Abstract
In this study, we used a novel imaging technique, DTI (diffusion tensor imaging)-driven tensor-based morphometry, to investigate brain anatomy in subjects diagnosed with Moebius syndrome (n = 21), other congenital facial weakness disorders (n = 9) and healthy controls (n = 15). First, we selected a subgroup of subjects who satisfied the minimum diagnostic criteria for Moebius syndrome with only mild additional neurological findings. Compared to controls, in this cohort, we found a small region of highly significant volumetric reduction in the paramedian pontine reticular formation and the medial longitudinal fasciculus, important structures for the initiation and coordination of conjugate horizontal gaze. Subsequently, we tested if volume measurements from this region could help differentiate individual subjects of the different cohorts that were included in our study. We found that this region allowed discriminating Moebius syndrome subjects from congenital facial weakness disorders and healthy controls with high sensitivity (94%) and specificity (89%). Interestingly, this region was normal in congenital facial weakness subjects with oculomotor deficits of myopathic origin, who would have been classified as Moebius on the basis of purely clinical diagnostic criteria, indicating a potential role for diffusion MRI morphometry for differential diagnosis in this condition. When the entire Moebius syndrome cohort was compared to healthy controls, in addition to this ‘landmark’ region, other areas of significantly reduced volume in the brainstem emerged, including the location of the nuclei and fibres of cranial nerve VI (abducens nerve), and fibres of cranial nerve VII (facial nerve), and a more rostral portion of the medial longitudinal fasciculus. The high sensitivity and specificity of DTI-driven tensor-based morphometry in reliably detecting very small areas of volumetric abnormality found in this study suggest broader applications of this analysis in personalized medicine to detect hypoplasia or atrophy of small pathways and/or brainstem nuclei in other neurological disorders.
Keywords: brainstem, DTI, DTBM, magnetic resonance imaging, quantitative
This study focuses on providing brain-imaging of Moebius syndrome, a rare disorder characterized by limited abduction of one or both eyes and facial palsy. Sadeghi et al. report that the volume of pontine tegmentum region which includes structures that are involved in horizontal eye movement could be used as an imaging marker for Moebius syndrome subjects.
Graphical Abstract
Graphical Abstract.
Introduction
Moebius syndrome (MBS) is a rare birth defect with minimum diagnostic criteria of congenital limitation of ocular abduction and congenital non-progressive facial weakness (Von Graefe, 1880; Möbius, 1888, 2008; Miller, 2007). While some MBS subjects have no additional clinical issues, many present with additional clinical manifestations (Verzijl et al., 2003; Carta et al., 2011; MacKinnon et al., 2014; Rucker et al., 2014; Bell et al., 2019), which can include adduction deficit or bilateral horizontal gaze palsy, other cranial nerve dysfunction, craniofacial and limb deformities (e.g. Poland syndrome and clubfoot), mirror movements, sleep disorders, seizures and neurocognitive and social impairments (Verzijl et al., 2003; Carta et al., 2011; Webb et al., 2014).
Although MBS is associated with agenesis or hypoplasia of the abducens (CN VI) and facial (CN VII) cranial nerves or nuclei (Towfighi et al., 1979; Verzijl et al., 2005a, b), its etiology and pathogenesis are largely unknown. There are several reported hypotheses including mutations of genes involved in organization and development of the brainstem (Tomas-Roca et al., 2015), ischemia causing interruption of blood supply in the brainstem (Bavinck et al., 1986; Charles et al., 1993; D’Cruz et al., 1993), effects of environmental factors such as toxins (Pastuszak et al., 1998; Bandim et al., 2003; Puvabanditsin et al., 2005; Miller et al., 2009), or mechanical forces affecting development of brainstem structures.
The National Institutes of Health (NIH) study on Moebius syndrome and other congenital facial weakness disorders (clinicaltrials.gov identifier: NCT 02055248) began in 2014 as a collaborative effort between the NIH Intramural Program, Icahn School of Medicine at Mount Sinai and Boston Children’s Hospital and Harvard Medical School with the goal of providing deep phenotyping of subjects affected by MBS and other congenital facial weakness disorders (CFW) and elucidating potential genetic causes. Multisystem phenotyping included ophthalmological, audiological, craniofacial, neurophysiological, neurocognitive and neuroimaging assessments. Here, we present brain imaging data to address the following specific questions: (i) Can we identify specific anatomical abnormalities associated with MBS and with the other CFW disorders? (ii) Are the abnormalities localized only to the brainstem or do they involve other brain regions? (iii) Do we see different abnormalities associated with subcategories of MBS, for example in subjects with limb deformities or mirror movements? (iv) Can accurate imaging phenotyping provide information useful for individual subject assessment and perhaps shed light on the aetiopathogenesis of these disorders?
To assess the degree of agenesis or hypoplasia of cranial nerves VI and VII and potential involvement of other cranial nerves, we acquired 3D balanced fast field echo (BFFE) high-resolution structural brainstem MRI images in which the emergence of nerves is evident against the cerebral spinal fluid (CSF) background. However, in these images, the brainstem appears as a homogenous region, so they are unsuitable for characterization of the brain parenchyma. Diffusion tensor MRI (DTI) (Basser et al., 1994) can provide anatomical characterization and microstructural evaluation of brain tissue (Pierpaoli et al., 1996). DTI enables identification of different white matter pathways in regions that appear homogenous by conventional MRI. For example, the corticospinal tract can be easily differentiated from the transverse pontine fibres in the directionally encoded colour (DEC) map of the brainstem (Pajevic and Pierpaoli, 1999). This advantage is relevant for studying neurodevelopmental disorders where brain circuit formation and maturation might be altered and potentially result in aplastic, hypoplastic or aberrant white matter pathways. With recent development of morphometric studies based on diffusion data, in addition to assessing microstructural changes by analysing diffusion-derived metrics, we can use methods such as DTI-driven tensor-based morphometry (DTBM) (Sadeghi et al., 2018) to assess morphological changes of white matter tracts with higher specificity. The first step in DTBM analysis is spatial normalization in which all the brain images are aligned to allow for voxel-wise comparison. To achieve this, DTBM takes advantage of both scalar and directional information of diffusion tensor data as this enables a better alignment of brain structures, especially in areas that appear homogeneous in the other imaging modalities, and subsequently leads to a better identification of local volume differences with respect to a normative population.
Given the unique ability of diffusion MRI to provide both microstructural/architectural and morphometric information about brain tissue, our study focused on using diffusion MRI for our imaging phenotyping goals in the parenchyma. Typically, diffusion-weighted images (DWIs) are acquired with echo planar imaging (EPI) (Turner and Le Bihan, 1990) and suffer from susceptibility-induced distortions and ghosting, especially in regions near air-tissue interfaces such as the temporal lobe and brainstem. Given the particular importance of the brainstem in this study and challenges of obtaining high-quality diffusion measurements in this region, we adopted a novel acquisition and data processing strategy that entails acquiring DWIs in multiple phase-encoding directions and processing all data together to obtain a final diffusion tensor dataset with correct morphology that also is more immune from ghosting artefacts.
Materials and methods
All data were collected at the NIH Clinical Center in Bethesda, MD. Patient studies were approved by the National Human Genome Research Institute Institutional Review Board and performed in compliance with the Helsinki Declaration. Participants or their parents/legal guardians provided informed consent.
Participants
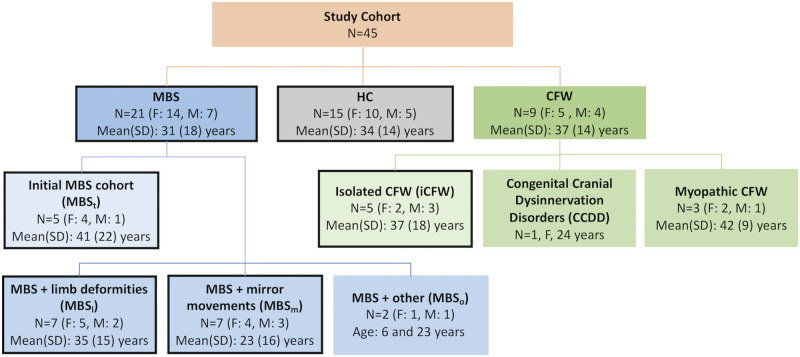
Participants included 21 subjects diagnosed with MBS, nine subjects diagnosed with CFW, and 15 healthy controls (HC) with no history of neurological disorders (see Fig. 1 for demographic detail).
Figure 1.
Demographic information of the study cohort. HC = healthy controls; MBS = Moebius syndrome; CFW = congenital facial weakness. Boxes highlighted by a black outline are subcategories used in group comparison.
All the subjects with MBS met minimum diagnostic criteria for MBS, which is impairment of ocular abduction of one or both eyes and congenital, non-progressive facial weakness (MacKinnon et al., 2014; Rucker et al., 2014). The initial MBS cohort used as a training dataset for a machine-learning classifier (MBSt) (n = 5) included patients with a spectrum of clinical severity, but without limb defects (other than clubfoot), notable mirror movements or severe neurological or other system involvement. The remaining MBS subjects were grouped into: (i) MBS with limb deformities other than clubfoot, which included uni- or bilateral brachy-/syndactyly, Poland anomaly or transverse limb reduction defects (MBSl) (n = 7), (ii) MBS with severe mirror movements—involuntary movement of one side of body with intentional movement of the other side (MBSm) (n = 7) and (iii) MBS with other neurological and system involvement (MBSo) (n = 2), including neurological symptoms, seizures, dysautonomia, autism, cleft palate and/or vocal cord paralysis.
The CFW cohort included isolated CFW (iCFW) (n = 5) subjects whose main phenotype was facial weakness. Other CFW subjects include a subject with a congenital cranial dysinnervation disorder not otherwise specified (CCDD NOS) affecting CNs I and VII and presenting with limb deformities, and three myopathic CFW cases.
The myopathic cases were documented by electrodiagnostic studies that included facial nerve, blink reflex, peripheral sensory and motor nerve conduction (NC) studies, as well as needle electromyography performed on the face (orbicularis oculi and orbicularis oris) and limbs (biceps brachii, tibialis anterior). The three myopathic CFW subjects included one subject with Carey–Fineman–Ziter syndrome (CFZ) with Robin sequence, arthrogryposis, scoliosis and myopathy (Carey et al., 1982; Di Gioia et al., 2017), another subject with external ophthalmoplegia, and one last subject with a CFZ-like myopathy and oculomotor apraxia. Interestingly, while these three individuals met the current minimum diagnostic criteria for Moebius syndrome, the combination of their EMG/NC findings (short duration, polyphasic motor units with early recruitment and no membrane irritability or abnormal spontaneous activity in the EMG, suggesting a primary muscle disorder) together with the evaluation of BFFE images and neuro-ophthalmology and neurology assessments led us to assign these subject to a myopathic CFW group.
Clinical severity score
Each subject was evaluated by a multidisciplinary team for cranial nerve dysfunction, craniofacial and limb deformities, as well as neurocognitive and social impairments. Table 1 shows the clinical characterization of our study cohort. In addition, each subcategory was scored to calculate a ‘clinical severity score’, as follows: facial palsy (0 no palsy, 1 unilateral, 2 bilateral mild, 3 bilateral severe); limited horizontal eye movements (0 no abduction deficit, 1 unilateral, 2 bilateral abduction deficit, 3 bilateral horizontal gaze palsy); limited vertical eye movements (0 no palsy, 1 unilateral, 2 bilateral mild, 3 bilateral severe); limb deformities (0 normal, 1 clubfoot, 2 unilateral limb defect other than clubfoot, 3 bilateral limb defects other than clubfoot); intellectual impairment (0 normal, 1 borderline, 2 mild, 3 moderate) graded based on thorough age-appropriate neurocognitive testing. The scores for these subcategories were normalized to give the maximum score of 1 prior to calculating the clinical severity score. Additional neurological signs/symptoms were each given a score of 1 if present and included sleep disorders, mirror movements, severe difficulty with balance, autism spectrum disorder (ASD), attention-deficit/hyperactivity disorder (ADD/ADHD), mood disorder, seizures, dysautonomia and syringomyelia. Additional system involvement were each given a score of 1 if present and included syncopal episodes, cleft palate, contractures, vocal cord paralysis, endocrine anomalies and congenital heart defects. The scores from each category were added together to calculate the clinical severity score and are detailed in Supplementary Table 1.
Table 1.
Summary of clinical characterization of Moebius syndrome (MBS; rows in blue) and congenital facial weakness (CFW; rows in green) subjects The colour of column headings refer to what means the information was obtained by: grey, clinical evaluation; orange, radiologist reading of BFFE images; purple, morphometric analysis of DTI images (DTBM)

|
R = right, L = left, a = amblyopia, cp = cleft palate, min = minimal, ni = not informative due to artefacts.
Image acquisition and processing
All participants were scanned on a Philips Achieva 3.0 T system with an 8-channel head coil. DWIs were acquired with a single-shot spin-echo EPI sequence. Repetition time (TR) was 10.5 s, echo time (TE) was 85 ms, 90 slices, the voxel size was 2 × 2 × 2 mm, zero-filled at the scanner to 1 × 1 × 2 mm. For each phase encoding direction, the acquisition consisted of two series: the first included b = 0 and fifteen directions with b = 1100 s/mm2; and the second was multishell with b = 50, 200, 400, 600 and 1100 s/mm2 each with six directions. The two series combined produced a DTI set for each phase encoding direction of 46 volumes with 21 unique directions in the b = 1100 shell. This acquisition was repeated four times with anterior–posterior (AP), posterior–anterior (PA), left–right (LR), right–left (RL) phase-encoding directions. A fat-suppressed T2-weighted image (T2WI) was also acquired with 1 × 1 × 2 mm resolution (TR: 7.6 s, TE: 120 ms) to enable better correction of EPI distortion (Irfanoglu et al., 2015). In addition, 3D-balanced fast-field echo (BFFE) high-resolution brainstem scans were acquired with voxel size of 0.4 × 0.4 × 0.8, zero-filled at the scanner to a 0.3 × 0.3 × 0.4 mm resolution. TR was 5.6 ms, TE was 2.2 ms and 30 degrees flip angle.
Diffusion-weighted volumes (DWIs) were processed with the diffprep module of TORTOISE software (Pierpaoli et al., 2010; Irfanoglu et al., 2017). DWIs were corrected for motion and eddy-currents distortions with appropriate rotations to the b-matrix (Rohde et al., 2004). All DWIs were reoriented to a common standard orientation with the midsagittal plane as the vertical plane of symmetry and the axial plane encompassing the anterior and posterior commissures lying horizontally. Alignment to a structural image (T2WI) and EPI distortion correction was done in three steps: (i) AP/PA data were merged into one distortion corrected dataset using the DR-BUDDI software (Irfanoglu et al., 2015) and the diffusion tensor (DT) was computed for the corrected DWIs using the nonlinear tensor estimation of diffcalc module of TORTOISE software (Pierpaoli et al., 2010); (ii) RL/LR were merged into one corrected dataset in a similar manner as AP/PA dataset; (iii) diffeomorphic tensor-based DR-TAMAS registration (Irfanoglu et al., 2016) was used to create a subject specific final morphologically accurate DT from DTAP/PA and DTRL/LR.
Spatial normalization and construction of Jacobian maps
HC subjects’ DTs were used to create a study-specific control template using the diffeomorphic tensor-based DR-TAMAS registration method (Irfanoglu et al., 2016). Subsequently, all subjects’ DTs were registered to the control template. Diffusion metrics such as fractional anisotropy (FA) and mean diffusivity (MD) were derived from spatially normalized DTs.
The natural log of determinant of the Jacobian (lnJ) of the transformations that map each individual to the control template were calculated in each voxel, for each subject. These lnJ maps are informative on the relative size of anatomical structures with respect to the average size of the corresponding structures in the control template. For each subject, a negative value of lnJ indicates local volume reduction (atrophy or hypoplasia), while a positive value indicates local volume expansion.
In addition, we created a study-specific template for spatial normalization made of 27 subjects with equal representation across groups (9 MBS, 9 CFW and 9 HC). This alternative template was used to verify that the measured morphometric differences were not largely affected by the specific choice of the target for spatial normalization.
Statistical analysis
We used various statistical approaches to answer different questions.
Individual classification
Can we find specific anatomical abnormalities in MBSt? To address this question, we first considered MBSt and compared their FA, MD and lnJ maps to healthy controls (HC) using randomized tool (Winkler et al., 2014) with 10 000 permutations. To be highly specific and to reduce false positives, we used a very conservative threshold for voxelwise analysis with no clustering, considering voxels to be significantly different from controls if their familywise error corrected (FWE corrected) p value was less than 0.01.
If such region exists is it shared among all MBS subjects, including other MBS subgroups? We used linear discriminant analysis (LDA) (package MASS of R software), a supervised machine-learning algorithm, to examine whether the region/voxels identified could serve as an imaging marker at an individual subject level. In supervised learning, the algorithm is presented with training data: a set of images and their associated labels (i.e. MBS, HC), the algorithm ‘learns’ a mapping from the training data which can then be used to predict the label of a new image (test data). We use HC and MBSt subjects as training data, and the rest of MBS subjects as testing data.
Does this region discriminate between MBS and CFWs? The same discriminant analysis that we used to test MBS subjects was applied to the CFW cohort.
Group comparison
Do we find specific abnormalities in different subcategories of MBS? Two other primary subgroups, MBSl and MBSm could be identified in addition to MBSt. Both of these subgroups were compared to HC to assess their differences in FA, MD as well as in lnJ maps using similar technique to the previous group comparison.
Comprehensive MBS and iCFW population analysis. To assess overall differences between MBS group and HC, group differences in FA, MD as well as in lnJ maps were assessed using similar technique that was used in the subgroup analysis. In addition, the effect size maps were computed using the following formula: ([mean of patients] − [mean of HC])/pooled standard deviation. As the two groups are dissimilar in size, we used pooling of weighted standard deviation such that each group’s standard deviation is weighted by its sample size (Hedges, 1981). Similar analysis was also performed to assess isolated CFW subjects compared to HC.
Individual subject and abnormality burden assessment
MBS and CFW include a large range of clinical manifestations (Table 1). This heterogeneity may be reflected into anatomical variability within the group, potentially resulting in no differences being detected at the group level analysis, while individual subjects may have unique anatomical abnormalities of remarkable magnitude. Therefore, we also evaluated each individual subject separately using z-score calculated by the following formula: ([subject’s data] − [mean of HC])/standard deviation of HC. Shapiro–Wilk test of normality (Shapiro and Wilk, 1965) [R implementation based on the code described in Royston (1995)] was used to only include voxels where assumption of normality was not violated (voxels with P < 0.01 were excluded in the z-score analysis). We also defined a ‘volumetric abnormality index’ as the total number of abnormal voxels (z-score < −4, voxels indicative of volume loss) in the brain, and calculated this volumetric abnormality index for the brainstem, cerebellum, and the rest of the brain. Brainstem and cerebellum masks were drawn using the snake tool, a semi-automated tool in ITK-SNAP 3.4.0-rc1 software (Yushkevich et al., 2006), and the rest of the brain include the brain parenchyma excluding the brainstem and cerebellum. One-tailed Pearson correlation was used to test whether there is a significant correlation between clinical severity score and volumetric abnormality index of the brainstem, cerebellum and the rest of the brain.
Data availability
The imaging data used in this study are available via request to the corresponding authors.
Results
Salient feature of MBS
Can we find specific anatomical abnormalities in MBSt?
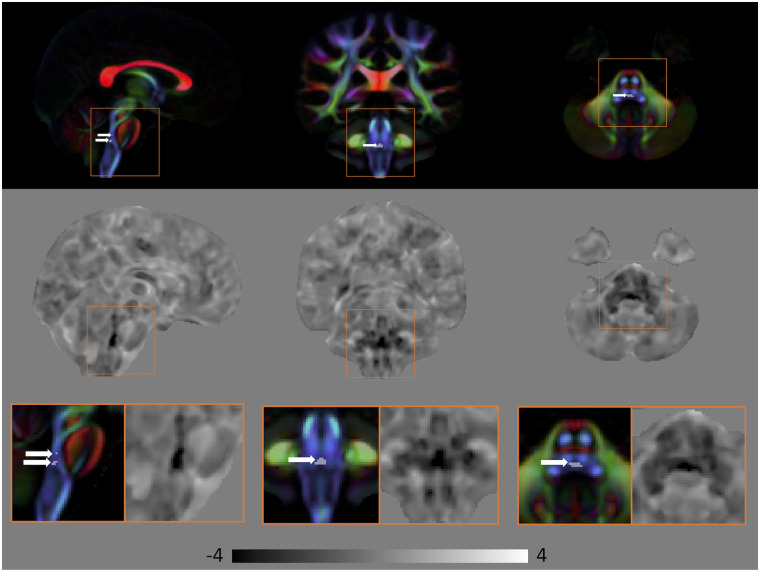
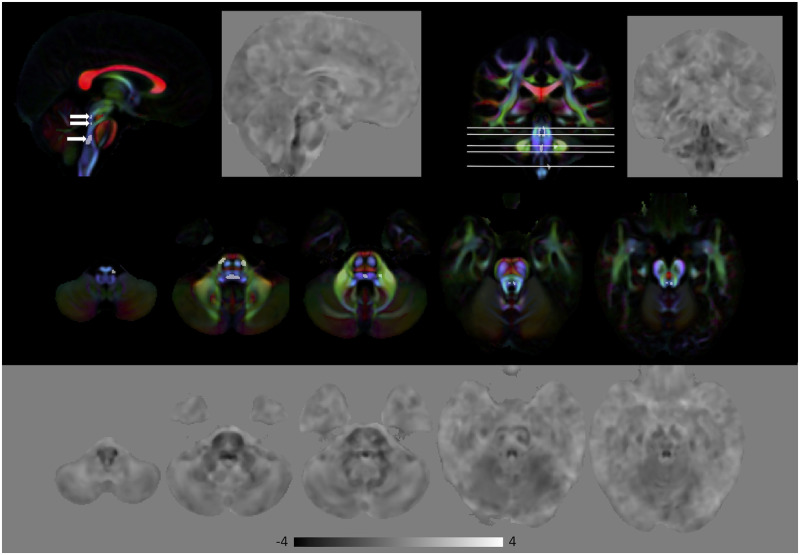
Comparing MBSt to HC, there were no group differences in diffusivity measures, FA and MD, at FWE corrected P < 0.01. In contrast, voxel-wise analysis of the lnJ maps revealed volumetric reduction in the pontine tegmentum region of the MBSt subjects relative to HC. Figure 2 shows the area of volumetric reduction (FWE corrected P < 0.01) for MBSt compared to HC overlaid on the DEC map of the HC template. The anatomical coordinates of this region are provided in Supplementary Table 2. This region is consistent with the location of the medial longitudinal fasciculus (MLF) and the paramedian pontine reticular formation (PPRF). The MLF is an important structure facilitating conjugate horizontal gaze by connecting the nuclei of CN VI to CN III, and PPRF is a collection of cells in the pons directly involved in generation of horizontal saccadic eye movements. The middle and bottom rows of Fig. 2 show the effect size of lnJ maps, where negative values (darker regions) indicate areas that are smaller in the patients with respect to the HC, and positive values (brighter regions) indicate regions that are larger in the patients compared to the HC. The dark region extends in the inferior–superior direction in the pontine tegmentum in a region consistent with the location of the MLF, PPRF, CN VII and CN VI. The effect size map shows that the region of volume reduction extends beyond what is detected in the significance map computed using the very conservative threshold of FWE corrected P < 0.01. The caudal portion of the volume reduction region also covers the location of the nuclei of CN VI and fibres of CN VII which wrap around dorsal aspect of nuclei of CN VI (facial colliculus), as well as the expected intraparenchymal location of the fibres of CN VI.
Figure 2.
Volumetric differences in subjects with MBSt compared to HC. Top: Areas of significant volumetric reduction (indicated by the arrows) in subjects with MBSt compared to HC (FWE corrected P < 0.01) superimposed on the directionally encoded color (DEC) map. Middle: Effect size of lnJ maps. Bottom: Areas in the orange boxes are enlarged. Areas of reduced volume can be clearly seen in the brainstem. In the effect size map, dark areas indicate regions that are smaller in MBSt subjects, whereas bright areas indicate areas that are larger in these subjects. Black corresponds to −4, white to +4, and the grey background is equal to 0.
Is this region of reduced volume, an imaging marker shared among all MBS subjects?
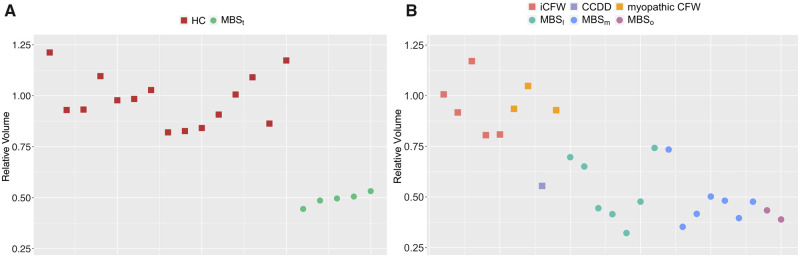
Next, we examined whether the region of volume reduction that we detected with high statistical significance in the MBSt (shown in Fig. 2) group is an imaging marker shared by all MBS subjects. Figure 3 reports for each subject the value of ratio of the volume of this region and the volume of the same region in the HC template. We used the MBSt and HC data to train a classifier (LDA algorithm). Subsequently, we tested the classifier on the other sixteen MBS subjects. We achieved 94% sensitivity (15 of 16 MBS subjects were correctly classified as MBS); all MBS subjects except one (Subject 14 in Table 1, with mild phenotype and unilateral facial weakness) were correctly classified as MBS.
Figure 3.
The average value of the Jacobian (relative volume) for the region of volume reduction detected with high significance in the MBSt versus HC comparison for each subject. The data reported in the left panel (A) is used for training the machine learning algorithm while the data used in the right panel (B) is used in performing individual classification.
Does this region discriminate between MBS and CFW?
Given that the region of significantly reduced volume co-localizes with structures involved in ocular functions (MLF and PPRF), we hypothesized that the iCFW subjects should not have abnormalities in this region, and therefore would be classified as HC if the volume of this region would be probed by the classifier that included only two classes: HC and MBS. Indeed, no iCFW subjects were classified as MBS. In addition, myopathic CFW subjects, who had abduction deficit of muscular origin (subject 28, 29, 30 in Table 1), were not classified as MBS. Only the CCDD NOS subject (subject 27 with CFW and additional CN involvement) was misclassified as MBS. We achieved 89% specificity (eight of nine CFW subjects were correctly classified as not MBS).
Therefore, we conclude that this highly significant tegmental brainstem region of reduced volume is an imaging marker of MBS. Moreover, this region discriminates well between MBS and CFW.
Group comparison
Do we find specific abnormalities in two subcategories of MBS: MBS with limb deformities (MBSl) and MBS with mirror movements (MBSm)? We found no significant differences in the entire brain in both MD and FA when comparing MBSl versus HC and MBSm versus HC (FWE corrected P < 0.01).
Comparing volumes of MBSm versus HC, we found a pattern of abnormality similar to MBSt in the pontine tegmentum of the brainstem. No volumetric differences at the FWE corrected P < 0.01 significance level were found in the rest of the brain in both groups.
We noticed an abnormal decussation of the pyramids in the medulla during visual inspection of DEC maps of individual MBSm subjects. However, since this region is at the border of our imaging volume and coverage is inconsistent, we have initiated a new study to specifically image the lower brainstem and study decussation of pyramids in MBS subjects.
Comprehensive MBS population analysis
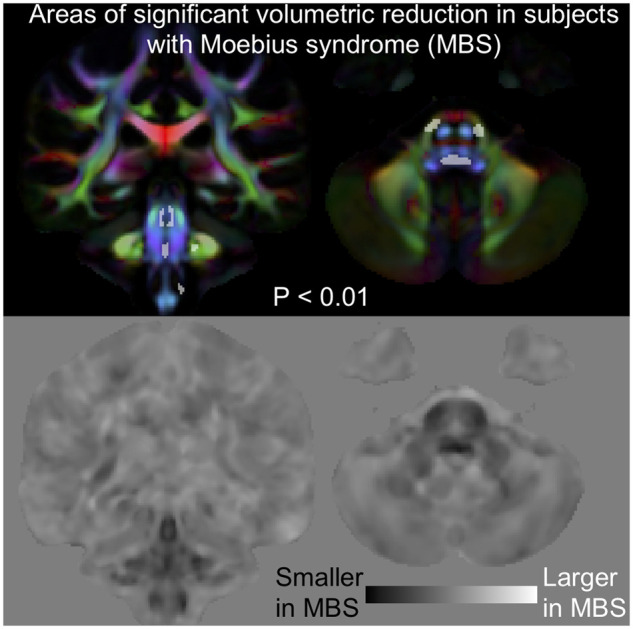
We examined whether additional regions of more subtle abnormalities would emerge when considering all MBS subjects compared to HC. Figure 4 shows a few voxels indicative of significant decrease in FA and increase in MD at FWE corrected P < 0.01. The coordinates for the anatomical locations are provided in Supplementary Table 3. Except for these few voxels, we found no additional regions of FA nor MD differences between MBS and HC. Figure 5 and Supplementary Fig. 1 show the area of volume reduction (FWE corrected P < 0.01) for all MBS subjects compared to HC overlaid on the control template DEC map, and the coordinates for the anatomical locations are provided in Supplementary Table 4. While the area of significant volume reduction is still concentrated in the brainstem, the area detected as significant is larger compared to when considering just the MBSt subgroup. Additional areas of significant volumetric reduction now also include the inferior olives, the middle cerebellar peduncle, and the transverse pontine fibres. In Supplementary Fig. 2, we show that similar results are obtained using either template (control template or a template made of MBS, CFW and HC subjects) as a reference for the spatial normalization.
Figure 4.
Differences in diffusion metrics in subjects with MBS compared to HC. A few voxels (indicated by the arrows) are areas of significant increase in MD (top) and decrease in FA (bottom) in subjects with MBS compared to HC (FWE corrected P < 0.01) superimposed on the MD and FA maps, respectively.
Figure 5.
Volumetric differences in subjects with MBS compared to HC. Top: Areas of significant volumetric reduction (indicated by the arrows in the sagittal view) in subjects with MBS compared to HC (FWE corrected P < 0.01) superimposed on the directionally encoded color (DEC) map. Bottom: Effect size of lnJ maps. Dark areas indicate regions that are smaller in MBS subjects, whereas bright areas indicate areas that are larger. In the effect size map black corresponds to −4, white to +4, and the grey background is equal to 0. Areas of reduced volume can be clearly seen in the brainstem.
The effect size maps showed similar areas of volume reduction for all MBS subjects compared to MBSt, indicating similar areas are being affected in all MBS subjects. There is a relatively large area of volumetric reduction in the brainstem (dark regions in the effect size map), however, since we used a very conservative statistical threshold (FWE corrected P < 0.01) only a few areas in the brainstem reached significance. The effect size map, however, showed a few other local abnormalities. We examined regions that have effect size of less than negative one and show abnormalities that are bilateral and appear as an anatomically meaningful cluster. Using these criteria, three salient areas were detected: (i) the parahippocampal portion of cingulum (Supplementary Fig. 3), (ii) a region in centrum semiovale (Supplementary Fig. 3) that when seeded for DTI-based tractography on the HC template gave tracts projecting to the postcentral gyrus and (iii) cerebellum lobules V and VI (Supplementary Fig. 3), which are cerebellar regions functionally connected with sensorimotor cortices (Grodd et al., 2001; Buckner et al., 2011).
iCFW versus HC
Similar to the MBS analysis, we compared FA, MD and lnJ of iCFW subjects to HC. There were no significant differences in all of these metrics for this cohort of subjects. Figure 6 shows the effect size map of lnJ comparing iCFW subjects to HC, where there are no apparent volumetric differences for this group of subjects. Note the clear difference in the effect size maps of Fig. 5 for the MBS group, where volume reduction is evident in the brainstem, and those shown in Fig. 6 for the iCFW group where no volumetric abnormalities are evident.
Figure 6.
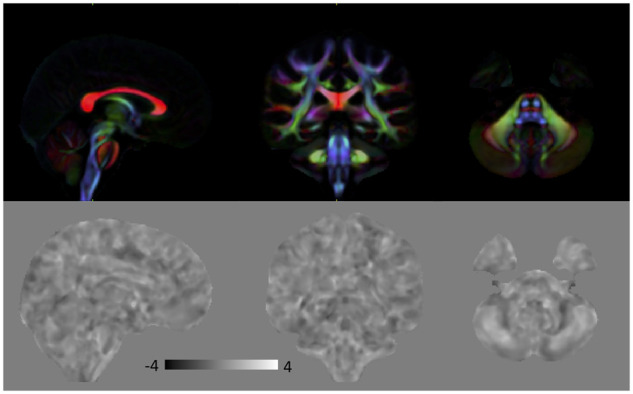
Top: Directionally encoded color (DEC) map for anatomical reference. Bottom: Effect size of lnJ maps comparing iCFW subjects to HC. Dark areas indicate regions that are smaller in iCFW subjects, whereas bright areas indicate areas that are larger in these subjects. There are no regions of significant volumetric difference between iCFW subjects and HC. In the effect size map, black corresponds to −4, white to +4 and the grey background is equal to 0.
Abnormality burden and individual subject assessment
Table 1 shows the clinical characterization of our cohort and highlights variable presentation and severity of the disorder. Among the 21 subjects diagnosed as MBS based on minimal criteria, 18 had tongue hypoplasia and/or high palate (86%), 15 had adduction deficit (71%), 10 had clubfoot (48%), 8 had limited upgaze (38%) and 6 had intellectual impairment (29%).
The iCFW subjects form a more homogenous group as the main phenotype for these subjects is facial weakness, except for the CFW subjects with additional cranial nerve involvement (CN I) but normal CN VI or with extraocular muscle weakness of myopathic origin who have more complex clinical manifestations (refer to Table 1).
Overall, MBS subjects compared to CFW subjects had higher clinical severity score: 5.16 (range: 1.67 to 11) MBS subjects compared to 2.37 (range: 0.67 to 5) CFW subjects.
Table 1 shows the volumetric abnormality index for the brainstem, cerebellum and the rest of the brain in relation to the clinical characteristics. CFW subjects had an overall lower abnormality index compared to MBS subjects for all the three regions (brainstem: 0.17% compared to 2.92%, cerebellum: 0.27% compared to 0.63%, and the rest of the brain: 0.11% compared to 0.30%). It is interesting to note that the abnormality index for the brainstem of MBS subjects is 17-fold higher than the CFW subjects abnormality index, whereas the abnormality index for the cerebellum and the rest of the brain are of similar magnitude for both of these groups.
For MBS subjects, a highly significant correlation between volumetric abnormality index and clinical severity score was found in the brainstem (r = 0.77, P < 0.0001). No significant correlation was observed in the cerebellum (r = 0.09, P = 0.36) or the rest of the brain (r = 0.25 P = 0.14).
On average, the MBSm and MBSl subgroups showed similar volumetric abnormality index for the brainstem, 2.81% (range: 0.01–7.21%) and 2.20% (range: 0.03–9.19%), respectively; however, MBSl had a higher abnormality index for the cerebellum 0.77% (range: 0.11–3.02%) compared to MBSm abnormality index of 0.39% (range: 0.07–0.94).
Discussion
Identification of an imaging marker for MBS and classification in individual brains
The main finding of this work is that DTBM analysis of diffusion MRI data revealed a small region of reduced volume in the brainstem of MBS subjects localized in the paramedian pontine reticular formation (PPRF) and the medial longitudinal fasciculus (MLF), important structures that are involved in the initiation and coordination of conjugate horizontal gaze. In addition, the caudal portion of the volume reduction region also adjoins the location of the nuclei of CN VI and fibres of CN VI and CN VII. This region of reduced volume is also a very powerful imaging marker for discriminating between MBS and CFW. All iCFW subjects and myopathic CFW subject were classified correctly as not MBS. The classifier was able to correctly diagnose the myopathic CFW cases that had similar limitations of horizontal gaze to MBS, that only through electromyography (EMG), and not clinical examination, was determined to be myopathic and not neurogenic. In contrast to typical quantitative neuroimaging studies that often find differences only at the population level, we were also able to classify individuals accurately. This was achieved using quite stringent criteria, such as not using typical cross-validation methods, but rather by dividing the imaging data into training and test datasets, thereby treating the test data as a true unseen dataset.
Typically, the hallmark of MBS is limited abduction due to agenesis or hypoplasia of CN VI in addition to facial weakness due to agenesis or hypoplasia of CN VII. However, we note that 71% of our MBS subjects also have bilateral horizontal gaze palsy (BHGP) which is consistent with an involvement of MLF (Supplementary Fig. 1). Other studies have also reported BHGP in many of their MBS subjects, for example a study by Verzijl et al. (2003) reported 48% of their subjects had BHGP. A recent study by Rucker et al. (2014) reported 93% of the MBS subjects to have BHGP.
Topological distribution of parenchymal abnormalities in MBS
The MBS ‘landmark’ region that was identified in the MBSt group is smaller than the area of significant abnormality when group analysis is performed including the entire MBS cohort. This is not surprising because increased statistical power is expected with a larger number of subjects. In the entire MBS cohort, regions of significantly reduced volume include larger portions of PPRF and MLF structures in addition to CN VI and CN VII. The area of volumetric reduction extends further along the superior direction encompassing more of the MLF and reaching the level of the oculomotor nuclei. Impairment of the connections between the nuclei of the CN VI and CN III may explain common features observed in the MBS subjects (i.e. adduction and/or upgaze limitations in addition to abduction limitation). We also found multiple regions with reduced volume located in the rhombencephalon in addition to the pontine tegmentum region (Fig. 5), including inferior olives, transverse pontine fibres and middle cerebellar peduncles. The corticopontocerebellar pathway, a bundle composed of major afferent fibres through the middle cerebellar peduncle, is involved in the communication between the cerebellum and the prefrontal cortex for the coordination and planning of motor tasks. The inferior olives are also involved in the timing and learning of movement. The abnormality in these regions could be at the basis of abnormal motor coordination and impaired balance, as assessed by tandem gait performance and application of the Berg balance scale (Berg et al., 1989), found on clinical examination in some of the MBS subjects.
In addition, we observed regions of volumetric reduction in the parahippocampal region of cingulum. The cingulum bundle is comprised of both short and long fibres connecting different regions of the brain. While the cingulum is typically depicted as a uniform bundle in DTI-based studies, it contains multiple fibre populations. Furthermore, animal studies have indicated different subdivisions contain different population of white matter fibres (Mufson and Pandya, 1984). Recently, Jones et al. (2013) suggested subdividing the cingulum tract into three subdivisions: ‘parahippocampal’, ‘retrosplenial’ and ‘subgenual’. Using the proposed subdivision, we observed volumetric reduction exclusively in the parahippocampal region of cingulum; this region contains not only fibres from posterior cingulate, connecting the cingulum gyrus to the hippocampal formation (Mufson and Pandya, 1984), but also fibres that project to the medial temporal lobe from visual areas in the occipital lobe (Suzuki and Amaral, 1994). The multiple abnormalities we found in the brainstem are in agreement with (Verzijl et al., 2003, 2005b) who proposed that MBS is a developmental anomaly of the rhombencephalon, not just a cranial nerve disorder. However, our finding of additional regions of reduced volume in the forebrain suggests that abnormalities can also extend beyond the brainstem, albeit with very selective anatomical locations. The topological distribution of parenchymal abnormalities in MBS contrasts with that of iCFW subjects, in whom no region of statistical significance was observed in the whole-brain parenchyma. In addition, no bilateral continuous clusters of volume reduction were observed in the effect size maps. The z-score maps also indicate that the parenchymal volumetric abnormality index in MBS is higher than that of the CFW group. Even though a detailed individual subject evaluation is beyond the scope of this paper, there are a few main takeaways from the z-score analysis: in general, the clinically more severe cases had higher volumetric abnormality index, and the CFW group had a lower volumetric abnormality index compared to the MBS group. The majority of the abnormal voxels for the MBS group were located in the brainstem, and there was a high correlation between the volumetric abnormality index for the brainstem and the clinical severity score. Of note, subjects with limb defects and no significant additional system involvement (subjects #13–15) had very low clinical severity and imaging z-score.
Importance of DTBM analysis to detect abnormality in these cohorts
A finding in this study was the paucity of abnormalities revealed by the analysis of typical DTI metrics, such as FA and MD, in the MBS group. Diffusion derived metrics, especially FA, have been extensively used to study normal brain development (Lebel and Beaulieu, 2011; Sadeghi et al., 2013, 2015, 2017), aging (Kochunov et al., 2012), disease and disease progression (Assaf and Pasternak, 2008). One of the main reasons of the popularity of FA as a metric is that it is very sensitive to the underlying microstructural tissue features such as coherence of axon orientation, oedema, gliosis and to a lesser extend myelination. In our MBS cohort, however, even when all subjects were examined, no significant differences in FA or MD were found between MBS subjects and HC except a few voxels in the pontine tegmentum region (Fig. 4). In contrast, DTBM analysis revealed the presence of prominent and consistent morphometric abnormalities. Essentially, the anatomical deficit we identified in the brainstem tegmental region in MBS is volumetric, not microstructural. It is interesting to note that this is an example of how different analytical approaches of the same diffusion data can be informative of different aspects of the underlying anatomical abnormality.
In addition to the localization of morphometric abnormalities to small tracts, this study has shown DTBM can be used for individual subject assessment, which is relevant for precision medicine. While radiologic MRI reports of MBS generally indicate a qualitative outcome of flattened 4th ventricle, this is the first DTBM study of MBS and clearly extends existing observations by localizing the morphometric change to the pontine tegmentum specifically, in a region consistent with the location of the MLF, PPRF, CN VI and CN VII. Notably, the MLF was not selected a priori for investigation in this study but was consistently found in virtually all MBS subjects in an unsupervised exploratory DTBM analysis.
Methodological considerations for achieving a reliable DTBM assessment
Diffusion data are typically acquired with an EPI sequence (Turner and Le Bihan, 1990), since it is an acquisition method which is relatively insensitive to motion achieving high signal-to-noise ratio per unit time. However, EPI images suffer from geometric distortions along the phase-encoding direction caused mainly by the B0 field inhomogeneities (Jezzard and Balaban, 1995). These distortions are worse in areas of tissue/air boundary such as the brainstem region. Acquisition of additional data are usually needed for EPI distortions correction algorithms. We acquired full DWI datasets with opposite phase encoding in two perpendicular directions (AP/PA and RL/LR). This approach allowed us to obtain a final diffusion tensor dataset in each subject with correct morphology and immunity from ghosting artifacts. This is an experimental design that, to our knowledge, is unprecedented in clinical research applications. However, if we would not have used this design, we would have been unable to detect the anatomical abnormalities we report.
The second challenge for performing DTBM is the accurate alignment of different brain areas such that each voxel is compared to its corresponding voxel in a different subject. Here, we used DR-TAMAS, a diffeomorphic diffusion tensor-based registration method that uses all scalar and directional information provided by diffusion tensor, achieving good overall quality of registration for all tissue types including white matter, grey matter and CSF regions, and also local alignment of fibre tracts within white matter regions (Irfanoglu et al., 2016). Image registration using the full diffusion tensor information is also a relatively unusual approach, but it was key in our study to achieve the results we presented.
In this study, the diffusion images were acquired at 2 × 2 × 2 mm with final diffusion maps up-sampled to 1 × 1 × 1. The brainstem is a small and highly complex structure, therefore, as technology improves, acquisition of data at higher resolution is desirable to elucidate more anatomical details potentially leading to a better localization of anatomical differences between MBS and HC.
Conclusions and future work
We have identified an imaging marker that has proved to be both sensitive and specific and can be used as a complementary tool to aid accurate diagnosis and further define the aetiopathogenesis of MBS. This study shows with proper acquisition, processing and novel analysis methods, we can take full advantage of diffusion data for detecting both microstructural and volume changes with high sensitivity and specificity. The DTI protocol and proposed analysis method used here can be applied to other disorders to gain a better understanding of anatomical differences at the population and single subject level.
Supplementary material
Supplementary material is available at Brain Communications online.
Supplementary Material
Acknowledgments
We thank Dr Joelle Sarlls for help on the implementation of the DTI protocol. We also thank Ms Amritha Nayak, Mr Timothy Wood and Drs Catherine J. Stoodley, Okan M. Irfanoglu, Laura Reyes, Cibu Thomas for useful comments and discussions. We also thank the subjects for their participation in this research and the Moebius Syndrome Foundation for their support.
Glossary
- ADD =
attention deficit disorder
- ADHD =
attention-deficit/hyperactivity disorder
- ASD =
autism spectrum disorder
- AP =
anterior–posterior
- BFFE =
balanced fast field echo
- BHGP =
bilateral horizontal gaze palsy
- CCDD NOS
= congenital cranial dysinnervation disorders not otherwise specified
- CFW =
congenital facial weakness disorders
- CFZ =
Carey–Fineman–Ziter syndrome
- CN =
cranial nerve
- DEC =
directionally encoded color map
- DTBM =
DTI-driven tensor-based morphometry
- DT =
diffusion tensor
- DTI =
diffusion tensor imaging
- DWIs =
diffusion weighted images
- EOM =
extra-ocular muscles
- EPI =
echo planar imaging
- FA =
fractional anisotropy
- FEW =
familywise error
- HC =
healthy controls
- iCFW =
isolated congenital facial weakness
- LDA =
linear discriminant analysis
- lnJ =
natural log of the Jacobian
- LR =
left–right
- MBS =
Moebius syndrome
- MBSl =
Moebius syndrome with limb deformities
- MBSm =
Moebius syndrome with mirror-movements
- MBSo =
Moebius syndrome with other neurological symptoms and system involvement
- MBSt =
Moebius syndrome cohort used as a training dataset for a machine learning classifier
- MD =
mean diffusivity
- MLF =
medial longitudinal fasciculus
- NC =
nerve conduction
- PA =
posterior–anterior
- PPRF =
paramedian pontine reticular formation
- RL =
right–left
- T2WI =
T2-weighted image
- TR =
repetition time
- TE =
echo time
Appendix
Membership of the Moebius Syndrome Collaborative Research consortium:
Icahn School of Medicine at Mount Sinai, New York City, New York, USA—group
Monica Erazo,1 Tamiesha Frempong,2 Ke Hao,1,3 Ethylin Wang Jabs,1,4,5 Thomas P. Naidich,6 Janet C. Rucker,5,7 Bryn D. Webb1,5 and Zhongyang Zhang1,3
1Department of Genetics and Genomic Sciences, Icahn School of Medicine at Mount Sinai, New York, New York
2Department of Ophthalmology, Icahn School of Medicine at Mount Sinai, New York, New York
3Icahn Institute for Data Science and Genomic Technology, Icahn School of Medicine at Mount Sinai, New York, New York
4Cell, Developmental and Regenerative Biology, Icahn School of Medicine at Mount Sinai, New York, New York
5Pediatrics, Icahn School of Medicine at Mount Sinai, New York, New York
6Departments of Radiology, Neurosurgery, and Pediatrics, Icahn School of Medicine at Mount Sinai, New York, New York
7Department of Neurology, New York University School of Medicine, New York, New York
Boston Children’s Hospital and Harvard Medical School, Boston, MA—group
Caroline Andrews,1,2,3,4 Brenda J. Barry,1,2,3,4 Silvio Alessandro DiGioia,1,2,3 Elizabeth Engle,1,2,3,4,5,6,7,8 David G.Hunter,6,7 Sarah E. MacKinnon,6 Caroline Robson9 and Matthew Rose1,5,2,8,10
1Department of Neurology, Boston Children’s Hospital, Boston, MA
2F.M. Kirby Neurobiology Center, Boston Children’s Hospital, Boston, MA
3Department of Neurology, Harvard Medical School, Boston, MA
4Howard Hughes Medical Institute, Chevy Chase, MD
5Department of Pathology, Boston Children’s Hospital, Boston, MA
6Department of Ophthalmology, Boston Children’s Hospital, Boston, MA
7Department of Ophthalmology, Harvard Medical School, Boston, MA
8Medical Genetics Training Program, Harvard Medical School, Boston, MA
9Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, MA
10Department of Pathology, Brigham and Women’s Hospital, Boston, MA
National Institutes of Health, Bethesda, MD—Group
Barbara B. Biesecker,1 Lori L. Bonnycastle,2 Carmen C. Brewer,3 Brian P. Brooks,4 John A. Butman,5 Wade W. Chien,3 Peter S. Chines,2 Francis S. Collins,2,6 Flavia Facio,2 Kathleen Farrell,7 Edmond J. FitzGibbon,4 Andrea L. Gropman,8 Elizabeth Hutchinson,9 Mina S. Jain,7 Kelly A. King,3 Tanya J. Lehky,10 Janice Lee,11 Denise K. Liberton,11 Irini Manoli,2 Narisu Narisu,2 Scott M. Paul,7 Carlo Pierpaoli,9 Neda Sadeghi,9 Joseph Snow,12 Beth Solomon,7 Angela Summers,12 Amy J. Swift,2 Camilo Toro,13 Audrey Thurm,14 Carol Van Ryzin2 and Chris K. Zalewski3
1Social and Behavioral Research Branch, National Human Genome Research Institute, Bethesda, MD
2Medical Genomics and Metabolic Genetics Branch, National Human Genome Research Institute, Bethesda, MD
3Audiology Unit, Otolaryngology Branch, National Institute of Deafness and other Communications Disorders, Bethesda, MD
4Ophthalmic Genetics & Visual Function Branch, National Eye Institute, Bethesda, MD
5Radiology and Imaging Sciences, Clinical Research Center, Bethesda, MD
6Office of the Director, National Institutes of Health, Bethesda, MD
7Rehabilitation Medicine Department, Clinical Research Center, Bethesda, MD
8George Washington University and Children’s National Medical Center, Washington, DC
9Quantitative Medical Imaging Section, National Institute of Biomedical Imaging and Bioengineering, Bethesda, MD
10Electromyography Section, National Institute of Neurological Disorders and Stroke, Bethesda, MD
11Office of the Clinical Director, National Institute of Dental and Craniofacial Research, Bethesda, MD
12Office of the Clinical Director, National Institute of Mental Health, Bethesda, MD
13NIH Undiagnosed Diseases Program, Common Fund, National Human Genome Research Institute, Bethesda, MD
14Pediatrics and Developmental Neuroscience Branch, National Institute of Mental Health, Bethesda, MD
Contributor Information
Moebius Syndrome Research Consortium:
Monica Erazo, Tamiesha Frempong, Ke Hao, Ethylin Wang Jabs, Thomas P Naidich, Janet C Rucker, Bryn D Webb, Zhongyang Zhang, Caroline Andrews, Brenda J Barry, Silvio Alessandro DiGioia, Elizabeth Engle, David G Hunter, Sarah E MacKinnon, Caroline Robson, Matthew Rose, Barbara B Biesecker, Lori L Bonnycastle, Carmen C Brewer, Brian P Brooks, John A Butman, Wade W Chien, Peter S Chines, Francis S Collins, Flavia Facio, Kathleen Farrell, Edmond J FitzGibbon, Andrea L Gropman, Elizabeth Hutchinson, Mina S Jain, Kelly A King, Tanya J Lehky, Janice Lee, Denise K Liberton, Irini Manoli, Narisu Narisu, Scott M Paul, Carlo Pierpaoli, Neda Sadeghi, Joseph Snow, Beth Solomon, Angela Summers, Amy J Swift, Camilo Toro, Audrey Thurm, Carol Van Ryzin, and Chris K Zalewski
Funding
This research was supported by the Intramural Research Programs of the National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Human Genome Research Institute (NHGRI), Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), National Institute of Health Clinical Center, and National Eye Institute (NEI). Support for this work included funding from U01 HD079068 grant and the Moebius Syndrome Foundation.
Competing interests
The authors report no competing interests.
References
- Assaf Y, Pasternak O.. Diffusion tensor imaging (DTI)-based white matter mapping in brain research: a review. J Mol Neurosci 2008; 34: 51–61. [DOI] [PubMed] [Google Scholar]
- Bandim JM, Ventura LO, Miller MT, Almeida HC, Costa A.. Autism and Möbius sequence: an exploratory study of children in northeastern Brazil. Arq Neuro-Psiquiatr 2003; 61: 181–5. [DOI] [PubMed] [Google Scholar]
- Basser PJ, Mattiello J, LeBihan D.. Estimation of the effective self-diffusion tensor from the NMR spin echo. J Magn Reson 1994; 103: 247–54. [DOI] [PubMed] [Google Scholar]
- Bavinck JNB, Weaver DD, Opitz JM, Reynolds JF.. Subclavian artery supply disruption sequence: hypothesis of a vascular etiology for Poland, Klippel-Feil, and Mobius anomalies. Am J Med Genet 1986; 23: 903–18. [DOI] [PubMed] [Google Scholar]
- Bell C, Nevitt S, McKay VH, Fattah AY.. Will the real Moebius syndrome please stand up? A systematic review of the literature and statistical cluster analysis of clinical features. Am J Med Genet Part A 2019; 179: 257–65. [DOI] [PubMed] [Google Scholar]
- Berg K, Wood-Dauphine S, Williams JI, Gayton D.. Measuring balance in the elderly: preliminary development of an instrument. Physiother Canada 1989; 41: 304–11. [Google Scholar]
- Buckner RL, Krienen FM, Castellanos A, Diaz JC, Yeo B.. The organization of the human cerebellum estimated by intrinsic functional connectivity. J Neurophysiol 2011; 106: 2322–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey JC, Fineman RM, Ziter FA.. The Robin sequence as a consequence of malformation, dysplasia, and neuromuscular syndromes. J Pediatr 1982; 101: 858–64. [DOI] [PubMed] [Google Scholar]
- Carta A, Mora P, Neri A, Favilla S, Sadun AA.. Ophthalmologic and systemic features in Möbius syndrome: an Italian case series. Ophthalmology 2011; 118: 1518–23. [DOI] [PubMed] [Google Scholar]
- Charles SS, Dimario FJ Jr,, Grunnet ML. Möbius sequence: further in vivo support for the subclavian artery supply disruption sequence. Am J Med Genet 1993; 47: 289–93. [DOI] [PubMed] [Google Scholar]
- D’Cruz OF, Swisher CN, Jaradeh S, Tang T, Konkol RJ.. Möbius syndrome: evidence for a vascular etiology. J Child Neurol 1993; 8: 260–5. [DOI] [PubMed] [Google Scholar]
- Di Gioia SA, Connors S, Matsunami N, Cannavino J, Rose MF, Gilette NM, et al., Moebius Syndrome Research Consortium. A defect in myoblast fusion underlies Carey-Fineman-Ziter syndrome. Nat Commun 2017; 8: 16077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grodd W, Hülsmann E, Lotze M, Wildgruber D, Erb M.. Sensorimotor mapping of the human cerebellum: fMRI evidence of somatotopic organization. Hum Brain Mapp 2001; 13: 55–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges LV. Distribution theory for Glass’s estimator of effect size and related estimators. J Educ Stat 1981; 6: 107–28. [Google Scholar]
- Irfanoglu MO, Modi P, Nayak A, Hutchinson EB, Sarlls J, Pierpaoli C.. DR-BUDDI (diffeomorphic registration for blip-up blip-down diffusion imaging) method for correcting echo planar imaging distortions. Neuroimage 2015; 106: 284–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irfanoglu MO, Nayak A, Jenkins J, Hutchinson EB, Sadeghi N, Thomas CP, et al. DR-TAMAS: diffeomorphic registration for tensor accurate alignment of anatomical structures. Neuroimage 2016; 132: 439–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irfanoglu MO, Nayak A, Jenkins J, Pierpaoli C, TORTOISE v3: Improvements and new features of the NIH diffusion MRI processing pipeline. In: The ISMRM 25th Annual Meeting & Exhibition. Hawaii, USA; 2017.
- Jezzard P, Balaban R.. Correcting for geometric distortion in echo-planar images from B0 field distortions. Magn Reson Med 1995; 34: 65–73. [DOI] [PubMed] [Google Scholar]
- Jones DK, Christiansen KF, Chapman RJ, Aggleton JP.. Distinct subdivisions of the cingulum bundle revealed by diffusion MRI fibre tracking: implications for neuropsychological investigations. Neuropsychologia 2013; 51: 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Williamson DE, Lancaster J, Fox P, Cornell J, Blangero J, et al. Fractional anisotropy of water diffusion in cerebral white matter across the lifespan. Neurobiol Aging 2012; 33: 9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel C, Beaulieu C.. Longitudinal development of human brain wiring continues from childhood into adulthood. J Neurosci 2011; 31: 10937–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKinnon S, Oystreck DT, Andrews C, Chan W, Hunter DG, Engle EC.. Diagnostic distinctions and genetic analysis of patients diagnosed with Moebius syndrome. Ophthalmology 2014; 121: 1461–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G. The mystery of the missing smile. Science 2007; 316: 826–7. [DOI] [PubMed] [Google Scholar]
- Miller MT, Ventura L, Strömland K.. Thalidomide and misoprostol: ophthalmologic manifestations and associations both expected and unexpected. Birth Defects Res Part A Clin Mol Teratol 2009; 85: 667–76. [DOI] [PubMed] [Google Scholar]
- Möbius PJ. Ueber angeborene doppelseitige Abducens-Facialis-Lahmung. MMW Munch Med Wochenschr 1888; 35: 91–4. [Google Scholar]
- Möbius PJ. About congenital bilateral abducens and facialis palsy. Strabismus 2008; 16: 39–44. [DOI] [PubMed] [Google Scholar]
- Mufson EJ, Pandya DN.. Some observations on the course and composition of the cingulum bundle in the rhesus monkey. J Comp Neurol 1984; 225: 31–43. [DOI] [PubMed] [Google Scholar]
- Pajevic S, Pierpaoli C.. Color schemes to represent the orientation of anisotropic tissues from diffusion tensor data. Magn Reson Med 1999; 42: 526–40. [PubMed] [Google Scholar]
- Pastuszak AL, Koren G, Schüler L, Schwarz IVD, Larrandaburu M, Speck-Martins CE, et al. Use of misoprostol during pregnancy and Mobius’ syndrome in infants. N Engl J Med 1998; 338: 1881–5. [DOI] [PubMed] [Google Scholar]
- Pierpaoli C, Jezzard P, Basser PJ, Barnett A, Di Chiro G.. Diffusion tensor MR imaging of the human brain. Radiology 1996; 201: 637–48. [DOI] [PubMed] [Google Scholar]
- Pierpaoli C, Walker L, Irfanoglu M, Barnett A, Chang L-C, Koay C, et al. TORTOISE: an integrated software package for processing of diffusion MRI data. In: The ISMRM 18th Annual Meeting & Exhibition. Stockholm, Sweden; 2010.
- Puvabanditsin S, Garrow E, Augustin G, Titapiwatanakul R, Kuniyoshi KM.. Poland-Möbius syndrome and cocaine abuse: a relook at vascular etiology. Pediatr Neurol 2005; 32: 285–7. [DOI] [PubMed] [Google Scholar]
- Rohde GK, Barnett AS, Basser PJ, Marenco S, Pierpaoli C.. Comprehensive approach for correction of motion and distortion in diffusion-weighted MRI. Magn Reson Med 2004; 51: 103–14. [DOI] [PubMed] [Google Scholar]
- Royston P. Remark AS R94: A remark on algorithm AS 181: The W-test for normality. J R Stat Soc Ser C (Applied Stat) 1995; 44: 547–51. [Google Scholar]
- Rucker JC, Gaspar H, Jabs EW, Naidich TP, Webb BD, Frempong T.. Characterization of ocular motor deficits in congenital facial weakness: Moebius and related syndromes. Brain 2014; 137: 1068–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghi N, Arrigoni F, D'Angelo MG, Thomas C, Irfanoglu MO, Hutchinson EB, et al. Tensor-based morphometry using scalar and directional information of diffusion tensor MRI data (DTBM): application to hereditary spastic paraplegia. Hum Brain Mapp 2018; 39: 4643–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghi N, Gilmore JH, Gerig G.. Twin–singleton developmental study of brain white matter anatomy. Hum Brain Mapp 2017; 38: 1009–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghi N, Nayak A, Walker L, Irfanoglu MO, Albert PS, Pierpaoli C.. Analysis of the contribution of experimental bias, experimental noise, and inter-subject biological variability on the assessment of developmental trajectories in diffusion MRI studies of the brain. Neuroimage 2015; 109: 480–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghi N, Prastawa M, Fletcher PT, Wolff J, Gilmore JH, Gerig G.. Regional characterization of longitudinal DT-MRI to study white matter maturation of the early developing brain. Neuroimage 2013; 68: 236–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro SS, Wilk MB.. An analysis of variance test for normality (complete samples). Biometrika 1965; 52: 591–611. [Google Scholar]
- Suzuki WA, Amaral DG.. Topographic organization of the reciprocal connections between the monkey entorhinal cortex and the perirhinal and parahippocampal cortices. J Neurosci 1994; 14: 1856–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomas-Roca L, Tsaalbi-Shtylik A, Jansen JG, Singh MK, Epstein JA, Altunoglu U, et al. De novo mutations in PLXND1 and REV3L cause Möbius syndrome. Nat Commun 2015; 6: 7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towfighi J, Marks K, Palmer E, Vannucci R.. Möbius syndrome. Acta Neuropathol 1979; 48: 11–7. [DOI] [PubMed] [Google Scholar]
- Turner R, Le Bihan D.. Single-shot diffusion imaging at 2.0 Tesla. J Magn Reson 1990; 86: 445–52. [Google Scholar]
- Verzijl H, Valk J, De Vries R, Padberg GW.. Radiologic evidence for absence of the facial nerve in Möbius syndrome. Neurology 2005; 64: 849–55. [DOI] [PubMed] [Google Scholar]
- Verzijl H, Van der Zwaag B, Cruysberg JRM, Padberg GW.. Möbius syndrome redefined: a syndrome of rhombencephalic maldevelopment. Neurology 2003; 61: 327–33. [DOI] [PubMed] [Google Scholar]
- Verzijl H, van der Zwaag B, Lammens M, ten Donkelaar HJ, Padberg GW.. The neuropathology of hereditary congenital facial palsy vs Möbius syndrome. Neurology 2005; 64: 649–53. [DOI] [PubMed] [Google Scholar]
- Von Graefe A. Diagnostik der Augenmuskellähmungen and Aetiologie und Pathogenese der Augenmuskellähmungen. Handb Der Gesamten Augenheilkunde (Handb Ophthalmol) 1880; 6: 60–7. [Google Scholar]
- Webb BD, Frempong T, Naidich TP, Gaspar H, Jabs EW, Rucker JC.. Mirror movements identified in patients with Moebius syndrome. Tremor Other Hyperkinet Mov 2014; 4: 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler AM, Ridgway GR, Webster MA, Smith SM, Nichols TE.. Permutation inference for the general linear model. Neuroimage 2014; 92: 381–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, et al. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 2006; 31: 1116–28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The imaging data used in this study are available via request to the corresponding authors.