BloodPAC is a public–private consortium that develops standards and best practices, organizes and coordinates research studies through its members, and operates a data commons to support the liquid biopsy research community. Data from the studies it organizes are contributed to the BloodPAC Data Commons. BloodPAC developed recommendations for 11 preanalytical attributes called the Minimum Technical Data Elements (MTDEs) that are recommended for studies that it sponsors and for data contributed to the BloodPAC Data Commons.
Background
Liquid biopsies are samples of nonsolid biospecimens, such as blood, that may be used for molecular or cellular analysis. These biospecimens offer a number of important clinical benefits relative to more traditionally obtained single‐site biopsies. First they are safer. Second, they are more likely to be representative of molecular alterations present from multiple metastatic sites. Third, involves the ease of acquisition on a repeated basis to monitor disease over time with limited patient risk. As a result, this mode of sample acquisition and analysis has become a top priority of diagnostic and pharmaceutical companies, who are looking for translational approaches focused on the development of liquid biopsy biomarkers to guide treatment selection, assess treatment efficacy, and understand mechanisms of acquired resistance after an initial response to therapy.
Additional advantages of a liquid biopsy‐based testing approach include: specimen availability within a routine clinical practice setting, the ability to control most, if not all, preanalytical steps, and the potential for short turnaround times to inform medical decision making.
One of the principles adopted by the BloodPAC members was that not only should the consortium be data driven, but that it should create a data resource for the liquid biopsy community. BloodPAC has developed just such a resource called the BloodPAC Data Commons, which follows the principles that its data be FAIR (findable, attributable, interoperable, and reusable).
An analysis of the data contributed by members to the BloodPAC Data Commons during late 2016 and early 2017 played an important role in motivating the members to initiate the effort to develop the minimum technical (preanalytical) data elements (MTDEs) for any data submitted to the commons. With the focus being on standardization of fields, BloodPAC undertook the critical task of identifying and selecting the MTDEs for the preanalytical variables most commonly associated with cell‐free DNA (cfDNA) test design and development.
BloodPAC believes that if it is to succeed in its mandate, it will need to provide all test developers, including translational researchers in academia, pharmaceutical and diagnostic testing centers, regulators, pathologists, and clinicians, with guidance regarding factors that influence the performance of the assay itself in the laboratory that may affect final assay result. These include defining the factors that may affect the limit of detection and the variables required to ensure reproducibility/repeatability for each phase or aspect of test development.
This begins with the definition of preanalytical variables and continues to analytical variables and the patient context variables that will drive clinical validation.
This paper describes the steps taken by the BloodPAC Preanalytical Working Group to develop a comprehensive list of preanalytical variables relevant to cfDNA‐based tests. This list was developed with input from all BloodPAC members. By leveraging the strength of the diverse BloodPAC membership and in collaboration with the US Food and Drug Administration's (FDA’s) Center for Devices and Radiological Health (CDRH) and the College of American Pathologists’ (CAP's) Preanalytics for Precision Medicine Project team, we aligned on a list of 11 preanalytical MTDEs. Use of these MTDEs by investigators and researchers in the field will enable standardization of data input into the BloodPAC Data Commons, which is necessary to enable cross‐assay comparisons and other joint analysis of its data by its members. We describe the process BloodPAC used for selecting preanalytical variable MTDEs, along with the final list of 11 preanalytical MTDEs, with the hope that the research community embraces these standards for robust cfDNA assay development.
Preanalytical MTDE for Liquid Biopsies
Version 2.0 of the preanalytical MTDEs are listed in Table 1. These MTDEs were approved by the BloodPAC Consortium on September 26, 2017, received FDA input on November 3, 2017, and were approved by CAP on June 6, 2018. Figure 1 contains a graphical summary of the MTDEs.
Table 1.
A summary of the minimum technical data elements (MTDE)
| # | Data element | Data model element | Type | Description |
|---|---|---|---|---|
| 1 | Blood collection tube type | blood_tube_type |
Controlled vocabulary from:
|
The kind of tube used to collect the sample(s) taken from a biological entity for testing, diagnostic, propagation, treatment, or research purposes. |
| 2 | Sample composition | Composition |
Controlled vocabulary from:
|
Sample type describing the cellular composition of the sample, as specified from a controlled vocabulary, containing clinical, contrived, and other terms. |
| 3 | Shipping temperature | shipping_temperature | Float | The temperature, in centigrade, at which the biospecimen was kept while it was being transported from the procurement site to its processing destination. |
| 4 | Blood fractionalization method | blood_fractionation_method | String | The name or description of the method used to obtain the blood fraction sample. (e.g., Ficoll Method, Novartis Protocol #001, 2,000 g centrifuge at 4°C with gentle deceleration). Alternatively, if you have provided a detailed protocol, enter its file_name here. |
| 5 | Time to fractionation | hours_to_fractionation_upper, hours_to_fractionation_lower – |
|
The upper/lower limit on the amount of time, in hours, between the blood draw and the fractionation into its components. If the exact time is known, make this value equal to that of the lower limit. If the time is completely unknown, enter Unknown. If no fractionation was performed on this sample, enter Not Applicable. |
| 6 | Analyte isolation method | analyte_isolation_method | String | The name or general description of the method used to isolate the analyte. Alternatively, if you have provided a protocol, put the file_name here. |
| 7 | Time to freezer | hours_to_freezer_upper, hours_to_freezer_lower |
|
The upper/lower limit on the amount of time, in hours, that it took between the sample being fractionated and the aliquot being frozen or otherwise preserved. If the exact time is known, make this value equal to that of the lower limit. If the time is completely unknown, enter Unknown. If no fractionation was performed on this sample, enter Not Applicable. |
| 8 | Storage temperature | storage_temperature | Float | The temperature, in centigrade, at which the aliquot was preserved and/or stored. |
| 9 | Concentration: cellular concentration or molecular concentration | molecular_concentration or cellular_concentration | Float | If the analyte is a molecule (e.g., DNA or RNA), report the observed concentration in nanograms per microliter (for molecular concentration). If the measurement is a cell count, then this is reported as cells per microliter (cellular_concentration) |
| 10 | Assay method | assay_method |
Controlled vocabulary from:
|
General name or description of the method used to characterize the analyte. |
| 11 | Time to assay | days_to_assay | Integer | The amount of time, in days, between the date used for index and the assay used to address this analyte. |
Figure 1.
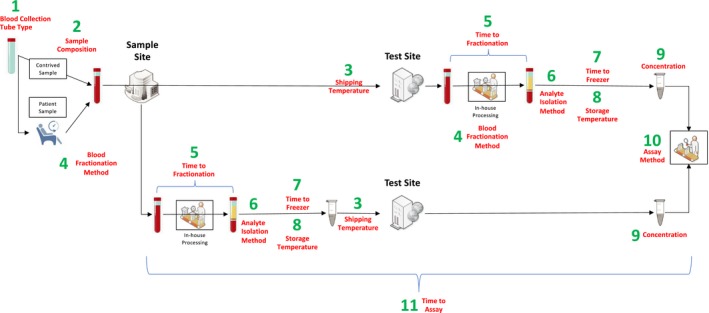
A graphical workflow view of the 11 Minimum Technical Data Elements (MTDEs).
Fifty‐two data elements that are specific to cfDNA‐based tests were discussed by the BloodPAC Consortium, including 26 preanalytical data elements that are relevant for this paper. The 11 data elements in Table 1 were consistently ranked by BloodPAC members as “important and required.” Other categories included “important and useful,” if the consortium members saw these as potentially affecting cfDNA assay results, and “useful but not required.” This latter category included variables that may be useful to collect for rigorous research purposes, but that should not be required when submitting data to resources such as the BloodPAC Data Commons. For example, the variable “Temperature of Sample During Centrifugation” fell into this ranking, since it is important data to have when available, but may not be captured in every data set. Finally, some of the data elements were ranked as “not important and not useful” by members and were not considered to be required for data collection or upload.
When selecting the preanalytical MTDEs, BloodPAC’s PreAnalytical Working Group wanted to balance preanalytical variables that could contribute to changes in molecular results and could be readily obtained vs. those that might not be readily obtained in a real‐world setting. The selection process was also informed by preanalytical data elements contributed by members and the experience of those that have led and/or participated in preanalytical validation studies profiling molecular and cellular components of blood.1
Preanalytic Standards for Biospecimens
Common preanalytical variable standards were established for the handling and processing of histopathology samples that predated the implementation of molecular testing (reviewed in ref. 2). The collection, handling, and processing of biospecimens have long been recognized to contribute to assay variability and challenges of assay validation.3, 4, 5, 6, 7, 8 In fact, overlooking preanalytical variables can have negative consequences for diagnostic development.9
In the context of assay development, the preanalytical steps pertain to everything related to the sample before any assay is run. The preanalytical phases include the patient phase (whereby variables are difficult to control) and the collection phase (whereby variables can be more easily controlled). Nevertheless, patient‐context factors that can influence preanalytical variables, such as age, gender, comorbidities, medications, pregnancy, exercise, and diurnal cycles, are becoming more important in the development and execution of molecular assays, and where possible these variables are being factored into the development of newer molecular assays. Common preanalytical elements include, but are not limited to, collection containers, specimen temperature, sample preparations/stabilizations along with time in transit and storage until a sample is tested.
In addition, we are seeing examples of local coverage determination for cfDNA panels offered by single commercial laboratories. While these are the early entries into clinical care, given the logistical and safety benefits of blood‐based vs. tissue‐based testing and growing confidence in the ability for cfDNA tests to have clinically appropriate sensitivity and specificity, more tests are being developed in Clinical Laboratory Improvement Amendments (CLIA)‐approved and CAP‐approved laboratory settings. As such, undoubtedly, there will be an increasing number of tests submitted for regulatory approval; such is the case for the Guardant 360 assay (Guardant Health, Redwood City, CA) and FoundationOne Liquid Assay (Foundation Medicine, Cambridge, MA), both of which have received breakthrough designation status.
BloodPAC’s Process for Defining MTDE
At the beginning of 2017, the Sample Working Group within the larger BloodPAC Consortium initiated a collaborative and iterative process to identify, define, standardize, and prioritize a list of variables to be required as annotations to each submission of liquid biopsy sample data into the BloodPAC Data Commons. The initial focus of the Sample Working Group’s objective was to identify preanalytical variables specifically related to the collection and processing of samples for cfDNA analysis.
The process of identifying, defining, and standardizing key cfDNA preanalytical variables began with a review of protocols for sample collection and processing that were submitted by BloodPAC members at the time of the initial deposition of sample data into the prototype BloodPAC Data Commons at the end of 2016.10 A total of nine protocols representative of submissions by diagnostic companies, the pharmaceutical industry, and academic institutions and that spanned multiple assay platforms were reviewed. As a result of this initial review, a total of 26 variables relevant to cfDNA sample collection, storage, handling, and processing were identified. This list of variables was then further reviewed by the BloodPAC cochairs and Sample Working Group members who refined the list to 11 variables that were proposed by the cochairs as the preanalytical MTDEs, or the minimal descriptive variables required for the annotation of any data into the BloodPAC Data Commons. This list of 11 MTDEs was then presented to all BloodPAC members at the Q2 2017 meeting held in Boston, Massachusetts in June 2017. Based on member consensus, this list was incorporated into Data Model 2.0, and consensus definitions were included in the BloodPAC Data Dictionary. See Figure S1 in the Supplementary Materials. As a result, all new and existing sample data submissions were made compliant to the inclusion of these 11 MTDEs.
After the consensus list of 11 initial preanalytical MTDEs was defined and implemented, the working group continued this iterative process of identifying and prioritizing relevant cfDNA preanalytical variables through an expanded protocols review, initial FDA consultation, and additional rounds of cochair review. As a result of this process, the list of data variables was expanded to the 52 variables. The 11 MTDEs identified were included in this second round of review based on the cochairs’ consensus on their importance. The Sample Working Group then continued to refine and prioritize the list.
The Sample Working Group’s rankings were averaged to assign an importance ranking to each of the 52 variables and then presented to the entire BloodPAC Consortium at the Q3 2017 all‐member meeting held in New York, New York in September 2017.
After the consensus rankings were compiled, the prioritized list as well as the iterative process followed to identify MTDEs were presented to the FDA at a second consultation meeting held in November 2017. As a result of FDA feedback and alignment, the 11 MTDEs were finalized as minimally important variables to describe preanalytical conditions relevant to cfDNA analyses. Figure S1 in the Supplementary Materials contains a summary of this process.
Note that the final MTDEs are designed to cover all analytes, not just cfDNA. For this reason, during the past 18 months, we have updated the names of some of the MTDEs, such as "DNA_concentration," which was renamed "molecular_concentration" in order to cover both molecular and cellular concentrations.
Funding
Funding for this project was provided by the BloodPAC Consortium and its members.
Conflict of Interest
The work described here was done through the BloodPAC Consortium, which is a not‐for‐profit consortium consisting of members from industry, academia, not‐for‐profits, and US government agencies, including companies that sell liquid biopsy assays, companies that use liquid biopsy assays as companion diagnostics, organizations that do research related to liquid biopsies, organizations that conduct clinical trials involving liquid biopsies, and agencies that develop policies and procedures related to liquid biopsies. In addition, some of the authors are employed by companies in the liquid biopsy field, employed by companies that have projects and partnerships with liquid biopsy companies, have stock in companies in the liquid biopsy field, or consult with companies in the liquid biopsy field. The authors worked together collaboratively to develop consensus recommendations on data elements for the liquid biopsy field as a whole and the authors do not have any particular or specific conflict with the work described in this paper, beyond those just enumerated.
Disclaimer
This article reflects the views of the authors and should not be construed to represent the FDA’s policies.
Supporting information
Figure S1. The collaborative and iterative process that the BloodPAC Sample Working Group used to develop the Minimum Technical Data Elements (MTDEs).
Acknowledgments
The authors wish to acknowledge the contributions to the BloodPAC Project of Darya Chudova, Guardant Health; Maneesh Kumar, Breast Cancer Research Foundation; Tracy Lively, National Cancer Institute; Doug Lowy, National Cancer Institute; Craig Shriver, Department of Defense; and Judith Wolf, Provista Diagnostics.
References
- 1. Kang, Q. et al Comparative analysis of circulating tumor DNA stability In K3EDTA, Streck, and Cell Save blood collection tubes. Clin. Biochem. 49, 1354–1360 (2016). [DOI] [PubMed] [Google Scholar]
- 2. Hewitt, S.M. , Badve, S.S. & True, L.D. Impact of preanalytic factors on the design and application of integral biomarkers for directing patient therapy. Clin. Cancer Res. 18, 1524–1530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schilsky, R.L. , Doroshow, J.H. , LeBlanc, M. & Conley, B.A. Development and use of integral assays in clinical trials. Clin. Cancer Res. 18, 1540–1546 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Poste, G. , Carbone, D.P. , Parkinson, D.R. , Verweij, J. , Hewitt, S.M. & Jessup, J.M. Leveling the playing field: bringing development of biomarkers and molecular diagnostics up to the standards for drug development. Clin. Cancer Res. 18, 1515–1523 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ransohoff, D.F. & Gourlay, M.L. Sources of bias in specimens for research about molecular markers for cancer. J. Clin. Oncol. 28, 698–704 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Agrawal, L. , Engel, K.B. , Greytak, S.R. & Moore, H.M. eds. Understanding preanalytical variables and their effects on clinical biomarkers of oncology and immunotherapy. Semin. Cancer Biol. 52, 26–38 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Merker, J.D. et al Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists joint review. Arch. Pathol. Lab. Med. 142, 1242–1253 (2018). [DOI] [PubMed] [Google Scholar]
- 8. Compton, C.C. et al Preanalytics and precision pathology: pathology practices to ensure molecular integrity of cancer patient biospecimens for precision medicine. Arch. Pathol. La. Med. 143, 1346–1363 (2019). [DOI] [PubMed] [Google Scholar]
- 9. Chau, C.H. , Rixe, O. , McLeod, H. & Figg, W.D. Validation of analytic methods for biomarkers used in drug development. Clin. Cancer Res. 14, 5967–5976 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grossman, R.L. et al Collaborating to compete: Blood Profiling Atlas in Cancer (BloodPAC) Consortium. Clin. Pharmacol. Ther. 101, 589–592 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The collaborative and iterative process that the BloodPAC Sample Working Group used to develop the Minimum Technical Data Elements (MTDEs).


