., 2 h) followed by washing. Initiation by the aptamer of a nanotrain formation was confirmed by AFM (Fig. 2B). The nanotrains have a worm-like shapes up to 100’s of nanometers. Images of the GACTZP nanotrains were similar to those of nanotrains built from standard nucleotides.[8]
Figure 2.
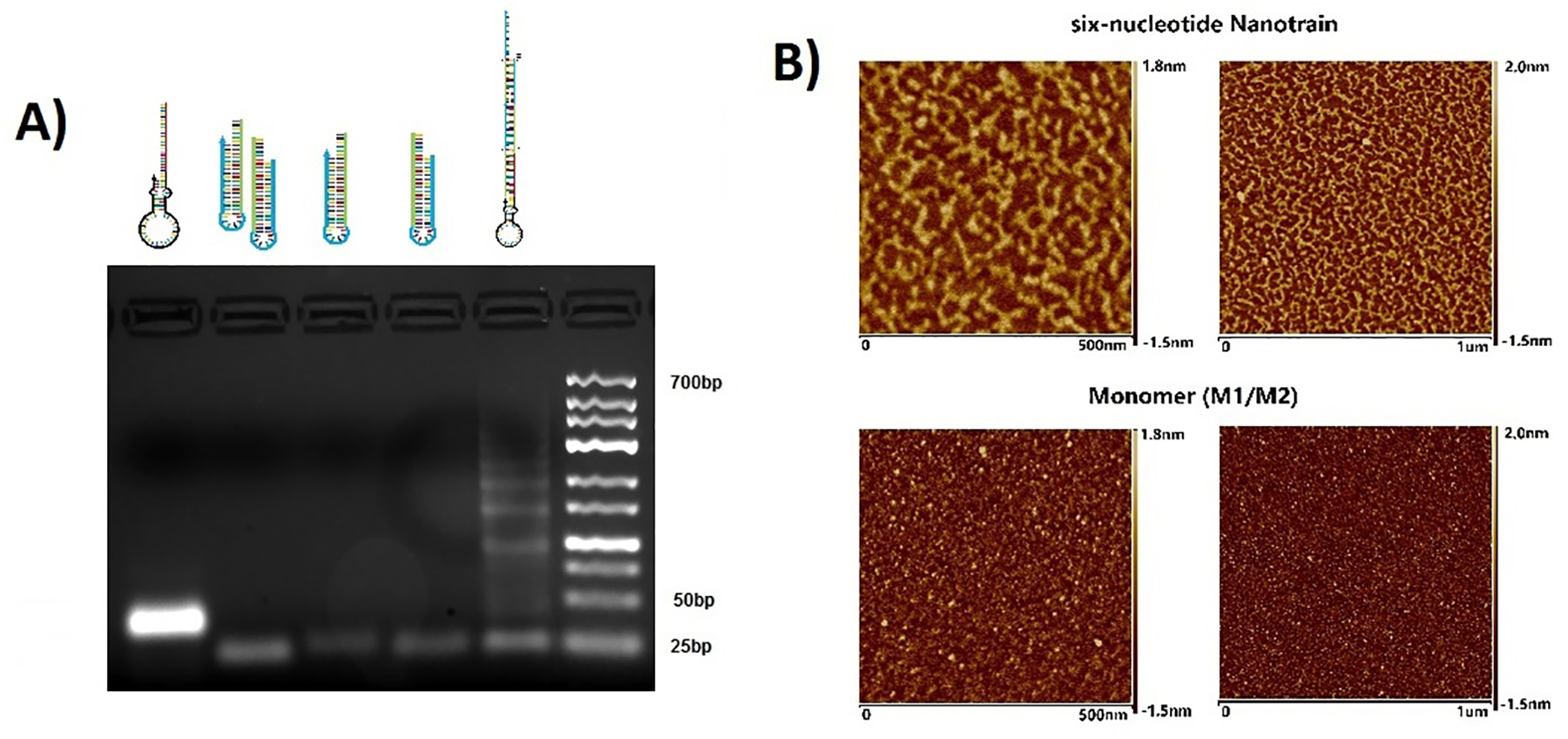
Characterization of nanotrain assembly. (A) Agarose gel image shows migration of (left to right) 6N-Apt-trigger, mixture of 6N-M1 and 6N-M2, 6N-M1, 6N-M2, 6N-Apt-NTrs, DNA ladders. (B) Atomic force microscopy images show the formation of 6-letter nanotrains (top two images) compared to the 6-letter monomers (bottom two images).
We next evaluated the stability of 6N-LZH5B-NTr-Dox complexes through a drug diffusion experiment using a dialysis units. Results showed negligible independent diffusion of the drug once it was bound to the 6N-LZH5B-NTrs. This result stands in contrast to fast diffusion of the drug presented as free in solution (PBS, with 5 mM Mg2+). This indicated that the stability of 6N-LZH5B-NTr-Dox com-plexes is adequate to prevent cargo leakage (Fig. 3A). Interestingly, 6N-LZH5B-NTr-Dox showed better stability than an LZH5B-initiated nanotrain built from standard 4-letter DNA carrying doxorubicin (LZH5B-NTr-Dox, PBS with 10% fetal bovine serum) (Fig. 3B). This may arise from the non-invadability of duplexes containing P:Z pairs.
Figure 3.
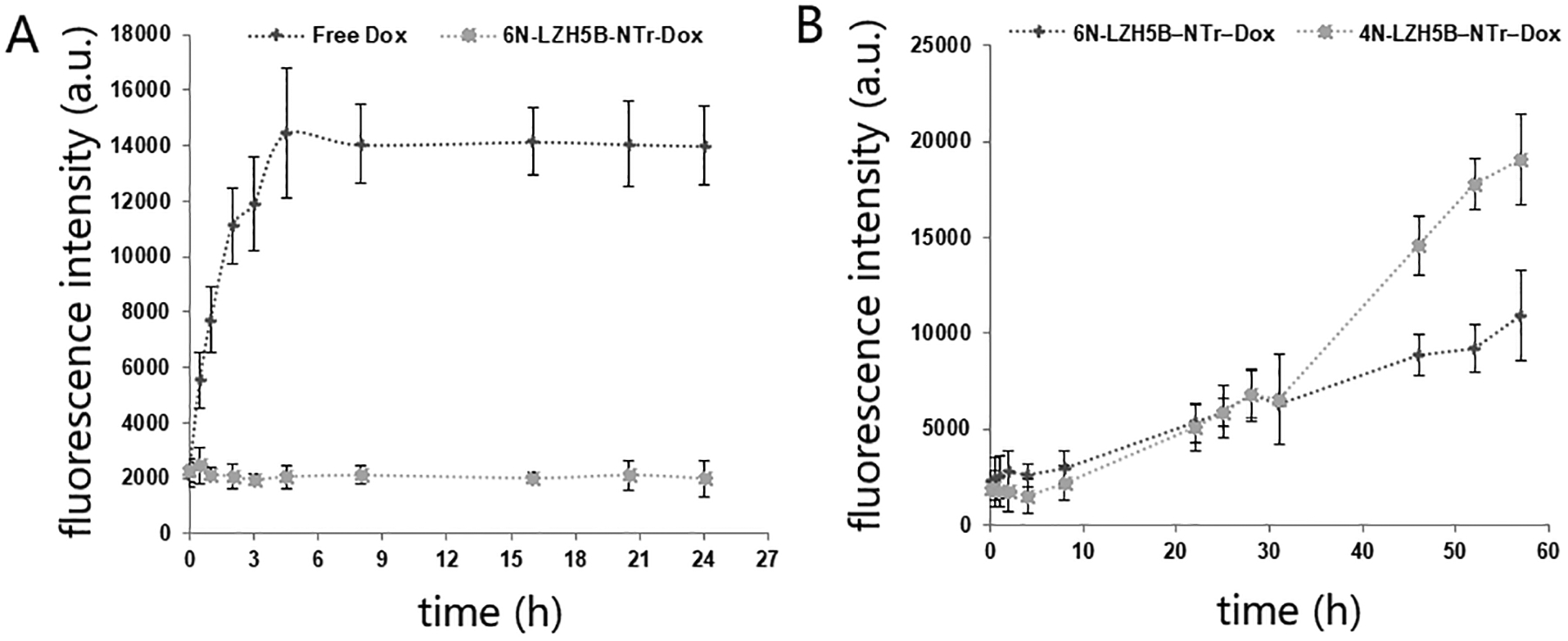
Loading and releasing of doxorubicin. Fluorescence of dialyzed doxorubicin was detected to represent the concentration of diffused doxorubicin. (A) Drug diffusion of free doxorubicin and doxorubicin carried by 6-nucleotide nanotrain in PBS with 5 mM Mg2+. (B) Drug diffusion of doxorubicin carried by 6-nucleotide nanotrain and 4-nucleotide nanotrain in PBS with 10% fetal bovine serum.
Experiments then tested the ability of the nanoassembly built from this expanded genetic alphabet to selectively kill cells that the aptamer recognizes. This design requires that (i) doxorubicin be effectively sequestered by its intercalation into the nanotrain, and therefore not “free” to be toxic (as free doxorubicin is), but (ii) the doxorubicin bound to an aptamer-nanotrain assembly be internalized into a cell after the aptamer binds to the cell surface, where (iii) doxorubicin is released into the cell in a toxic form after the nucleotides that sequester it are digested. Thus, while free doxorubicin is expected to be toxic to any cells,[25] doxorubicin bound to this aptamer-nanotrain complexed should not kill any cells except those that are bound by the LZH5B aptamer.
To confirm this, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) was used to assess the survival of cells after they were exposed to free doxorubicin compared to the aptamer-nanotrain-Dox nanostructure. In this assay, both HepG2 cells, which bind the aptamer, and control Hu1545 cells, which do not, were treated with free Dox and LZH5B-NTr-Dox complex.
Free doxorubicin showed the standard dose-dependent cytotoxicity in both HepG2 cells and Hu1545 cells. [26] In contrast, doxorubicin bound in LZH5B-NTrs for transport to target HepG2 cells (Fig. 4) displayed a dose-dependent cytotoxicity similar to that of free doxorubicin, but only against HepG2 cells. Essentially, no toxicity was seen with non-transformed liver cells that had been used as the counter-selection in the GACTZP LIVE experiment that generated the LZH5B aptamer. This showed a robust cytotoxic efficacy of LZH5B-NTr-Dox against target cells and excellent selective cytotoxicity of this molecular drug transported by aptNTrs. The lack of cytotoxicity of LZH5B-NTrs in Hu1545V cells indicates the biocompatibility of these transporters under our experimental conditions (Fig. 4).
Figure 4.
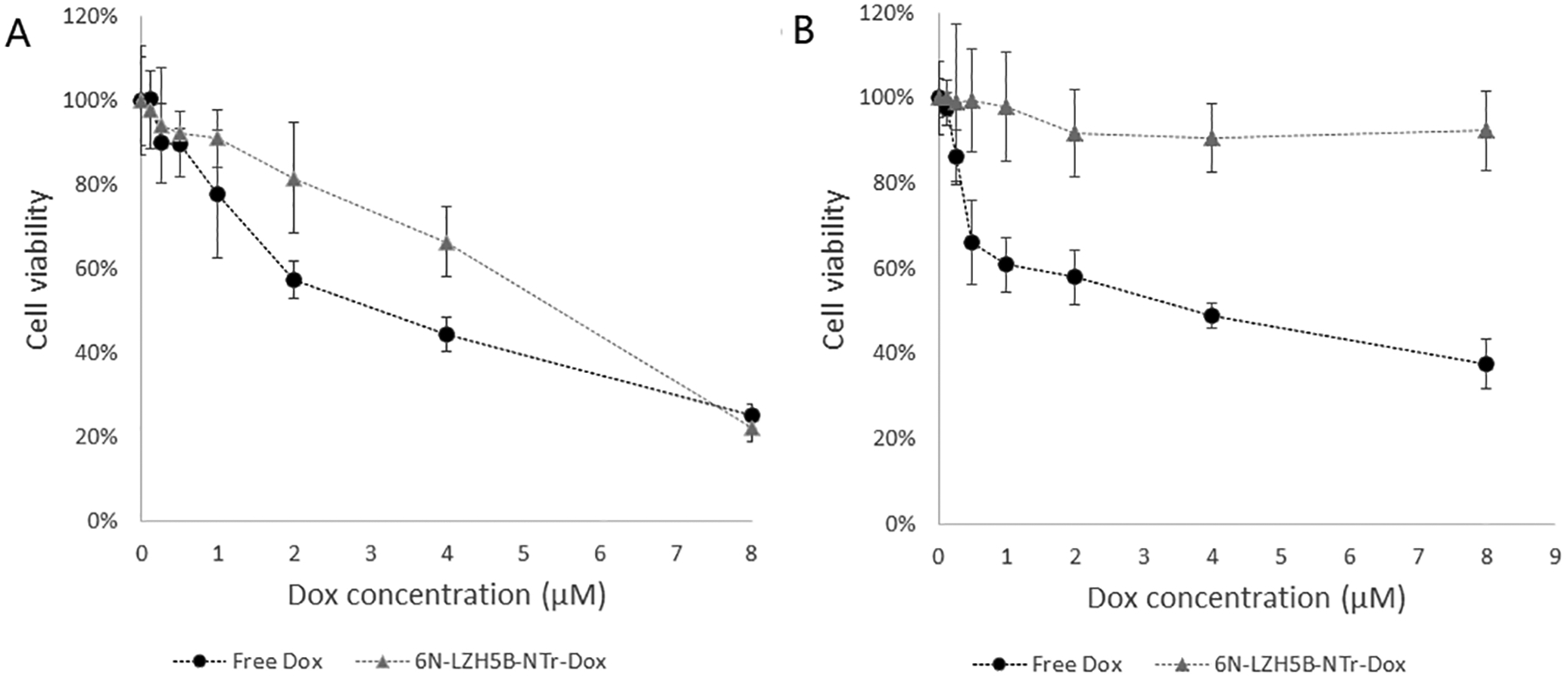
Selective cytotoxicity of molecular drugs (Dox) transported by aptNTrs. (A and B) MTS assay results showing that Dox transported by LZH5B-NTrs (LZH5B-NTr-Dox) selectively induced potent cytotoxicity and inhibited cell proliferation in target HepG2 cells (A), but not in non-target Hu1545 cells (B), in contrast to nonselective cytotoxicity induced by free Dox in both target and non-target cells.
Discussion
The results combine four recent themes in applied biomolecular sciences:
An emerging class of medicines that use protein derivatization chemistry to attach drugs to antibodies to target to specific cells compounds that are too toxic to administer without targeting (e.g. trastuzumab emtansine and brentuximab vedotin).[27]
The desire to replace antibodies by aptamers, reflecting a concern that antibodies are “a major driver of what has been deemed a reproducibility crisis” in medicine.[28]
Using DNA molecules emerging from expanded genetic alphabets to improve the performance of classical aptamers made from only the four standard nucleotides.
The programmability of DNA to form nanostructures.
In this work, a toxic drug (doxorubicin) was appended to an aptamer that has been improved by expanding the genetic alphabet. However, the attachment exploited the ability of expanded DNA to self-assemble; no conjugation chemistry was required. The self-assembling nanostructure likewise benefits from the expanded DNA alphabet because it cannot be invaded by standard DNA or RNA, both abundant in a biological background. Thus, GACTZP nanostructures can assemble in stable form in complex biological systems without invasion by natural DNA or RNA.
Perhaps surprising is the observation that replacing standard nucleotide pairs by P:Z pairs did not alter, and possibly improved, the affinity of the DNA for doxorubicin. Available structures of DNA containing P:Z pairs show that they distort very little the standard duplex structure. Indeed, the A:T pair, joined by only two hydrogen bonds, provides a greater distortion.
The versatility of the GACTZP system is especially illustrated by its dual use in these nanostructures. First, it was used to create the aptamer that binds specifically to liver cancer cells. Especially for cell-targeted LIVE, the literature suggests that the GACTZP-expanded alphabet generates cell-specific aptamers with higher affinity and does so faster than standard GACT LIVE. By exemplifying new ways of approaching current problems in applied nanotechnology, further examination of such systems is warranted.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health (R42GM115130 to S.A.B., and R01GM128186 to S.A.B.). The W.T. laboratory is indebted to the NIH Maximizing Investigators’ Research Award R35GM127130 and NSF 1645215, and by NSFC grants (NSFC 21827811). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH, NSF, or any other funding agency.
Footnotes
Experimental Section
Additional details regarding the materials and methods may be found in SI Appendix.
Supporting Information
Supporting Information contains a detailed description of the nanotrain assembly and data from affinity measurement experiments.
References
- [1].Bushnell S; Budde J; Catino T; Cole J; Derti A; Kelso R; Collins ML; Molino G; Sheridan P; Monahan J; Urdea M, ProbeDesigner: for the design of probesets for branched DNA (bDNA) signal amplification assays. Bioinformatics 1999, 15 (5), 348–55. [DOI] [PubMed] [Google Scholar]
- [2].Tyagi S; Kramer FR, Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol 1996, 14 (3), 303–8. [DOI] [PubMed] [Google Scholar]
- [3].Seeman NC, DNA in a material world. Nature 2003, 421 (6921), 427–31. [DOI] [PubMed] [Google Scholar]
- [4].Zhao Z; Yan H; Liu Y, A route to scale up DNA origami using DNA tiles as folding staples. Angew Chem Int Ed Engl 2010, 49 (8), 1414–7. [DOI] [PubMed] [Google Scholar]
- [5].Klug SJ; Famulok M, All you wanted to know about SELEX. Mol Biol Rep 1994, 20 (2), 97–107. [DOI] [PubMed] [Google Scholar]
- [6].Tuerk C; Gold L, Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249 (4968), 505–10. [DOI] [PubMed] [Google Scholar]
- [7].Bartel DP; Szostak JW, Isolation of new ribozymes from a large pool of random sequences. Science 1993, 261 (5127), 1411–8. [DOI] [PubMed] [Google Scholar]
- [8].Zhu G; Zheng J; Song E; Donovan M; Zhang K; Liu C; Tan W, Self-assembled, aptamer-tethered DNA nanotrains for targeted transport of molecular drugs in cancer theranostics. Proc Natl Acad Sci U S A 2013, 110 (20), 7998–8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hu Q; Li H; Wang L; Gu H; Fan C, DNA Nanotechnology-Enabled Drug Delivery Systems. Chem Rev 2019, 119(10), 6459–6506. [DOI] [PubMed] [Google Scholar]
- [10].Tao W; Zeng X; Wu J; Zhu X; Yu X; Zhang X; Zhang J; Liu G; Mei L, Polydopamine-based surface modification of novel nanoparticle-aptamer bioconjugates for in vivo breast cancer targeting and enhanced therapeutic effects. Theranostics 2016, 6(4), 470–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Xiao Z; Farokhzad OC, Aptamer-functionalized nanoparticles for medical applications: challenges and opportunities. ACS Nano 2012, 6(5), 3670–3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Levy-Nissenbaum E; Radovic-Moreno AF; Wang AZ; Langer R; Farokhzad OC, Nanotechnology and aptamers: applications in drug delivery. Trends in Biotechnology 2008. 26(8), 442–449. [DOI] [PubMed] [Google Scholar]
- [13].Jayasena SD, Aptamers: an emerging class of molecules that rival antibodies in diagnos-tics. Clin Chem 1999, 45 (9), 1628–50. [PubMed] [Google Scholar]
- [14].Abbott JA; Francklyn CS; Robey-Bond SM, Transfer RNA and human disease. Front Genet 2014, 5, 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Carrigan MA; Ricardo A; Ang DN; Benner SA, Quantitative analysis of a RNA-cleaving DNA catalyst obtained via in vitro selection. Biochemistry 2004, 43 (36), 11446–59. [DOI] [PubMed] [Google Scholar]
- [16].Wang L; Yang CJ; Medley CD; Benner SA; Tan W, Locked nucleic acid molecular beacons. J Am Chem Soc 2005, 127 (45), 15664–5. [DOI] [PubMed] [Google Scholar]
- [17].Hoshika S; Leal NA; Kim MJ; Kim MS; Karalkar NB; Kim HJ; Bates AM; Watkins NE Jr.; SantaLucia HA; Meyer AJ; DasGupta S; Piccirilli JA; Ellington AD; SantaLucia J Jr.; Georgiadis MM; Benner SA, Hachimoji DNA and RNA: A genetic system with eight building blocks. Science 2019, 363 (6429), 884–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sheng P; Yang Z; Kim Y; Wu Y; Tan W; Benner SA, Design of a novel molecular beacon: modification of the stem with artificially genetic alphabet. Chem Commun (Camb) 2008, (41), 5128–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bain JD; Switzer C; Chamberlin AR; Benner SA, Ribosome-mediated incorporation of a non-standard amino acid into a peptide through expansion of the genetic code. Nature 1992, 356 (6369), 537–9. [DOI] [PubMed] [Google Scholar]
- [20].Benner SA, Understanding nucleic acids using synthetic chemistry. Acc Chem Res 2004, 37 (10), 784–97. [DOI] [PubMed] [Google Scholar]
- [21].Sefah K; Yang Z; Bradley KM; Hoshika S; Jimenez E; Zhang L; Zhu G; Shanker S; Yu F; Turek D; Tan W; Benner SA, In vitro selection with artificial expanded genetic information systems. Proc Natl Acad Sci U S A 2014, 111 (4), 1449–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhang L; Yang Z; Le Trinh T; Teng IT; Wang S; Bradley KM; Hoshika S; Wu Q; Cansiz S; Rowold DJ; McLendon C; Kim MS; Wu Y; Cui C; Liu Y; Hou W; Stewart K; Wan S; Liu C; Benner SA; Tan W, Aptamers against cells overexpressing Glypican 3 from expanded genetic systems combined with cell engineering and laboratory evolution. Angew Chem Int Ed Engl 2016, 55 (40), 12372–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Biondi E; Lane JD; Das D; Dasgupta S; Piccirilli JA; Hoshika S; Bradley KM; Krantz BA; Benner SA, Laboratory evolution of artificially expanded DNA gives redesignable aptamers that target the toxic form of anthrax protective antigen. Nucleic Acids Res 2016, 44 (20), 9565–9577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhang L; Yang Z; Sefah K; Bradley KM; Hoshika S; Kim MJ; Kim HJ; Zhu G; Jimenez E; Cansiz S; Teng IT; Champanhac C; McLendon C; Liu C; Zhang W; Gerloff DL; Huang Z; Tan W; Benner SA, Evolution of functional six-nucleotide DNA. J Am Chem Soc 2015, 137 (21), 6734–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bagalkot V; Farokhzad OC; Langer R; Jon S, An aptamer-doxorubicin physical conjugate as a novel targeted drug-delivery platform. Angew Chem Int Ed Engl 2006, 45 (48), 8149–52. [DOI] [PubMed] [Google Scholar]
- [26].Cagel M; Grotz E; Bernabeu E; Moretton MA; Chiappetta DA, Doxorubicin: nanotechnological overviews from bench to bedside. Drug Discov Today 2017, 22 (2), 270–281. [DOI] [PubMed] [Google Scholar]
- [27].Diamantis N; Banerji U; Antibody-drug conjugates. An emerging class of cancer treatment. British J. Cancer 2016, 114(4), 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Baker M Blame it on the antibodies. Nature 2015, 521, 274–276. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


