Abstract
Transmission in hospital settings of seasonal influenza viruses and novel agents such as the Middle East respiratory syndrome coronavirus (MERS-CoV) is well-described but poorly understood. The characterization of potentially infectious bio-aerosols in the healthcare setting remains an important yet ill-defined factor in the transmission of respiratory viruses. Empiric data describing the distribution of bio-aerosols enable discernment of potential exposure risk to respiratory viruses. We sought to determine the distribution of influenza virus RNA emitted into the air by participants with laboratory-confirmed influenza, and whether these emissions had the potential to reach healthcare workers’ breathing zones. Two-stage cyclone bio-aerosol samplers from the Centers for Disease Control and Prevention – National Institute for Occupational Safety and Health were placed 0.5–1.0 m (near field) and 2.1–2.5 m (far field) from infected patient participants, as well as in the corridor immediately outside their rooms. In addition, healthcare worker participants providing care to infected participants were recruited to wear a polytetrafluoroethylene (PTFE) filter cassette in their breathing zones. Viral RNA was detected from the air emitted by 37.5% of the 16 participants infected with influenza virus and distributed both in near and far fields and in all tested particle sizes (<1 µm, 1–4 µm, and >4 µm). Viral RNA was recovered in droplet nuclei and beyond 1 m from naturally-infected participants in the healthcare setting and from the breathing zone of one healthcare worker. There was no correlation between patient participant nasal viral load and recovery of viral RNA from the air, and we did not identify any significant association between RNA detection from the air and patient demographics or clinical presentation. A more substantial study is required to identify patient determinants of virus emission into the air and delineate implications for evidence-based policy for prevention and control.
Keywords: Bio-aerosols, exposure, healthcare worker, respiratory virus
Introduction
Patients with acute infectious respiratory illness may emit bio-aerosols containing pathogens capable of infecting susceptible hosts, including other patients and healthcare workers (HCWs). There is a lack of insight into the determinants of host bio-aerosol emission and spread of established and emerging pathogens. The importance of identifying means by which to control potentially infectious bio-aerosols was underscored by the severe acute respiratory syndrome-associated coronavirus (SARS-CoV) outbreak in 2003, where the burden of illness and death was largely borne by healthcare workers.[1,2] Part of the chaos which characterized the SARS-CoV epidemic can be attributed to a lack of knowledge regarding the route(s) of transmission of SARS-CoV and the appropriate use of personal protective equipment (PPE). Unfortunately, a similar scenario unfolded more recently in Saudi Arabia and South Korea where Middle East respiratory syndrome coronavirus (MERS-CoV) spread within healthcare institutions.[3,4] Also, the first case of confirmed MERS-CoV in North America was in a healthcare worker.[5] The attack rate for HCWs working in units caring for MERS patients was 8% in one study, with radiology technologists, respiratory therapists, and nurses being at highest risk; 90% of these individuals reported being within 2 m of an infected patient.[6] Extensive environmental contamination during the MERS-CoV outbreak in South Korea was demonstrated,[7] which may in part explain super-spreading events for MERS-CoV and SARS-CoV.[8]
Similarly, the annual spread of seasonal influenza virus within healthcare institutions poses an ongoing risk for patients, their families and healthcare workers. Approximately 17% of influenza cases are healthcare-associated, with 40% of these acquired in acute care settings.[9] Influenza virus RNA has been recovered from the breathing zones of 42% of participating emergency room workers, with nurses and nursing assistants experiencing the highest frequency of positivity, and viral RNA was detected from the personal breathing zones of all ten of the participant nurses involved in aerosol-generating procedures.[10] Influenza virus RNA has also been recovered from ambient air in acute care units and outpatient clinics, underscoring the potential for exposure across healthcare systems.[10–14]
The conventional paradigm is to classify respiratory pathogen transmission as droplet vs. airborne, with clear policies and procedures for each purported mode of transmission. Where there is doubt, both droplet and airborne precautions are generally employed. Large respiratory droplets are >10 µm in diameter and are involved in short-range (≤2 m) droplet spread. Droplet nuclei are ≤5 µm and are responsible for short- or long-range (>2 m) airborne transmission;[15] these respirable particles are small enough to be inhaled into the alveoli. The relative contribution of each route to overall transmission of influenza is unknown, leading to debate regarding the important mode(s) of transmission and appropriate means of transmission prevention.[16,17] In this paper, we use the term bio-aerosol to denote particles of any size presumed to originate from the respiratory tract.
Our primary goal was to determine the distribution of viral RNA by particle size and distance in an inpatient setting and establish whether influenza virus RNA could be recovered from HCWs’ breathing zones. We also sought to identify host determinants for the recovery of viral RNA from the air.
Methods
Participants
Participants were identified through daily review of respiratory virus testing logs from December 1, 2015–April 30, 2016, in a combined 900-bed acute care teaching hospital and a 500-bed long-term care facility. As part of routine care, mid-turbinate (MT) swabs (Copan, Murrieta, CA, USA) were collected from individuals with fever and either cough and/or sore throat, and (a) requiring admission to hospital or (b) are residents in long-term care. These were submitted for analysis using the xTAG RVP FAST v2 (Luminex, Toronto, ON, Canada) multiplex PCR assay for respiratory viruses, which includes targets for influenza A (subtypes H1N1 and H3N2) and B viruses. Inclusion criteria included laboratory-confirmed influenza with PCR assay median fluorescence intensity (MFI) of >500, <48 hr since MT swab obtained, and ability to consent. The minimum MFI was arbitrarily set at >500 to optimize air sampling yield, given the limited number of instruments and personnel available. Clinical data pertaining to past medical history, history of presenting illness, management, and outcomes were collected through chart review and patient interview.
In addition, nurses (HCW participants) who provided direct care to recruited individuals with laboratory-confirmed influenza (patient participants) were approached. HCW participants were excluded if they reported current upper respiratory tract symptoms, and MT swabs were obtained to exclude contamination of air samples from asymptomatic shedding. This study was conducted with the approval of the Sunnybrook Human Research Protections Program’s Research Ethics Board and informed consent was obtained from patient and HCW participants.
Air sampling
Patients were in single-patient rooms and droplet precautions (routine practices, use of gloves, gowns, eye protection, and procedural or surgical masks and daily cleaning of high-touch surfaces) were in place as per institutional protocol. Air handling was through isolated heating, ventilation and cooling (HVAC) systems by unit location, all of which were HEPA-filtered and maintained on regular filter changes. These provided a minimum of four to six air exchanges per hour, with variable proportions of recycled air depending on unit location. Negative pressure, isolated HEPA-filtration and increased air exchange were not routinely applied. Area air samples in proximity to patients were collected using the NIOSH two-stage cyclone bio-aerosol sampler (National Institute for Occupational Safety and Health, Morgantown, WV, USA).[18] This instrument consists of stages which collect particles >4 µm into 15 mL conical tubes (stage 1) or 1–4 µm into 1.5 mL conical tubes (stage 2); particles <1 µm were recovered on a 3.0 µm pore size, 37 mm, polytetrafluoroethylene (PTFE) membrane filter (Millipore, Etobicoke, ON, Canada) housed in a three-piece cassette. Prior to each use, two-stage cyclone sampling devices were connected to battery-powered pumps (SKC Inc., Eighty-Four, PA, USA) and calibrated to an average flow rate of 3.5 L/min ± 5% using the BIOS DC-1 DryCal flow calibrator (SKC Inc., Eighty-Four, PA, USA).
The instruments were then positioned on a cart at distances of 0.5–1 m and 2.1–2.5 m from the participant’s head, and in the corridor immediately outside their rooms; the height range was 0.8–0.85 m above the floor (bedside level). During the 2-hour collection period there were no restrictions on patient, visitor or HCW activity. Two-stage cyclone samplers were then placed in coolers for transportation to the on-site laboratory and processed immediately as described below. Postsampling, pump flow rates were verified again to ensure they did not exceed 5% of the target. Percent relative humidity (%RH) and temperature were recorded at each sampler location (at 0.5–1 m and 2.1–2.5 m from participants, and in the corridor) at the beginning of the sampling period. In addition, negative air sampling controls were taken near the end of influenza season (March–June ending and November beginning, 2016) on three inpatient units where sampling had occurred during influenza season: a total of six air samples were collected in the corridor using the NIOSH bio-aerosol sampler (N = 5) and PTFE cassette (N = 1) at 3.5 L/min ± 5% for 2 hr. One more air sample was collected within our laboratory using the PTFE cassette.
The HCW breathing zone air samples were collected using 1.0 µm pore size, 37 mm, PTFE cassettes (SKC Inc., Eighty-Four, PA, USA) clipped to participants’ collars and attached to a calibrated portable battery-powered pump (GilAir, Sensidyne, St. Petersburg, FL, USA) worn in a back or hip pack. Sampling was performed at a nominal flow rate of 3.0 L/min ± 5% for up to 4 hr during the HCW’s shift. During this time, HCWs performed their usual assigned duties inside and outside participants’ rooms, including providing care to other patients. At the end of the sampling period, HCWs completed a brief questionnaire estimating the time spent in the room, presence during aerosol-generating procedures, and caring for other patients with acute respiratory symptoms. The PTFE cassettes were placed in coolers at the end of the sampling period and processed on-site by research personnel the same evening or following morning as described in the following sections.
PCR
Viral RNA was eluted from stage 1, stage 2, and the filter of the two-stage cyclone sampler by vortexing for 1 min with 1 mL, 0.5 mL, and 2 mL of viral transport medium (DMEM F12 with 0.5% BSA and 1X penicillin-streptomycin) respectively, and stored at −80 °C.[18,19] Total RNA from 200 µL of MT swabs and air samples was extracted using the MagMax Viral RNA Isolation Kit (ThermoFisher Scientific, Mississauga, ON, Canada).[18] One-step qRT-PCR was carried out using the Superscript III Platinum One-Step qRT-PCR kit (ThermoFisher Scientific, Mississauga, ON, Canada) to quantify influenza A and influenza B virus RNA on the ABI 7500 FAST by absolute quantitation (ThermoFisher Scientific, Mississauga, ON, Canada); RNA transcripts were utilized to develop standards for the curve.[20,21] The 25-µL reaction volume contained 2.5 µL extracted RNA template, 1 µM primer, and 0.1 µM probe. The following cycling conditions were used: reverse transcription at 50 °C for 30 min, initial denaturation at 95 °C for 2 min, followed by 45 amplification cycles at 95 °C for 15 sec and 55 °C for 1 min, and final extension at 72 °C for 5 min. Targets for influenza A and B viruses were to the matrix gene and nucleoprotein gene respectively. Influenza A virus primers and probe sequences were: forward 5′-GGAATGGCTAAAGACAAGACCAAT-3′, reverse 5′-GGGCATTTTGGACAAAGCGTCTAC-3′ and probe 5′-AGTCCTCGCTCACTGGGCACGGTG-3′.[22] Influenza B virus primers and probe sequences were: forward 5’-TCCTCAACTCACTCTTCGAGCG-3′, reverse 5’-CGGTGCTCTTGACCAAATTGG-3′ and probe 5′-CCAATTCGAGCAGCTGAAACTGCGGTG-3′.[23] The limit of detection of this assay was 3.56 copies/µL and 1.01 copies/µL for influenza A and B viruses, respectively, with limits of quantitation of 10 copies/µL and 50 copies/µL and efficiencies of 98.82% and 101.5%. PCR positivity for influenza virus was based on a threshold cycle (CT) of ≤40, with quantitation provided for CT ≤37 in the presence of robust amplification curves. Viral loads from MT swabs and air samples were recorded as log10 copies/mL and copies/L of air sampled respectively. RNAse P was used as a reference gene for the former; absolute quantitation was used for the latter in the absence of a reference gene for air. RNase P qRT-PCR was performed as a multiplex under the same cycling conditions as described for influenza A and B viruses. RNase P primer and probe sequences were: forward 5′-AGATTTGGACCTGCGAGCG-3′, reverse 5′-GAGCGGCTGTCTCCACAAGT-3′ and probe 5′-TTCTGACCTGAAGGCTCTGCGCG-3′.[24] Air volumes were calculated as the product of sampling time and the calibrated flow rate.
Statistical analysis
Dependent variables were identified as the MT swab viral load (log10 copies/mL) and detection of viral RNA from one or more participant air samples (positive) vs. all negative air samples. The Wilcoxon-Mann-Whitney, Kruskal Wallis tests and Spearman correlation were used to assess the presence of statistically significant correlations or distributions between independent variables and the MT swab viral load, while the Wilcoxon-Mann-Whitney and Fisher’s exact tests were used for air samples. Statistical analysis was performed using SAS University Edition (SAS Institute, Cary, NC, USA).
Results
Sixteen patients and seven HCWs were recruited between January 14 and April 27, 2016. Most cases of influenza were community-acquired. The median length of stay was 7 days, and the mean number of days was 15.5 (range 3–56) days. Two patients were admitted to intensive care, one of whom required mechanical ventilation. No deaths were attributed to influenza (Table 1).
Table 1.
Demographic, clinical, and outcome data for hospitalized patients with laboratory confirmed influenza based on the recovery of influenza virus RNA from the air.
| Negative air* (n = 10) | Positive air (n = 6) | Overall (n = 16) | P-value | |
|---|---|---|---|---|
| Age-years, median (range) | 58 (49–92) | 68 (28–80) | 58.5 (28–92) | 0.91 |
| Sex, n male (%) | 4 (40.0) | 1 (16.7) | 5 (31.3) | 0.59 |
| Number of symptoms, mean (SD) | 7 (2.5) | 9 (2.5) | 8 (2.5) | 0.34 |
| Documented fever, n (%) | 6 (60.0) | 6 (100.0) | 12 (75) | 0.23 |
| Pharyngitis, n (%) | 2 (20.0) | 3 (50.0) | 5 (31.3) | 0.30 |
| Need for O2 therapy, n (%) | 5 (50.0) | 4 (66.7) | 9 (56.3) | 0.63 |
| Influenza vaccination, n (%)† | 5 (55.6) | 3 (50.0) | 8 (53.3) | 1.0 |
| Past or current smoker, n (%)† | 6 (66.7) | 3 (50.0) | 9 (60) | 0.62 |
| Chest X-ray changes, n (%) | 4 (40.0) | 4 (66.7) | 8 (50) | 0.61 |
| Mid-turbinate swab viral load, log10 copies/mL mean (SD)‡ | 3.8 (2.7) | 4.8 (1.6) | 4.1 (2.4) | 0.54 |
| Community-acquired influenza, n (%) | 7 (70.0) | 4 (66.7) | 11 (68.8) | 1.0 |
| Influenza A virus, n (%) | 7 (70.0) | 6 (100) | 13 (81.3) | 0.25 |
| H1N1, n (%) | 5 (50.0) | 4 (66.7) | 9 (56.3) | 0.63 |
| H3N2, n (%) | 2 (20.0) | 2 (33.3) | 4 (25) | 0.60 |
| Influenza B virus, n (%) | 3 (30.0) | 0 (0) | 3 (18.8) | 0.25 |
| Length of stay – days, mean (range) | 15.3 (3–56) | 15.8 (3–47) | 15.5 (3–56) | 0.79 |
| Admission to ICU, n (%) | 2 (20.0) | 0 (0) | 2 (12.5) | 0.5 |
Group includes 9 patients with negative air samples and 1 with insufficient quantity for PCR.
n = 15; unable to complete interview for one participant in negative air group.
n = 14; insufficient sample for MT viral load for two participants with positive air samples.
Four and nine patient participants were infected with influenza A(H3N2) and influenza A(H1N1) viruses respectively; three participants were infected with influenza B virus. There was insufficient MT sample for quantification from one patient with influenza A(H1N1) virus and one patient with influenza B virus. The mean and median log10 copies/mL for MT swab viral load in patients with available samples was 4.08 (SD 2.39) and 4.13 (IQR 2.93–6.08) respectively. In patients with positive influenza A virus swabs, the mean and median log10 copies/mL for MT swab viral load were 4.71 (SD 2.21) and 4.83 (IQR 2.98–6.12). In patients with swabs positive for influenza A(H1N1) virus, the mean and median MT swab viral loads were 4.79 (SD 2.37) and 4.83 (IQR 3.85–6.12) log10 copies/mL. The mean and median MT swab viral load in patients with influenza A(H3N2) were identical at 4.32 (SD 1.97, IQR 2.93–5.71) log10 copies/mL. Finally, the mean and median MT swab viral load in patients with positive influenza B virus swabs was 1.78 (SD 1.60) and 2.24 (IQR 0–3.11) log10 copies/mL. There was no statistically significant difference between MT viral load and age, sex, number of symptoms, pharyngitis, need for oxygen, vaccination status, smoking status, or chest X-ray changes. However, there was a statistically significant association between higher viral load and fever (p-value 0.032).
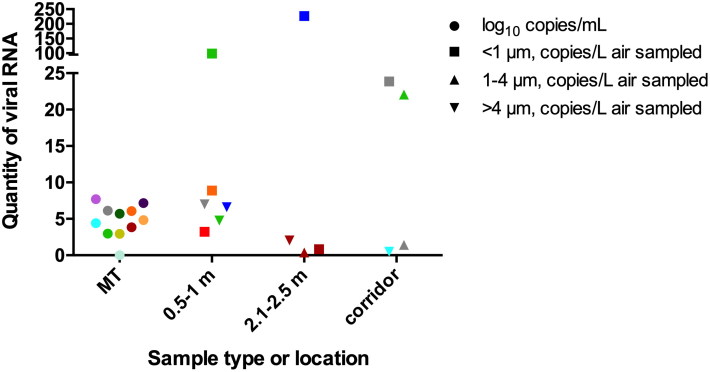
Four participants were sampled in the emergency department, and 12 on inpatient units, including one in critical care. Influenza virus RNA was recovered from one or more room air samples from 6 of 13 (46%) participants with influenza A (4 of 9 H1N1, 2 of 4 H3N2) in at least one of the three size ranges (<1 µm, 1–4 µm and >4 µm), at <1 m in five cases, >2 m in two cases, and from the corridor in three cases (Figure 1). In two of these, there was insufficient quantity for repeat PCR for all the ranges/distances, one of which included a small quantity of RNA detected in the corridor only; for the statistical analysis this was included in the negative air sample group, although it is included in Figure 1. No detectable viral RNA was recovered from air emitted by the three participants with influenza B virus infection. Average percent relative humidity (%RH) was 21.3% (SD 7.9, range 11.0–40.9%) and average temperature was 23.8 °C (SD 1.4, range 20.3–27.9 °C). There was no correlation between patient demographic or clinical characteristics and air sample results (Table 1). There was also no association between median relative humidity and bio-aerosol positivity.
Figure 1.
Colors indicate individual patient participants with laboratory-confirmed influenza and include viral RNA load from mid-turbinate (MT) swabs (log10 copies/mL), denoted in circles, and from air sampled at 0.5–1 m (<1 m), 2.1–2.5 m (>2 m) and the corridor immediately outside participants’ rooms (copies/L air sampled). Squares represent the viral RNA copies per liter of air of particles <1 µm in size, upright triangles represent viral RNA copies per liter of air of particles 1–4 µm in size and upside down triangles represent viral RNA copies per liter of air of particles >4 µm in size. For two patients with positive air samples at <1 m (red and blue), the original MT sample was not available. Thus, there are no corresponding data points for MT viral load.
Seven HCW participants were recruited, three of whom estimated they spent 30 min or less in patient-participants’ rooms, three estimated between 30 and 60 min spent, and one who estimated more than an hour was spent in the room with their patient participant while wearing the portable sampler. No HCW participants were present during an aerosol-generating procedure, and two cared for other patients with acute respiratory symptoms. Influenza A(H1N1) viral RNA was recovered from the breathing zone of one of seven HCWs providing care to a participant with influenza A(H1N1) virus infection. Particle size is unknown since the PTFE filters collect particles <40 µm and do not otherwise size-fractionate. This individual was not caring for another patient with symptoms, and estimated spending less than 30 min in their patient participant’s room; viral RNA was not recovered from air emitted by this patient participant by area sampling. All mid-turbinate swabs from participating HCWs were negative for respiratory viruses by the xTAG RVP FAST v2. Viral RNA was undetectable from all negative control air samples and there were no issues with pump flow rate noted on post-sampling re-calibration.
Discussion
We determined that 37.5% of participants presenting to an acute care facility with laboratory-confirmed influenza emitted viral RNA into the air. Viral RNA was detected in particles recovered from all size ranges of <1.0 µm, 1.0–4.0 µm, and >4 µm, with the majority emitted in the near field (0.5–1.0 m) of the participants. RNA was not detected from aerosol samples collected from participants with influenza B virus infection; however, this sample size was very small. The positivity rate of emitted aerosols from this study is similar to what was found by others,[12,25] although less than our previous study.[19] Killingley et al. also recovered viral RNA in all size ranges using the NIOSH cyclone sampler,[25] although others have not recovered RNA in the submicron range using the same instrument.[14] In this study, there was a significant association between fever and viral load from mid-turbinate swabs, but not between participant attributes, including nasal viral load, and detection of influenza virus RNA from the emitted air. Viral RNA was detected in one of seven HCW’s breathing zones after a 4-hr period of providing routine care to a participant with influenza.
Understanding respiratory pathogen transmission via bio-aerosols continues to be a pressing issue for endemic viruses such as seasonal influenza viruses, as well as novel agents such as avian influenza A(H7N9) and MERS-CoV. Spread in hospital settings of seasonal and pandemic influenza viruses and MERS-CoV may result in substantial amplification of transmission, illness, and death among patients as well as healthcare workers.
Several studies involving human subjects have revealed that most of influenza virus RNA was detected in smaller (<5 µm) aerosolized particles[26,27] and that these aerosols were present in the healthcare setting.[13] Infected subjects generated more particles per cough during infection compared to convalescence, and a substantial proportion of these particles were respirable (<5 µm).[28] In another study, 92% vs. 43% of naturally-infected volunteers with laboratory-confirmed influenza shed virus in fine (<5 µm) vs. coarse (>5 µm) exhaled particles.[29] In a study of infected volunteers, 53% and 42% emitted aerosols containing viable influenza virus during coughing and quiet breathing respectively,[30] and viable virus was isolated from the smallest particle fraction (0.3–8 µm).[31]
In the largest bedside air sampling study to date (N = 61), Bischoff et al.[12] determined that nasal viral load correlated with emission of viral RNA into the air (p = 0.01), particularly with expulsion events such as coughing or sneezing, and emitters in the emergency department reported greater severity of illness (p = 0.05) interfering with activities of daily living (p = 0.02). This study also demonstrated the presence of RNA-bearing droplet nuclei, which were more abundant in the near field compared to the far field (≤0.305–0.914 m vs. 1.829 m).[12] In another study examining the emissions of otherwise healthy college students, determinants of coarse and fine particle shedding included frequency of cough and body mass index.[32] Infectious virus was recovered from 39% of samples after exhaling for 30 min into the Gesundheit-II human source bio-aerosol sampler and up to 103 focus counts/sample were recovered.
We determined whether the median relative humidity correlated with air sampling results and found that there was no statistical significance, although the sample size was limited, impeding the ability to reach robust conclusions. In a similar study conducted in a two-bed isolation hospital room, relative humidity was continuously monitored over the duration of the sampling period and no correlation between RH and the total concentration of influenza virus RNA in the air was found, although when an outlier was removed then a positive association was observed.[14]
In addition to the small sample size, the current study has several limitations. The substantial variability in patient emissions of viral RNA compounded the limited sample size, further impeding the possibility of identifying significant associations. Individual host differences and sampling during differing points in patients’ clinical course depending on time of presentation likely contributed to this variability, despite sampling patients within a strict time frame relative to presentation to hospital. The time from onset of symptoms varied and was not included in the analysis. We recovered viral RNA from the breathing zone of one of seven participating HCWs, but did not record the specific activities undertaken during the sampling period; it is unknown exactly how much time HCW participants spent in close proximity with patient participants. Similarly, patient participant activities were not monitored and we did not record expulsion events such as coughing. No restrictions on visitors were placed during the sampling period, thus the possibility that household members or other visitors may have contributed viral RNA to the air is not excluded. We did not record the presence of other patients with influenza on the unit, or illness in staff, which may have contributed to detection of viral RNA, particularly in the corridor. Exact air exchanges/hour, ventilation direction, and flow rates were not determined and may have affected the particle distribution in the room and corridor. Also, although patient participants were on droplet precautions, consistency of door closure was not noted. Although these are important limitations, data collected under these uncontrolled conditions reflect that of the real world where other patient and HCW exposures actually occur. As with the majority of studies of this nature, we detected viral RNA, which is not a definitive indication of potential risk of infection after exposure. Experimental studies suggest that viable virus concentrations are several-fold lower than detected RNA copy concentrations.[33] In the clinical setting, differences may be due to technical or mechanical factors related to sampling and disruption of the viral envelope, or may be attributable to other biological factors, including the shedding of defective interfering viral particles (DI particles).[34] The role of DI particles in influenza virus transmission remains unclear, though there is evidence for a role in the transmission of other viruses.[35] As it stands, demonstration of infectious virus from aerosol samples remains the most convincing evidence for transmission potential. Even if viral RNA were to become a reliable indicator of exposure, translating these results to manage and reduce risk requires significantly more research, since little is known about transmission potential for given quantities of virus shed into the air.[36] Finally, this study represents a single center during a single influenza season. Although the sampling window included the influenza peak, the bulk of the sampling was done in February. Whilst H3N2 viruses predominated early in this particular season, H1N1 viruses emerged later and predominated the entire season. This is reflected in Table 1 showing that the majority of recruited participants were infected with H1N1 virus.
Delays in identifying potential emitters presenting for acute care and noncompliance with infection prevention and control precautions may pose significant risks to HCWs providing direct patient care. In one risk analysis, between 34,150 and 151,300 occupationally-acquired cases of influenza are estimated to occur annually in the United States.[37] Inconsistent application of infection prevention and control measures by both HCWs and patients has been studied, and there are opportunities for improvement among these groups, as well as from an administrative perspective.[38–40] The recovery of viral RNA in droplet nuclei and beyond 1 m from naturally-infected participants in the healthcare setting invites a concerted effort to address the issues of (a) virus viability and potential risk with exposure and (b) determinants of emission. The former poses methodological and technical challenges whereas the latter requires a collaborative, multidisciplinary and multicenter undertaking. A multicenter, multiseason study is required to determine the clinical, environmental, and viral factors associated with emission and dispersion of influenza virus. These issues can and should be addressed in order to generate the empiric data required to enhance evidence-based infection prevention and control and occupational health policies.
Funding Statement
We are grateful for support from the Ontario Ministry of Labour (grant number 13-R-052).
Acknowledgments
We are grateful for support from the hospital staff who enabled and participated in this study. We are also grateful to William Lindsley (NIOSH) for the use of the two-stage cyclone bio-aerosol samplers. The authors have no conflicts of interest to declare.
References
- 1.Koh D., Lim M.K., and Chia S.E.: SARS: Health care work can be hazardous to health. Occup. Med. (Lond) 53(4):241–243 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lim M.K., and Koh D.: SARS and occupational health in the air. Occup. Environ. Med. 60(8):539–540 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oboho I.K., Tomczyk S.M., Al-Asmari A.M., et al. : 2014 MERS-CoV outbreak in Jeddah—A link to health care facilities. N. Engl. J. Med. 372(9):846–854 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho S.Y., Kang J.M., Ha Y.E., et al. : MERS-CoV outbreak following a single patient exposure in an emergency room in South Korea: An epidemiological outbreak study. Lancet 388(10048):994–1001 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bialek S.R., Allen D., Alvarado-Ramy F., et al. : First confirmed cases of Middle East respiratory syndrome coronavirus (MERS-CoV) infection in the United States, updated information on the epidemiology of MERS-CoV infection, and guidance for the public, clinicians, and public health authorities - May 2014. MMWR Morb. Mortal. Wkly. Rep. 63(19):431–436 (2014). [PMC free article] [PubMed] [Google Scholar]
- 6.Alraddadi B.M., Al-Salmi H.S., Jacobs-Slifka K., et al. : Risk factors for Middle East Respiratory Syndrome Coronavirus infection among healthcare personnel. Emerg. Infect. Dis. 22(11):1915–1920 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim S.H., Chang S.Y., Sung M., et al. : Extensive viable Middle East Respiratory Syndrome (MERS) coronavirus contamination in air and surrounding environment in MERS isolation wards. Clin. Infect. Dis. 63(3):363–369 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chowell G., Abdirizak F., Lee S., et al. : Transmission characteristics of MERS and SARS in the healthcare setting: A comparative study. BMC Med. 13(1):210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor G., Mitchell R., McGeer A., et al. : Healthcare-associated influenza in Canadian hospitals from 2006 to 2012. Infect. Control. Hosp. Epidemiol. 35(2):169–175 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Rule A.M., Apau O., Ahrenholz S.H., et al. : Healthcare personnel exposure in an emergency department during influenza season. PLoS One 13(8):e0203223 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahrenholz S.H., Brueck S.E., Rule A.M., et al. : Assessment of environmental and surgical mask contamination at a student health center — 2012-2013 influenza season. J. Occup. Environ. Hyg. 15(9):664–675 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bischoff W.E., Swett K., Leng I., and Peters T.R.: Exposure to influenza virus aerosols during routine patient care. J Infect Dis 207(7):1037–1046 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Lindsley W.G., Blachere F.M., Davis K.A., et al. : Distribution of airborne influenza virus and respiratory syncytial virus in an urgent care medical clinic. Clin. Infect. Dis. 50(5):693–698 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Leung N.H., Zhou J., Chu D.K., et al. : Quantification of influenza virus RNA in aerosols in patient rooms. PLoS One 11(2):e0148669 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bridges C.B., Kuehnert M.J., and Hall C.B.: Transmission of influenza: Implications for control in health care settings. Clin. Infect. Dis. 37(8):1094–1101 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Brankston G., Gitterman L., Hirji Z., Lemieux C., and Gardam M.: Transmission of influenza A in human beings. Lancet Infect. Dis. 7(4):257–265 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Tellier R.: Review of aerosol transmission of influenza A virus. Emerg. Infect. Dis. 12(11):1657–1662 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao G., Noti J.D., Blachere F.M., Lindsley W.G., and Beezhold D.H.: Development of an improved methodology to detect infectious airborne influenza virus using the NIOSH bioaerosol sampler. J. Environ. Monit. 13(12):3321–3328 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mubareka S., Granados A., Naik U., et al. : Influenza virus emitted by naturally-infected hosts in a healthcare setting. J. Clin. Virol. 73:105–107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.World Health Organization : “Emergencies preparedness, response.” Available at http://www.who.int/csr/resources/publications/swineflu/realtimeptpcr/en/. (accessed August 4, 2017).
- 21.Granados A., Petrich A., McGeer A., and Gubbay J.B.: Measuring influenza RNA quantity after prolonged storage or multiple freeze/thaw cycles. J. Virol. Methods 247:45–50 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Lee H.K., Loh T.P., Lee C.K., Tang J.W., Chiu L., and Koay E.S.: A universal influenza A and B duplex real-time RT-PCR assay. J. Med. Virol. 84(10):1646–1651 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Selvaraju S.B., and Selvarangan R.: Evaluation of three influenza A and B real-time reverse transcription-PCR assays and a new 2009 H1N1 assay for detection of influenza viruses. J. Clin. Microbiol. 48(11):3870–3875 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu X., Holloway B., Dare R.K., et al. : Real-time reverse transcription-PCR assay for comprehensive detection of human rhinoviruses. J. Clin. Microbiol. 46(2):533–539 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Killingley B., Greatorex J., Digard P., et al. : The environmental deposition of influenza virus from patients infected with influenza A(H1N1)pdm09: Implications for infection prevention and control. J. Infect. Public Health 9(3):278–288 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Lindsley W.G., Blachere F.M., Thewlis R.E., et al. : Measurements of airborne influenza virus in aerosol particles from human coughs. PLoS One 5(11):e15100 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fabian P., McDevitt J.J., DeHaan W.H., et al. : Influenza virus in human exhaled breath: An observational study. PLoS One 3(7):e2691 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindsley W.G., Pearce T.A., Hudnall J.B., et al. : Quantity and size distribution of cough-generated aerosol particles produced by influenza patients during and after illness. J. Occup. Environ. Hyg. 9(7):443–449 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milton D.K., Fabian M.P., Cowling B.J., Grantham M.L., and McDevitt J.J.: Influenza virus aerosols in human exhaled breath: Particle size, culturability, and effect of surgical masks. PLoS Pathog. 9(3):e1003205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindsley W.G., Blachere F.M., Beezhold D.H., et al. : Viable influenza A virus in airborne particles expelled during coughs versus exhalations. Influenza Other Respir. Viruses 10(5):404–413 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindsley W.G., Noti J.D., Blachere F.M., et al. : Viable influenza A virus in airborne particles from human coughs. J. Occup. Environ. Hyg. 12(2):107–113 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan J., Grantham M., Pantelic J., et al. : Infectious virus in exhaled breath of symptomatic seasonal influenza cases from a college community. Proc. Natl. Acad. Sci. USA. 115(5):1081–1086 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown J.R., Tang J.W., Pankhurst L., et al. : Influenza virus survival in aerosols and estimates of viable virus loss resulting from aerosolization and air-sampling. J. Hosp. Infect. 91(3):278–281 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Saira K., Lin X., DePasse J.V., et al. : Sequence analysis of in vivo defective interfering-like RNA of influenza A H1N1 pandemic virus. J. Virol. 87(14):8064–8074 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ke R., Aaskov J., Holmes E.C., and Lloyd-Smith J.O.: Phylodynamic analysis of the emergence and epidemiological impact of transmissible defective dengue viruses. PLoS Pathog. 9(2):e1003193 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang J.W., Hoyle E., Moran S., and Pareek M.: Near-patient sampling to assist infection control — A case report and discussion. Int. J. Environ. Res. Public Health 15(2):238 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones R.M., and Xia Y.: Annual burden of occupationally-acquired influenza infections in hospitals and emergency departments in the United States. Risk. Anal. 38(3)442–453 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Turnberg W., Daniell W., Seixas N., et al. : Appraisal of recommended respiratory infection control practices in primary care and emergency department settings. Am. J. Infect. Control. 36(4):268–275 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.May L., Lung D., and Harter K.: An intervention to improve compliance with transmission precautions for influenza in the emergency department: successes and challenges. J. Emerg. Med. 42(1):79–85 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Longtin Y., Akakpo C., Rutschmann O.T., Pittet D., and Sax H.: Evaluation of patients' mask use after the implementation of cough etiquette in the emergency department. Infect. Control. Hosp. Epidemiol. 30(9):904–908 (2009). [DOI] [PubMed] [Google Scholar]