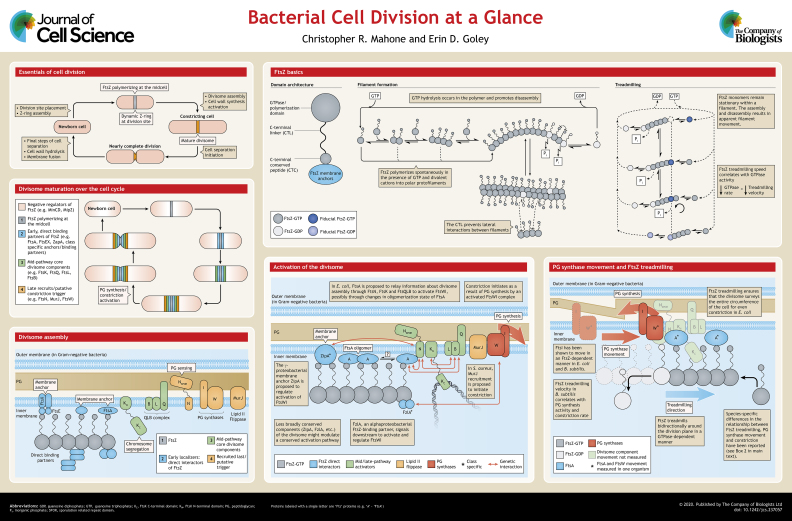
ABSTRACT
Bacterial cell division is initiated by the midcell assembly of polymers of the tubulin-like GTPase FtsZ. The FtsZ ring (Z-ring) is a discontinuous structure made of dynamic patches of FtsZ that undergo treadmilling motion. Roughly a dozen additional essential proteins are recruited to the division site by the dynamic Z-ring scaffold and subsequently activate cell wall synthesis to drive cell envelope constriction during division. In this Cell Science at a Glance article and the accompanying poster, we summarize our understanding of the assembly and activation of the bacterial cell division machinery. We introduce polymerization properties of FtsZ and discuss our current knowledge of divisome assembly and activation. We further highlight the intimate relationship between the structure and dynamics of FtsZ and the movement and activity of cell wall synthases at the division site, before concluding with a perspective on the most important open questions on bacterial cell division.
KEY WORDS: FtsZ, Bacteria, Bacterial cell biology, Cell division, Cytokinesis, Peptidoglycan
Summary: This Cell Science at a Glance article provides an overview of the initial stages of cytokinetic ring assembly, dynamics and activation during bacterial cell division.
Introduction
When observed by time-lapse microscopy, bacterial cell division looks very simple. Start with one bacterium; end up with two. As with most biological processes, however, things are more complicated under the surface. To initiate division, bacteria need to accomplish at least three tasks: mark the division site, recruit the division machinery (the divisome), and activate cell wall synthesis to drive constriction (see poster). Subsequent events, including membrane fusion and cell wall hydrolysis, are required for compartmentalization and physical separation of the cell into two daughters. This Cell Science at a Glance article and poster will focus on the initial stages of divisome assembly and constriction activation. To mark the division plane, almost all bacteria use a tubulin-like GTPase called FtsZ that polymerizes along the circumference of the cytoplasmic membrane at the future division site (Löwe and Amos, 1998; Bi and Lutkenhaus, 1991). Use of fluorescently labeled FtsZ has shown that FtsZ forms a ring-like structure called the Z-ring (Ma et al., 1996) that directly and indirectly recruits other division proteins to the midcell region (Adams and Errington, 2009). The Z-ring is a moving target, however, as recent studies have demonstrated that patches of FtsZ filaments treadmill at the division site and direct the dynamic movement of other division proteins (Bisson-Filho et al., 2017; Monteiro et al., 2018; Perez et al., 2019; Yang et al., 2017).
Although division begins with FtsZ, and there is in vitro evidence that FtsZ can deform membranes, the force generated by FtsZ alone is insufficient to initiate constriction. Instead, the driving force for constriction is thought to be cell wall synthesis (Coltharp et al., 2016; Daley et al., 2016). The peptidoglycan (PG) cell wall in bacteria comprises a meshwork of glycan strands crosslinked together by short peptides, and provides shape and protection against turgor pressure (Typas et al., 2011). The enzymes that polymerize the glycan strands are called glycosyltransferases (GTases), and the enzymes that crosslink the peptide side chains are called transpeptidases (TPases). During division, FtsW and a monofunctional penicillin-binding protein (FtsI) are the primary GTase and TPase, respectively (Adam et al., 1997; Ikeda et al., 1989; Spratt, 1975; Taguchi et al., 2019). However, localization of these enzymes to the division plane is not sufficient for PG synthesis and constriction. Instead, these enzymes require activating signals to trigger constriction (Lariviere et al., 2019; Rohs et al., 2018; Taguchi et al., 2019; Tsang and Bernhardt, 2015).
This Cell Science at a Glance article will synthesize our current understanding of the assembly, activation and dynamics of the bacterial cytokinetic machinery for the initial stages of cell division. We will pay particular attention to recent advances in understanding the links between the FtsZ cytoskeleton, and the activity and dynamics of the PG synthases that drive constriction.
FtsZ – the master regulator of bacterial division
FtsZ has three conserved domains: a polymerizing GTPase domain, a C-terminal conserved (CTC) peptide through which membrane anchors bind, and a disordered C-terminal linker (CTL) that connects the GTPase domain to the CTC (Vaughan et al., 2004) (see poster and Box 1). In addition, some organisms have a short extension at the extreme C-terminus of FtsZ called the C-terminal variable (CTV) region (Buske and Levin, 2012).
Box 1. Contribution of the C-terminal linker of FtsZ to polymer structure, dynamics and function.
Although the GTPase domain is the primary determinant of FtsZ assembly, the C-terminus of FtsZ has recently been demonstrated to impact FtsZ function and polymerization. The CTL is a disordered region that connects the GTPase domain and the CTC (see poster) and is highly variable in sequence and length across species (Vaughan et al., 2004). E. coli and B. subtilis are each tolerant of changes in the CTL sequence, but it must be disordered and close in length to the native CTL to function in division, implying a role as a flexible link to the membrane (Buske and Levin, 2013; Gardner et al., 2013). C. crescentus cells that produce FtsZ lacking its CTL (ΔCTL) exhibit dominant lethal cell bulging and lysis (Sundararajan et al., 2015), and B. subtilis cells producing ΔCTL rapidly lyse (Buske and Levin, 2013), suggesting downstream effects on PG metabolism. FtsZ variants bearing alterations to or deletion of the CTL have altered polymerization properties in vitro and altered filament superstructure in cells. C. crescentus ΔCTL forms hyperstable filament bundles in vitro and large non-ring assemblies in cells, implicating the CTL in regulating lateral interactions and polymer stability (Sundararajan and Goley, 2017; Sundararajan et al., 2015, 2018; Barrows et al., 2020). B. subtilis ΔCTL forms large extended bundles in cells, and in vitro the CTL impacts B. subtilis FtsZ interfilament spacing, suggesting a conserved role for the CTL in impacting FtsZ polymer superstructure (Buske and Levin, 2013; Huecas et al., 2017). The phenotypic outcomes of deleting or altering the CTL implicate this domain in regulation of PG metabolism (Buske and Levin, 2013; Gardner et al., 2013; Sundararajan et al., 2015). In addition to the CTL, the CTV of B. subtilis FtsZ is sufficient to induce lateral interactions in vitro that are important for formation of the Z-ring and, therefore, division in that organism (Buske and Levin, 2012).
The GTPase domain of FtsZ is required for polymerization and, like tubulin, the nucleotide-bound state influences the polymerization dynamics and structure of FtsZ filaments (Bramhill and Thompson, 1994; Erickson et al., 1996; Mukherjee and Lutkenhaus, 1994). In the presence of GTP and divalent cations, FtsZ spontaneously assembles into polymers in vitro (Bramhill and Thompson, 1994; Erickson et al., 1996; Mukherjee and Lutkenhaus, 1994). Once in a filament, FtsZ is competent to hydrolyze GTP, and nucleotide hydrolysis serves to favor depolymerization and to take the FtsZ filament from a more straight to a more curved conformation (Erickson et al., 1996) (see poster). Mutations or chemical perturbations that slow the GTP hydrolysis rate of FtsZ stabilize the polymer in vitro and in cells; as a consequence, the completion of constriction is slowed or prevented (Bisson-Filho et al., 2017; Monteiro et al., 2018; Perez et al., 2019; Stricker et al., 2002; Yang et al., 2017). FtsZ assembly in vitro occurs when it is above a critical concentration of ∼1 µM, and assembly is cooperative (Mukherjee and Lutkenhaus, 1998, 1999; Romberg et al., 2001). Recent structural work has demonstrated that the FtsZ monomer undergoes a conformational change from closed to open upon polymerization, which provides an explanation for the cooperative nature of FtsZ polymerization and the ability of filaments to treadmill (Wagstaff et al., 2017) (see poster).
In vitro under physiological pH and salt conditions, FtsZ forms mostly single protofilaments that are ∼120–200 nm long (Erickson et al., 2010; Romberg et al., 2001). Multiple FtsZ filaments can associate laterally into thicker bundles in vitro depending on the presence of crowding agents or binding partners, salt concentration and pH (Erickson et al., 1996; Huang et al., 2013). However, the structure(s) of FtsZ filaments in cells is not fully resolved. When imaged by electron cryotomography in vivo, FtsZ filaments appear to be of a width that reflects that of a single monomer, are ∼100 nm long and are at a stereotypical distance of ∼16 nm from the membrane (Li et al., 2007; Szwedziak et al., 2014; Yao et al., 2017). Super-resolution light microscopy of labeled Z-rings highlights loose filament clusters distributed around the division plane in a discontinuous structure that extends ∼50–100 nm radially into the cell (Fu et al., 2010; Holden et al., 2014; Strauss et al., 2012). Although bundling of FtsZ is reported under a variety of conditions in vitro, thick bundles of FtsZ have not been observed in wild-type bacteria, and induction of stable, large-scale bundling in vivo can lead to detrimental phenotypic changes (Buske and Levin, 2013; Durand-Heredia et al., 2012; Sundararajan et al., 2015; Barrows et al., 2020). However, mutation of residues that are implicated in lateral interactions between FtsZ filaments disrupts division, suggesting that transient lateral interactions are important to division progression (Guan et al., 2018). The CTL that connects the GTPase and CTC domain appears to have a major role in modulating lateral interactions (see Box 1).
In addition to its intrinsic polymerization properties, FtsZ has a host of interacting proteins that can modulate its assembly (Huang et al., 2013). Importantly, these include inhibitors of FtsZ that spatially and/or temporally regulate Z-ring assembly and proteins that contribute to formation of a focused Z-ring at the midcell region (see poster). In most well-studied organisms, spatial regulation of Z-ring assembly is primarily mediated by negative regulators of FtsZ polymerization. The Min proteins function in Escherichia coli (de Boer et al., 1989) and Bacillus subtilis (Levin et al., 1992) to inhibit FtsZ polymerization near the cell poles, and a functionally analogous protein called MipZ fulfills a similar function in α-proteobacteria (Thanbichler and Shapiro, 2006; Toro-Nahuelpan et al., 2019), with the ultimate result being the accurate placement of the Z-ring at the midcell. Additional negative (e.g. nucleoid occlusion factors; Bernhardt and De Boer, 2005; Wu and Errington, 2004) or positive (e.g. Zaps; Durand-Heredia et al., 2011, 2012; Gueiros-Filho and Losick, 2002; Marteyn et al., 2014) regulators of FtsZ polymerization and organization serve with Min proteins or MipZ to coordinate Z-ring assembly in time and space (see poster). As FtsZ is studied in additional organisms, novel modes of regulation are being recognized – for example, the primary role of positive regulation of Z-ring placement by PomX, PomY and PomZ in Myxococcus xanthus (Schumacher et al., 2017) or by MapZ in Streptococcus pneumoniae (Fleurie et al., 2014; Massidda et al., 2014). Collectively, intrinsic and extrinsic factors promote the nucleotide-dependent assembly of FtsZ into a dynamic Z-ring to establish the future division site.
Assembly and activation of the divisome
Although central to the process, FtsZ is not the only protein required for cell division; there are roughly a dozen conserved proteins that are required at the midcell for constriction (see poster). Once the Z-ring assembles, the remainder of the divisome is recruited in a roughly sequential manner. In E. coli and B. subtilis, the process occurs in a two-step fashion, with direct FtsZ interactors localizing first and other division components localizing in a second step (Aarsman et al., 2005; Gamba et al., 2009). In Caulobacter crescentus, cell synchronization experiments enabled the classification of divisome assembly into a series of seven functional modules (Goley et al., 2011). Across bacteria, the earliest arrivals to the division plane help to assemble a focused midcell Z-ring (Aarsman et al., 2005; Fleurie et al., 2015; Gamba et al., 2009; Goley et al., 2011; Schumacher et al., 2017). A subsequent wave (or waves) of protein recruitment brings factors in as their functions are required. Divisome components that localize just prior to initiation of constriction (FtsN in E. coli, FtsW in C. crescentus and MurJ in Staphylococcus aureus) have been proposed to trigger constriction through activation of cytokinetic cell wall synthesis (Aarsman et al., 2005; Goley et al., 2011; Monteiro et al., 2018) (see poster).
Simply localizing PG synthases to the midcell is not sufficient for constriction to begin, which implies a requirement for regulatory input into constriction activation. This makes sense, as improper timing of constriction could have drastic consequences for the cell. The GTPase FtsW requires its partner TPase FtsI to act as a PG polymerase in vitro, suggesting that it is inactive until engaged in a complex (Taguchi et al., 2019). Moreover, the fully assembled divisome in C. crescentus can be held in an inactive state by SidA or DidA, small protein inhibitors of constriction that bind the late divisome proteins FtsW and/or FtsN upon DNA damage (Modell et al., 2011, 2014). Mutations in FtsW or FtsI that bypass inhibition by SidA and DidA hyperactivate these PG synthases such that the cells constrict faster than wild type (Lambert et al., 2018; Lariviere et al., 2019; Modell et al., 2014). Similarly, hyperactivating mutations in the divisome proteins FtsL and FtsB were described in E. coli to cause premature initiation of constriction and/or cell shortening (Liu et al., 2015; Tsang and Bernhardt, 2015). Collectively, these observations imply the presence of inactive and active states of the PG synthases that promote constriction.
The most advanced – but still incomplete – model for constriction activation is derived from genetic studies in E. coli and includes a number of broadly conserved divisome components. In this model, the activating signal is proposed to initiate with FtsA, an actin homolog and conserved membrane anchor for FtsZ. Specifically, FtsA is thought to relay information about divisome assembly status by converting from an ‘off’ to an ‘on’ state through a mechanism that may involve FtsA transitioning from polymeric to monomeric state (Pichoff et al., 2012, 2015) (see poster). Although FtsA has been demonstrated to polymerize in vitro (Krupka et al., 2017; Szwedziak et al., 2012), its physiological polymerization state is not clear. Regardless of the mechanism, FtsA is proposed to be in a constriction activation pathway that includes FtsN and the complex formed by FtsQ, FtsL and FtsB (FtsQLB) (Liu et al., 2015; Pichoff et al., 2018). Variants of FtsA and FtsN identified in E. coli are also able to bypass loss of an essential, but γ-proteobacteria-specific, membrane anchor for FtsZ called ZipA that is thought to modulate interactions between FtsA, FtsZ and downstream signaling proteins like FtsN (Geissler et al., 2003; Pichoff et al., 2012, 2015; Schoenemann et al., 2018) (see poster). FtsN, which contains a sporulation-related repeat (SPOR) domain responsible for recognizing denuded glycans (the result of amidase activity) (Yahashiri et al., 2015), could direct the FtsW-FtsI PG synthetic complex (FtsWI) to locations where new PG material should be incorporated. FtsN has also been proposed through genetic studies to relay the polymerization status of FtsA downstream to FtsQLB (Liu et al., 2015; Pichoff et al., 2012; Tsang and Bernhardt, 2015). FtsQLB is a multimeric complex that is genetically implicated in activating FtsWI (Liu et al., 2015; Tsang and Bernhardt, 2015), though the details of its role in activation of FtsWI are unknown. Finally, FtsK, a bifunctional protein involved in division and chromosome segregation, is also genetically implicated in activation of constriction through its N-terminal domain (Dubarry et al., 2010). Such a role for FtsK could link chromosome segregation to PG synthase activation to ensure DNA is not trapped as the cell envelope constricts.
Although many of the proteins implicated in constriction activation in E. coli are broadly conserved, the divisome has also diversified across bacteria. This includes the incorporation of less broadly conserved participants in constriction activation that are, nevertheless, essential in their cognate organisms, such as ZipA in E. coli. In C. crescentus, recent work identified an activating signal that originates from a Z-ring-associated protein that is found only in α-proteobacteria. In that organism, an essential, direct binding partner of FtsZ called FzlA participates in activation of the downstream PG synthases (Goley et al., 2010; Lariviere et al., 2018, 2019) (see poster). Although fzlA is normally essential for division, hyper-activating mutations in ftsW and/or ftsI allow deletion of fzlA, indicating that the essential function of FzlA is to activate FtsWI (Lambert et al., 2018; Lariviere et al., 2019). Many of the proteins described in the E. coli activation pathway are present in C. crescentus; it is therefore likely that FzlA ultimately signals through FtsA, FtsK, FtsN and/or FtsQLB. We suspect that other bacteria similarly modulate a conserved core constriction activation pathway to suit their needs. Consistent with this prediction, in Staphylococcus aureus the MurJ lipid II flippase is proposed to trigger constriction, and its recruitment to the division site relies on the DivIB–DivIC–FtsL complex (the FtsQLB homologs in this organism) (Monteiro et al., 2018). In summary, the initiation of constriction during bacterial division requires assembly of the polymeric FtsZ ring, sequential recruitment of other divisome proteins and a poorly understood constriction activation step that promotes PG synthesis to drive constriction.
The dance of the divisome – dynamics of FtsZ and PG synthases
Cell division is a dynamic process, with the shape of the cell changing dramatically as constriction progresses. Perhaps unsurprisingly, the components of the divisome are also highly dynamic both before and during constriction. Early observations using fluorescence recovery after photobleaching indicated that FtsZ turns over rapidly within the Z-ring, with half-times of recovery for fluorescently tagged FtsZ on the order of tens of seconds (Anderson et al., 2004; Stricker et al., 2002). Subsequently, work in several bacterial systems including E. coli (Yang et al., 2017), B. subtilis (Bisson-Filho et al., 2017), S. aureus (Monteiro et al., 2018) and S. pneumoniae (Perez et al., 2019) demonstrated that FtsZ treadmills circumferentially around the division plane. That is, FtsZ patches appear to move directionally around the circumference of the cell, but individual monomers are stationary for their lifetime within the patch (Bisson-Filho et al., 2017; Perez et al., 2019; Yang et al., 2017). The apparent movement of FtsZ is mediated by net addition of FtsZ to one end of each patch and net loss from the other (see poster). The velocity of FtsZ treadmilling, at ∼30 nm/s on average, is remarkably consistent across species and is dependent on the GTPase activity of FtsZ (Bisson-Filho et al., 2017; Perez et al., 2019; Yang et al., 2017). Diminished FtsZ GTPase activity correlates with slower treadmilling speeds (Bisson-Filho et al., 2017; Perez et al., 2019; Yang et al., 2017). Conversely, FtsZ treadmilling is independent of PG synthesis; treatment of cells with PG synthesis inhibitors had no effect on treadmilling velocity (Bisson-Filho et al., 2017; Perez et al., 2019; Yang et al., 2017). At least in B. subtilis, the membrane anchor FtsA also moves along with FtsZ filaments (Bisson-Filho et al., 2017).
What is the purpose of FtsZ treadmilling? Two possibilities have gained recent experimental support. The first is that FtsZ treadmilling helps to organize divisome complexes, including the PG synthases, around the division plane for evenly distributed PG synthesis and constriction. Tracking of single molecules of PG synthases in the divisome in diverse species revealed that they move directionally around the division plane (Bisson-Filho et al., 2017; Yang et al., 2017) (see poster). In E. coli and B. subtilis, movement of the PG synthases is dependent upon FtsZ treadmilling, with the rates of movement correlating with treadmilling speed of FtsZ (Bisson-Filho et al., 2017; Yang et al., 2017). In E. coli, changing the treadmilling velocity of FtsZ did not change the rate of constriction, but changed the spatial distribution of PG synthesis such that septa were distorted when treadmilling was slowed (Yang et al., 2017). Collectively, these observations indicate that treadmilling FtsZ is important for the spatial distribution of PG synthesis during constriction. A second possibility, with support in B. subtilis, is that FtsZ treadmilling provides input into the activity of PG synthases. In that organism, changing treadmilling velocity causes a change in the rate of PG metabolism and subsequent constriction (Bisson-Filho et al., 2017). Genetic evidence implicating FtsA and FtsZ in regulation of PG metabolic activity is also consistent with the idea that FtsZ actively modulates PG metabolism, in addition to acting as a dynamic scaffold (Buske and Levin, 2013; Gardner et al., 2013; Mura et al., 2017; Sundararajan et al., 2015; Barrows et al., 2020; Varma and Young, 2004). Whether the differences in the relationship between FtsZ treadmilling and PG synthase activity reported in different bacterial species are due to technical or biological differences remains to be resolved (see Box 2). Nevertheless, it is clear that FtsZ polymer dynamics are linked to divisome dynamics and/or function across diverse bacterial species.
Box 2. FtsZ treadmilling and PG synthase movement across species.
Although FtsZ treadmilling and PG synthase movement or activity have been observed in four bacterial species to date, each species is reported to exhibit distinct properties from the others with respect to relationship between FtsZ treadmilling and movement or activity of PG synthases. In both E. coli and B. subtilis, FtsZ treadmilling velocity directly correlates with the rate of movement of the TPase FtsI (Bisson-Filho et al., 2017; Yang et al., 2017). However, there are key differences in the link between FtsZ dynamics and PG synthase activity. When FtsZ treadmilling is reduced in E. coli, the rate of PG incorporation is unchanged but the slowed movement of FtsI results in distorted, asymmetric septa (Yang et al., 2017). In contrast, in B. subtilis, PG synthase movement and activity correlates with FtsZ treadmilling – whether treadmilling velocity is decreased through mutation of FtsZ or increased by expression of an FtsZ inhibitor (Bisson-Filho et al., 2017). As FtsZ treadmilling velocity changes, the rates of PG incorporation and subsequent cell constriction change correspondingly.
S. aureus does not appear to fully follow the examples set above. FtsZ treadmilling and PG synthase activity appear to be correlated until the initiation of constriction. Specifically, pharmacological inhibition of FtsZ treadmilling prevents division if applied in the early stages of the cell cycle. However, if the cell has accumulated MurJ – the lipid II flippase and proposed division trigger in S. aureus – at the midcell prior to application of the inhibitor of FtsZ treadmilling, then the cell can carry out constriction independently of FtsZ movement (Monteiro et al., 2018). S. pneumoniae presents the most divergent picture of the link between FtsZ dynamics and PG synthases to date, in that the two processes appear to be independent of each other in that organism. PG synthases move at a slower speed than FtsZ and their speed is dependent on lipid II PG precursor availability rather than FtsZ treadmilling speed (Perez et al., 2019). Consistent with the FtsZ-independence of PG synthase movement, the PG synthesis rate during division in S. pneumoniae is also independent of FtsZ treadmilling rate, similar to what is seen in E. coli (Perez et al., 2019; Yang et al., 2017). Given the early stages in our understanding of divisome dynamics in any system, we cannot yet definitely say whether each of the species-specific differences reported are the result of biological diversification or arise from technical differences between studies.
Conclusions and perspectives
Bacterial cell division requires regulated polymerization of FtsZ into a dynamic cytokinetic ring that recruits roughly a dozen other division proteins, ultimately leading to activation of cell wall synthesis for constriction. The Z ring and other components of the divisome are highly dynamic, and these dynamics are apparently critical for efficient and accurate division. These recent exciting advances in understanding bacterial cell division have spurned new or renewed interest in old questions. With respect to activation of PG synthesis for constriction, it is not clear what is being sensed to license constriction initiation. Is it clearance of the chromosomal termini from the division plane, accumulation of a limiting component of the divisome or the substrate for PG synthesis, or changes in the assembly properties of FtsZ or FtsA? Once sensed, how are signals relayed between components of the division machinery to cause activation of FtsW and FtsI? Who are the players and how do they communicate with each other?
We are only at the beginning of understanding the relationship between FtsZ and both the movement and activity of the rest of the divisome. In E. coli (Yang et al., 2017) and B. subtilis (Bisson-Filho et al., 2017), the PG synthases rely on FtsZ treadmilling for their movement (see poster and Box 2). However, FtsZ and PG synthases have not been shown to directly interact, implying that PG synthase dynamics rely on additional molecules in the pathway. What are the protein–protein and protein–envelope interactions that control the dynamics of PG synthases and other divisome proteins? Does the divisome move as a whole or are there subcomplexes independently moving within it? Are processively moving PG synthase molecules actively synthesizing PG or are they being passively distributed? Is there more than one motile population representing different activation states of the divisome?
Although we are likely years away from a complete reconstitution of the divisome and its activity in vitro, there are short-term advancements that will shed light on the mechanisms of division. Continued technical advances in imaging, such as techniques allowing imaging of the divisome in cross-section or monitoring dynamics of multiple divisome components in the same cells by single-molecule tracking, will allow us to define division dynamics with greater precision and higher throughput. PG synthesis assays in vitro with reconstituted division components will demonstrate which divisome components activate or inhibit PG synthesis, and the order in which they provide their signals. Finally, continuing to study the mechanisms of cell division across diverse bacterial species will distinguish the conserved core mechanisms of division and identify the myriad ways bacteria adapt these mechanisms to replicate with distinct morphologies and in diverse niches.
Acknowledgements
We are grateful to Jordan Barrows, Allison Daitch, Erika Smith and Wanda Figueroa-Cuilan for helpful feedback on this article and poster. Discussions with members of the Super-Z community including the Xiao, Garner, Holden and Erickson laboratories helped inform our thinking of how bacterial cell division works. We particularly acknowledge Xinxing Yang, Ryan McQuillen and Joshua McCausland for helpful discussions.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
Work in the Goley Lab on FtsZ and cell division is funded by the National Institutes of Health through grant R01GM108640. Deposited in PMC for release after 12 months.
Cell science at a glance
A high-resolution version of the poster and individual poster panels are available for downloading at http://jcs.biologists.org/lookup/doi/10.1242/jcs.237057.supplemental
References
- Aarsman M. E. G., Piette A., Fraipont C., Vinkenvleugel T. M. F., Nguyen-Distèche M. and Den Blaauwen T. (2005). Maturation of the Escherichia coli divisome occurs in two steps. Mol. Microbiol. 55, 1631-1645. 10.1111/j.1365-2958.2005.04502.x [DOI] [PubMed] [Google Scholar]
- Adam M., Fraipont C., Rhazi N., Nguyen-Distèche M., Lakaye B., Frère J. M., Devreese B., Van Beeumen J., van Heijenoort Y., Van Heijenoort J. et al. (1997). The bimodular G57-V577 polypeptide chain of the class B penicillin- binding protein 3 of Escherichia coli catalyzes peptide bond formation from thiolesters and does not catalyze glycan chain polymerization from the lipid II intermediate. J. Bacteriol. 179, 6005-6009. 10.1128/JB.179.19.6005-6009.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams D. W. and Errington J. (2009). Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 7, 642-653. 10.1038/nrmicro2198 [DOI] [PubMed] [Google Scholar]
- Anderson D. E., Gueiros-filho F. J. and Erickson H. P. (2004). Assembly dynamics of FtsZ rings in bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. Society 186, 5775-5781. 10.1128/JB.186.17.5775-5781.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrows J. M., Sundararajan K., Bhargava A. and Goley E. D. (2020). FtsA regulates Z-ring morphology and cell wall metabolism in an FtsZ C-terminal linker-dependent manner Caulobacter crescentus. J. Bacteriol. 202, e00693-19 10.1128/JB.00693-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt T. G. and de Boer P. A. J. (2005). SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol. Cell 18, 555-564. 10.1016/j.molcel.2005.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E. and Lutkenhaus J. (1991). FtsZ ring structure associated with division in Escherichia col. Nature 354, 161-164. 10.1038/354161a0 [DOI] [PubMed] [Google Scholar]
- Bisson-Filho A. W., Hsu Y.-P., Squyres G. R., Kuru E., Wu F., Jukes C., Sun Y., Dekker C., Holden S., VanNieuwenhze M. S. et al. (2017). Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355, 739-743. 10.1126/science.aak9973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramhill D. and Thompson C. M. (1994). GTP-dependent polymerization of Escherichia coli FtsZ protein to form tubules. Proc. Natl. Acad. Sci. USA 91, 5813-5817. 10.1073/pnas.91.13.5813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buske P. J. and Levin P. A. (2012). Extreme C terminus of bacterial cytoskeletal protein FtsZ plays fundamental role in assembly independent of modulatory proteins. J. Biol. Chem. 287, 10945-10957. 10.1074/jbc.M111.330324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buske P. J. and Levin P. A. (2013). A flexible C-terminal linker is required for proper FtsZ assembly in vitro and cytokinetic ring formation in vivo. Mol. Microbiol. 89, 249-263. 10.1111/mmi.12272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coltharp C., Buss J., Plumer T. M. and Xiao J. (2016). Defining the rate-limiting processes of bacterial cytokinesis. Proc. Natl. Acad. Sci. USA 113, E1044-E1053. 10.1073/pnas.1514296113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley D. O., Skoglund U. and Söderström B. (2016). FtsZ does not initiate membrane constriction at the onset of division. Sci. Rep. 6, 33138 10.1038/srep33138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P. A. J., Crossley R. E. and Rothfield L. I. (1989). A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56, 641-649. 10.1016/0092-8674(89)90586-2 [DOI] [PubMed] [Google Scholar]
- Dubarry N., Possoz C. and Barre F.-X. (2010). Multiple regions along the Escherichia coli FtsK protein are implicated in cell division. Mol. Microbiol. 78, 1088-1100. 10.1111/j.1365-2958.2010.07412.x [DOI] [PubMed] [Google Scholar]
- Durand-Heredia J. M., Yu H. H., De Carlo S., Lesser C. F. and Janakiraman A. (2011). Identification and characterization of ZapC, a stabilizer of the FtsZ ring in Escherichia coli. J. Bacteriol. 193, 1405-1413. 10.1128/JB.01258-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand-Heredia J., Rivkin E., Fan G., Morales J. and Janakiraman A. (2012). Identification of ZapD as a cell division factor that promotes the assembly of FtsZ in Escherichia coli. J. Bacteriol. 194, 3189-3198. 10.1128/JB.00176-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson H. P., Taylor D. W., Taylor K. A. and Bramhill D. (1996). Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc. Natl. Acad. Sci. USA 93, 519-523. 10.1073/pnas.93.1.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson H. P., Anderson D. E. and Osawa M. (2010). FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol. Mol. Biol. Rev. 74, 504-528. 10.1128/MMBR.00021-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleurie A., Lesterlin C., Manuse S., Zhao C., Cluzel C., Lavergne J.-P., Franz-Wachtel M., MacEk B., Combet C., Kuru E. et al. (2014). MapZ marks the division sites and positions FtsZ rings in Streptococcus pneumoniae. Nature 516, 260-262. 10.1038/nature13966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleurie A., Lesterlin C., Manuse S., Zhao C., Cluzel C., Lavergne J. P., Franz-Wachtel M., Macek B., Combet C., Kuru E. et al. (2015). MapZ beacons the division sites and positions FtsZ-rings in Streptococcus pneumoniae. Nature 516, 259-262. 10.1038/nature13966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu G., Huang T., Buss J., Coltharp C., Hensel Z. and Xiao J. (2010). In Vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM). PLoS ONE 5, 1-16. 10.1371/journal.pone.0012680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamba P., Veening J.-W., Saunders N. J., Hamoen L. W. and Daniel R. A. (2009). Two-step assembly dynamics of the Bacillus subtilis divisome. J. Bacteriol. 191, 4186-4194. 10.1128/JB.01758-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner K. A. J. A., Moore D. A. and Erickson H. P. (2013). The C-terminal linker of Escherichia coli FtsZ functions as an intrinsically disordered peptide. Mol. Microbiol. 89, 264-275. 10.1111/mmi.12279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler B., Elraheb D. and Margolin W. (2003). A gain-of-function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc. Natl. Acad. Sci. USA 100, 4197-4202. 10.1073/pnas.0635003100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goley E. D., Dye N. A., Werner J. N., Gitai Z. and Shapiro L. (2010). Imaging-based identification of a critical regulator of FtsZ protofilament curvature in Caulobacter. Mol. Cell 39, 975-987. 10.1016/j.molcel.2010.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goley E. D., Yi-Chun Y., Hong S.-H., Fero M. J., Abeliuk E., McAdams H. H. and Shapiro L. (2011). Assembly of the Caulobacter cell division machine. Mol. Microbiol. 9, 19-22. 10.1111/j.1365-2958.2011.07677.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan F., Yu J., Yu J., Liu Y., Li Y., Feng X. H., Huang K. C., Chang Z. and Ye S. (2018). Lateral interactions between protofilaments of the bacterial tubulin homolog FtsZ are essential for cell division. Elife 7, 1-22. 10.7554/eLife.35578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueiros-Filho F. J. and Losick R. (2002). A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16, 2544-2556. 10.1101/gad.1014102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden S. J., Pengo T., Meibom K. L., Fernandez Fernandez C., Collier J. and Manley S. (2014). High throughput 3D super-resolution microscopy reveals Caulobacter crescentus in vivo Z-ring organization. Proc. Natl. Acad. Sci. USA 111, 4566-4571. 10.1073/pnas.1313368111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K., Durand-Heredia J. and Janakiraman A. (2013). FtsZ ring stability: of bundles, tubules, crosslinks, and curves. J. Bacteriol. 195, 1859-1868. 10.1128/JB.02157-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huecas S., Ramírez-Aportela E., Vergoñós A., Núñez-Ramírez R., Llorca O., Díaz J. F., Juan-Rodríguez D., Oliva M. A., Castellen P. and Andreu J. M. (2017). Self-organization of FtsZ polymers in solution reveals spacer role of the disordered C-terminal tail. Biophys. J. 113, 1831-1844. 10.1016/j.bpj.2017.08.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M., Sato T., Wachi M., Jung H. K., Ishino F., Kobayashi Y. and Matsuhashi M. (1989). Structural similarity among Escherichia coli FtsW and RodA proteins and Bacillus subtilis SpoVE protein, which function in cell division, cell elongation, and spore formation, respectively. J. Bacteriol. 171, 6375-6378. 10.1128/JB.171.11.6375-6378.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupka M., Rowlett V. W., Morado D., Vitrac H., Schoenemann K., Liu J. and Margolin W. (2017). Escherichia coli FtsA forms lipid-bound minirings that antagonize lateral interactions between FtsZ protofilaments. Nat. Commun. 8, 15957 10.1038/ncomms15957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert A., Vanhecke A., Archetti A., Holden S., Schaber F., Pincus Z., Laub M. T., Goley E. and Manley S. (2018). Constriction rate modulation can drive cell size control and homeostasis in C. crescentus. iScience 4, 180-189. 10.1016/j.isci.2018.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lariviere P. J., Szwedziak P., Mahone C. R., Löwe J. and Goley E. D. (2018). FzlA, an essential regulator of FtsZ filament curvature, controls constriction rate during Caulobacter division. Mol. Microbiol. 107, 180-197. 10.1111/mmi.13876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lariviere P. J., Mahone C. R., Santiago-Collazo G., Howell M., Daitch A. K., Zeinert R., Chien P., Brown P. J. B. and Goley E. D. (2019). An essential regulator of bacterial division links FtsZ to cell wall synthase activation. Curr. Biol. 29, 1460-1470.e4. 10.1016/j.cub.2019.03.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin P. A., Margolis P. S., Setlow P., Losick R. and Sun D. (1992). Identification of Bacillus subtilis genes for septum placement and shape determination. J. Bacteriol. 174, 6717-6728. 10.1128/JB.174.21.6717-6728.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Trimble M. J., Brun Y. V. and Jensen G. J. (2007). The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 26, 4694-4708. 10.1038/sj.emboj.7601895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Persons L., Lee L. and de Boer P. A. J. (2015). Roles for both FtsA and the FtsBLQ subcomplex in FtsN-stimulated cell constriction in Escherichia coli. Mol. Microbiol. 95, 945-970. 10.1111/mmi.12906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löwe J. and Amos L. A. (1998). Crystal structure of the bacterial cell-division protein FtsZ. Nature 391, 203-206. 10.1038/34472 [DOI] [PubMed] [Google Scholar]
- Ma X., Ehrhardt D. W. and Margolin W. (1996). Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc. Natl. Acad. Sci. USA 93, 12998-13003. 10.1073/pnas.93.23.12998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marteyn B. S., Karimova G., Fenton A. K., Gazi A. D., West N., Touqui L., Prevost M.-C., Betton J.-M., Poyraz O., Ladant D. et al. (2014). ZapE is a novel cell division protein interacting with FtsZ and modulating the Z-ring dynamics. MBio 5, 1-10. 10.1128/mBio.00022-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massidda O., Ulrych A., Buriánková K., Benada O., Kofroňová O., Molle V., Branny P., Doubravová L. and Holečková N. (2014). LocZ is a new cell division protein involved in proper septum placement in streptococcus pneumoniae. MBio 6, 1-13. 10.1128/mBio.01700-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modell J. W., Hopkins A. C. and Laub M. T. (2011). A DNA damage checkpoint in Caulobacter crescentus inhibits cell division through a direct interaction with FtsW. Genes Dev. 25, 1328 10.1101/gad.2038911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modell J. W., Kambara T. K., Perchuk B. S. and Laub M. T. (2014). A DNA damage-induced, SOS-independent checkpoint regulates cell division in caulobacter crescentus. PLoS Biol. 12, e1001977 10.1371/journal.pbio.1001977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro J. M., Pereira A. R., Reichmann N. T., Saraiva B. M., Fernandes P. B., Veiga H., Tavares A. C., Santos M., Ferreira M. T., Macário V. et al. (2018). Peptidoglycan synthesis drives an FtsZ-treadmilling-independent step of cytokinesis. Nature 554, 528-532. 10.1038/nature25506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A. and Lutkenhaus J. (1994). Guanine nucleotide-dependent assembly of FtsZ into filaments. J. Bacteriol. 176, 2754-2758. 10.1128/JB.176.9.2754-2758.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A. and Lutkenhaus J. (1998). Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 17, 462-469. 10.1093/emboj/17.2.462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A. and Lutkenhaus J. (1999). Analysis of FtsZ assembly by light scattering and determination of the role of divalent metal cations. J. Bacteriol. 181, 823-832. 10.1128/JB.181.3.823-832.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mura A., Fadda D., Perez A. J., Danforth M. L., Musu D., Rico A. I., Krupka M., Denapaite D., Tsui H.-C. T., Winkler M. E. et al. (2017). Roles of the essential protein FtsA in cell growth and division in streptococcus pneumoniae. J. Bacteriol. 199, e00608-16 10.1128/JB.00608-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez A. J., Cesbron Y., Shaw S. L., Bazan Villicana J., Tsui H.-C. T., Boersma M. J., Ye Z. A., Tovpeko Y., Dekker C., Holden S. et al. (2019). Movement dynamics of divisome proteins and PBP2x:FtsW in cells of Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 116, 3211-3220. 10.1073/pnas.1816018116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S., Shen B., Sullivan B. and Lutkenhaus J. (2012). FtsA mutants impaired for self-interaction bypass ZipA suggesting a model in which FtsA's self-interaction competes with its ability to recruit downstream division proteins. Mol. Microbiol. 83, 151-167. 10.1111/j.1365-2958.2011.07923.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S., Du S. and Lutkenhaus J. (2015). The bypass of ZipA by overexpression of FtsN requires a previously unknown conserved FtsN motif essential for FtsA-FtsN interaction supporting a model in which FtsA monomers recruit late cell division proteins to the Z ring. Mol. Microbiol. 95, 971-987. 10.1111/mmi.12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S., Du S. and Lutkenhaus J. (2018). Disruption of divisome assembly rescued by FtsN–FtsA interaction in Escherichia coli. Proc. Natl. Acad. Sci. USA 115, E6855-E6862. 10.1073/pnas.1806450115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs P. D. A., Buss J., Sim S. I., Squyres G. R., Srisuknimit V., Smith M., Cho H., Sjodt M., Kruse A. C., Garner E. C. et al. (2018). A central role for PBP2 in the activation of peptidoglycan polymerization by the bacterial cell elongation machinery. PLoS Genet. 14, 1-25. 10.1371/journal.pgen.1007726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romberg L., Simon M. and Erickson H. P. (2001). Polymerization of FtsZ, a bacterial homolog of tubulin. Is assembly cooperative? J. Biol. Chem. 276, 11743-11753. 10.1074/jbc.M009033200 [DOI] [PubMed] [Google Scholar]
- Schoenemann K. M., Krupka M., Rowlett V. W., Distelhorst S. L., Hu B. and Margolin W. (2018). Gain-of-function variants of FtsA form diverse oligomeric structures on lipids and enhance FtsZ protofilament bundling. Mol. Microbiol. 109, 676-693. 10.1111/mmi.14069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher D., Bergeler S., Harms A., Vonck J., Huneke-vogt S., Frey E., Søgaard-andersen L., Schumacher D., Bergeler S., Harms A. et al. (2017). The PomXYZ proteins self-organize on the bacterial nucleoid to stimulate cell division. Dev. Cell 41, 299-314.e12. 10.1016/j.devcel.2017.04.011 [DOI] [PubMed] [Google Scholar]
- Spratt B. G. (1975). Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc. Natl. Acad. Sci. USA 72, 2999-3003. 10.1073/pnas.72.8.2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss M. P., Liew A. T. F., Turnbull L., Whitchurch C. B., Monahan L. G. and Harry E. J. (2012). 3D-SIM super resolution microscopy reveals a bead-like arrangement for FtsZ and the division machinery: implications for triggering cytokinesis. PLoS Biol. 10, e1001389 10.1371/journal.pbio.1001389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker J., Maddox P., Salmon E. D. and Erickson H. P. (2002). Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc. Natl. Acad. Sci. USA 99, 3171-3175. 10.1073/pnas.052595099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararajan K. and Goley E. D. (2017). The intrinsically disordered C-terminal linker of FtsZ regulates protofilament dynamics and superstructure in vitro. J. Biol. Chem. 292, 20509-20527. 10.1074/jbc.M117.809939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararajan K., Miguel A., Desmarais S. M., Meier E. L., Huang K. C. and Goley E. D. (2015). The bacterial tubulin FtsZ requires its intrinsically disordered linker to direct robust cell wall construction. Nat. Commun. 6, 882-886. 10.1038/ncomms8281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararajan K., Vecchiarelli A., Mizuuchi K. and Goley E. D. (2018). Species- and C-terminal linker-dependent variations in the dynamic behavior of FtsZ on membranes in vitro. Mol. Microbiol. 110, 47-63. 10.1111/mmi.14081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwedziak P., Wang Q., Freund S. M. V. and Löwe J. (2012). FtsA forms actin-like protofilaments. EMBO J. 31, 2249-2260. 10.1038/emboj.2012.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwedziak P., Wang Q., Bharat T. A. M., Tsim M. and Löwe J. (2014). Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. Elife 3, e04601 10.7554/eLife.04601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi A., Welsh M. A., Marmont L. S., Lee W., Sjodt M., Kruse A. C., Kahne D., Bernhardt T. G. and Walker S. (2019). FtsW is a peptidoglycan polymerase that is functional only in complex with its cognate penicillin-binding protein. Nat. Microbiol. 4, 587-594. 10.1038/s41564-018-0345-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanbichler M. and Shapiro L. (2006). MipZ, a spatial regulator coordinating chromosome segregation with cell division in caulobacter. Cell 126, 147-162. 10.1016/j.cell.2006.05.038 [DOI] [PubMed] [Google Scholar]
- Toro-Nahuelpan M., Corrales-Guerrero L., Zwiener T., Osorio-Valeriano M., Müller F., Plitzko J. M., Bramkamp M., Thanbichler M. and Schüler D. (2019). A gradient-forming MipZ protein mediating the control of cell division in the magnetotactic bacterium Magnetospirillum gryphiswaldense. Mol. Microbiol. 112, 1423-1439. 10.1111/mmi.14369 [DOI] [PubMed] [Google Scholar]
- Tsang M.-J. and Bernhardt T. G. (2015). A role for the FtsQLB complex in cytokinetic ring activation revealed by an ftsL allele that accelerates division. Mol. Microbiol. 95, 925-944. 10.1111/mmi.12905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A., Banzhaf M., Gross C. A. and Vollmer W. (2011). From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 10, 123-136. 10.1038/nrmicro2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma A. and Young K. D. (2004). FtsZ collaborates with penicillin binding proteins to generate bacterial cell shape in Escherichia coli. J. Bacteriol. 186, 6768-6774. 10.1128/JB.186.20.6768-6774.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan S., Wickstead B., Gull K. and Addinall S. G. (2004). Molecular evolution of FtsZ protein sequences encoded within the genomes of archaea, bacteria, and eukaryota. J. Mol. Evol. 58, 19-29. 10.1007/s00239-003-2523-5 [DOI] [PubMed] [Google Scholar]
- Wagstaff J. M., Tsim M., Oliva M. A., García-Sanchez A., Kureisaite-Ciziene D., Andreu J. M. and Löwe J. (2017). A polymerization-associated structural switch in ftsz that enables treadmilling of model filaments. MBio 8, e00254-17 10.1128/mBio.00254-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L. J. and Errington J. (2004). Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell 117, 915-925. 10.1016/j.cell.2004.06.002 [DOI] [PubMed] [Google Scholar]
- Yahashiri A., Jorgenson M. A. and Weiss D. S. (2015). Bacterial SPOR domains are recruited to septal peptidoglycan by binding to glycan strands that lack stem peptides. Proc. Natl. Acad. Sci. USA 112, 11347-11352. 10.1073/pnas.1508536112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Lyu Z., Miguel A., McQuillen R., Huang K. C. and Xiao J. (2017). GTPase activity–coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science 355, 744-747. 10.1126/science.aak9995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Q., Jewett A. I., Chang Y. W., Oikonomou C. M., Beeby M., Iancu C. V., Briegel A., Ghosal D. and Jensen G. J. (2017). Short FtsZ filaments can drive asymmetric cell envelope constriction at the onset of bacterial cytokinesis. EMBO J. 36, 1577-1589. 10.15252/embj.201696235 [DOI] [PMC free article] [PubMed] [Google Scholar]