Abstract
Background
Diffuse midline glioma (DMG) is a pediatric malignancy with poor prognosis. Most children die less than one year after diagnosis. Recently, mutations in histone H3 have been identified and are believed to be oncogenic drivers. Targeting this epigenetic abnormality using histone deacetylase (HDAC) inhibitors such as panobinostat (PS) is therefore a novel therapeutic option currently evaluated in clinical trials.
Methods
BH3 profiling revealed engagement in an irreversible apoptotic process of glioma cells exposed to PS confirmed by annexin-V/propidium iodide staining. Using proteomic analysis of 3 DMG cell lines, we identified 2 proteins deregulated after PS treatment. We investigated biological effects of their downregulation by silencing RNA but also combinatory effects with PS treatment in vitro and in vivo using a chick embryo DMG model. Electron microscopy was used to validate protein localization.
Results
Scaffolding proteins EBP50 and IRSp53 were upregulated by PS treatment. Reduction of these proteins in DMG cell lines leads to blockade of proliferation and migration, invasion, and an increase of apoptosis. EBP50 was found to be expressed in cytoplasm and nucleus in DMG cells, confirming known oncogenic locations of the protein. Treatment of glioma cells with PS together with genetic or chemical inhibition of EBP50 leads to more effective reduction of cell growth in vitro and in vivo.
Conclusion
Our data reveal a specific relation between HDAC inhibitors and scaffolding protein deregulation which might have a potential for therapeutic intervention for cancer treatment.
Keywords: chick chorioallantoic membrane, diffuse midline glioma, EBP50, HDACi, IRSp53
Key Points.
HDAC inhibitor induces scaffolding protein EBP50 and IRSp53 expression.
EBP50 and IRSp53 are critical for cancer progression.
Blockade of EBP50 together with HDAC inhibitor could have therapeutic benefits.
Importance of the Study.
Panobinostat is in clinical trials for the treatment of DMG patients. Proteomic analysis revealed upregulation of 2 scaffolding proteins after treatment of DMG cells. To date, the role of EBP50 or IRSp53 in pediatric gliomas has not been investigated. This paper describes an oncogenic role of these scaffolding proteins in vitro and in vivo. A new DMG model based on chick chorioallantoic membrane assay has been used to study the effects of the inhibition of EBP50 on tumor growth. Combination of chemical inhibitor of EBP50 and panobinostat leads to a decrease in tumor proliferation. These two proteins could be potential targets for future treatment in combination with PS.
During cancer growth, changes in gene expression by alteration of epigenetic events including histone modifications are thought to have major impact on prognosis and therapeutic outcome. Controlling histone function by histone deacetylase inhibitors (HDACi) such as panobinostat (PS) (Farydak/LBH-589) is a promising therapeutic option for hematological and solid malignancies. Diffuse midline glioma (DMG; formerly called diffuse intrinsic pontine glioma [DIPG]1) is an incurable pediatric glioma localized in the brainstem. Survival time is less than one year for most patients. These tumors show little sensitivity to classical chemotherapeutic agents, and radiotherapy only has limited impact on disease progression. More than 80% of DMGs carry specific mutations in histone H3, which causes a substitution of lysine 27 to methionine (H3K27M), initiating an oncogenic signaling cascade. This discovery suggests that drugs targeting epigenetic abnormalities might be beneficial as an anticancer strategy for DMG. Indeed, recent studies provided evidence that HDACi using PS can significantly reduce DMG growth in vitro as well as in preclinical animal models.2,3 Panobinostat is actually being studied in numerous clinical trials against various malignancies, including DMG.4
It is therefore of importance to understand which biological processes are affected by PS treatment in DMG cells and how this drug achieves its antitumor effects. Previous studies using the BH3 profiling technique have shown that commitment to programmed cell death is initiated early after exposure to efficient cytotoxic drugs.5,6 We therefore used BH3 profiling to determine PS effects on DMG cells 16 hours after treatment. To gain insight into the molecular changes induced by PS treatment in DMG cells, we performed a proteomic study using 3 different DMG cell lines isolated from patient tissue.3,7 Even though HDACi treatment deregulated expression of many proteins, only 2 proteins were consistently upregulated in all 3 cell lines. Interestingly, both proteins are scaffolding proteins.
The first, EBP50, encoded by solute carrier family 9 isoform A3 regulatory factor 1 (SLC9A3R1) (or NHERF1; Na+/H+ exchange regulatory cofactor),8 plays critical roles in basic cellular functions by modulating signaling molecules such as β-catenin, platelet derived growth factor receptor, epidermal growth factor receptor, and phosphatase and tensin homolog. As a result, its functions are highly dependent on context and cellular compartment and vary between different tissues and cancer types.9
The second protein, IRSp53 (encoded by BAI1 associated protein 2 [BAIAP2]),10 is implicated in fundamental cellular processes such as migration and invasion by controlling actin formation.11 It is expressed in filopodia, interacts with cell division cycle 42 (CDC42) and Rac, and is thought to be critical for axon guidance and neurite outgrowth.12
In this study, we provide evidence that EBP50 and IRSp53 play important roles in DMG progression in vitro and in vivo and suggest that efficacy of anticancer therapy using HDACi could be enhanced by additional inhibition of protumoral proteins consistently induced by the treatment.
Materials and Methods
Cell Lines
Primary pediatric human glioma SU-DIPG-IV, SU-DIPG-VI, and SU-DIPG-XIII were a gift from M. Monje (Stanford University) and cultured as previously described.3 NEM163, NEM157, and NEM168 DMG primary (J. Grill, Gustave Roussy) were cultured as previously described.7 All cell lines have H3.3 K27M mutation status except for SU-DIPG-IV, which has H3.1 K27M mutation. All cell lines have been tested routinely for presence of mycoplasma by PCR. The genetic identity of the cell lines was verified by short tandem repeat profiling (LGC Standards).
Chemical Inhibitors
Panobinostat was purchased from Selleckchem (Euromedex). EBP50 inhibitor RS5517 was a gift from Prof R. Silvestri (Sapienza University of Rome, Institut Pasteur Italy).13
Small Interfering RNA Transfection
Small interfering (si)RNAs targeting SLC9A3R1 (5′-CGGCGAAAACGTGGAGAAG[dTdT]-3′) and BAIAP2 (5′-TTCCAGCTGATTCTGGATCTGCCTG[dTdT]-3′) were purchased from Eurofins (Genomics). AllStars negative control siRNA was purchased from Qiagen.
DMG Cell Immortalization
LoxP containing human immunodeficiency virus–derived lentiviral constructs expressed the SV40 T-Ag (pLOX-Ttag iresTK) or human telomerase (pLOX-TERT-iresTK) from an internal human cytomegalovirus immediate early promoter.14 Lentiviral vectors were produced by a triple transient transfection as previously described15 (see Supplementary Material).
EBP50 Overexpressing Cells
Subconfluent 293T cells were transfected with lentiviral vector (psPAX2),16 with an envelope coding plasmid (pMD2G-VSVG) and with GFP-NHER1 vector (H. J. Kreienkamp, University of Hamburg) constructs by calcium phosphate precipitation. Viruses were harvested 48 hours post-transfection and concentrated by ultracentrifugation. NEM157-i and SU-DIPG-IV-i were transduced with multiplicities of infection of 5, 10, and 20 (see Supplementary Material).
Production of VEGF165 Overexpressing NEM157-i Cells
The coding sequence of the codon optimized human vascular endothelial growth factor 165 (VEGF165) isolated from the plasmid PCCL.sin_PPT.SV40polyA.RFP.minhCMV.hPGK.hVEGF165.WPRE (gift from Nicholas D. Mazarkis) was cloned in bicistronic self-inactivating lentiviral vector17 (see Supplementary Material).
Cell Growth and Viability Assays
Cell growth was measured with the In Vitro Toxicology Assay Kit (sulforhodamine B, Sigma) according to manufacturer’s instructions. Cells were plated at a density of 5000 cells per well in 96-well plates in quadruplicates. Absorbance was measured at 565 nm using the CLARIOstar multiplate reader (BMG Labtech).
Cell Death Assays
BH3 profiling/annexin-V/propidium iodide (PI)
—BH3 profiling was performed as described.5 Ten thousand cells were used for all DIPG cell lines. Percentage of polarization was normalized with control dimethyl sulfoxide (DMSO) and percent of depolarization was calculated by the inverse of percent polarization.5 Apoptosis at 72 hours was confirmed using standard annexin-V/PI staining on a flow cytometer (see Supplementary Material).
Wound Healing Migration/Invasion Assays
For invasion assays, plates were pre-coated with Matrigel (growth factor reduced, Corning Life Sciences) at 100 µg/mL. After reaching confluence, the cellular layer was scratched using a plastic pipette tip and 50 µL of Matrigel at 8 mg/mL was added for invasion assay. The images were taken using a ZOE Fluorescent Cell Imager (Bio-Rad), and closure of the scratch was quantified using ImageJ (National Institutes of Health) and a wound healing macro (http://dev.mri.cnrs.fr/projects/imagej-macros/wiki/Wound_Healing_Tool).
Chemical Inhibitor/siRNA Combination Assays
For apoptosis assays, a reverse transfection was made with 12 nM of siRNA, and PS was added 24 hours after transfection. Assay was stopped for fluorescence activated cell sorting (FACS) analysis 24 hours after PS treatment. For proliferation assays, forward transfection was used after adherence of cells in 96-well plates according to manufacturer’s instructions (see Supplementary Material). For RS5517 treatment, cells were mixed at the same time with PS at different concentrations and proliferation was measured after 72 hours.
Proteomic Analysis
Analysis was performed by the Proteomics Core Facility at the University of Bordeaux (https://proteome.cgfb.u-bordeaux.fr/en). Sample preparation, protein digestion, and nano-liquid chromatography–tandem mass spectrometry analysis were performed as previously described.18 Parameters of label-free quantitative data analysis are listed in the Supplementary Material.
Western Blots
Protein extracts were prepared using radioimmunoprecipitation assay buffer plus proteinase inhibitors (Sigma). Protein concentrations were measured using Pierce bicinchoninic acid protein assays (Thermo Fisher Scientific) and cell extracts were loaded in 4–15 % precasted polyacrylamide gel (Bio-Rad). Proteins were immunoblotted on a nitrocellulose membrane (Transblot Turbo midi-size; Bio Rad) and detected with corresponding antibodies (see Supplementary Material). Membranes were scanned on the OdysseyFC (LI-COR Biosciences) or with Fusion FX (Vilber Lourmat).
Immunofluorescence Staining
Cells were seeded at 30 000 cells/well in 24 wells on a glass slide. After adherence, PS treatment or DMSO was added during 24 hours before fixation with 4% paraformaldehyde (PFA) during 10 minutes and washed with phosphate buffered saline (PBS). Permeabilization was made with 0.2% Triton X-100 for 10 minutes. After washing with PBS, cells were blocked with 4 % bovine serum albumin diluted in PBS for 10 minutes and probed with corresponding antibodies (see Supplementary Material).
Electron Microscopy
NEM157-i and SU-DIPG-VI-i were treated with 1 µM PS or 0.1% DMSO during 24 hours, prior to fixation and embedding in resin and processing for ultrathin sectioning.
For immunogold labeling, cells were incubated with rabbit polyclonal anti-EBP50 (1:50 dilution; ab3452, Abcam) followed by secondary immunoglobulin G antibodies coupled with 10 nm gold (Aurion). Images were collected with a TEM Hitachi H7650 and a Gatan Orius Camera (see Supplementary Material).
In Vivo Models
Chicken chorioallantoic membrane (CAM) assays
—Animal procedures were carried out in agreement with the European (directive 2010/63/UE) and French (decree 2013–118) guidelines. Briefly, embryos were received at the stage of segmentation and then incubated at 37.4°C at 70% humidity. At day 3 of development, the eggshell was opened on the top and the opening sealed with medical-grade Durapore tape. At day 10 of development, 5 × 105 NEM157-i plus 5 × 105 NEM157-i-VEGF cells were mixed with Matrigel and 40 µL were put directly on the CAM. Tumor growth was monitored under a stereomicrosope (DS-Fi2, Nikon /SMZ745T). Tumors were fixed with PFA 4% or collected for protein extraction after 6 or 7 days of implantation.
Xenopus laevis embryos
—For PS treatment, batches of 2-cell stage or gastrula stage embryos (10 embryos per batch) were incubated in 24-well plates containing 0.1-1-10-100 µM PS. Embryos were left in solution until control embryos (untreated embryos) reached stage 37–38 and then fixed in MEMFA for histology (see Supplementary Material).
Antibody Array
Cells were lysed and total proteins were extracted 48 hours after transfection. According to manufacturer’s protocol, 400 µg cell lysate was incubated on membrane with each human phosphokinase array (R&D Systems). Chemiluminescence was detected by using Fusion FX (Vilber Lourmat) and pixel densities were determined using ImageJ software.
Statistical Analysis
Statistical analysis was carried out using GraphPad Prism 5.0 (see the Supplementary Material). Experimental data reported were mean ± SEM of a minimum of 3 samples, n = independent experiments. A P-value of <0.05 was considered to be statistically significant. For all data in figures, *P < 0.05, **P < 0.01, and ***P < 0.001.
Results
Early Induction of Apoptosis After Panobinostat Treatment in DMG Cell Lines
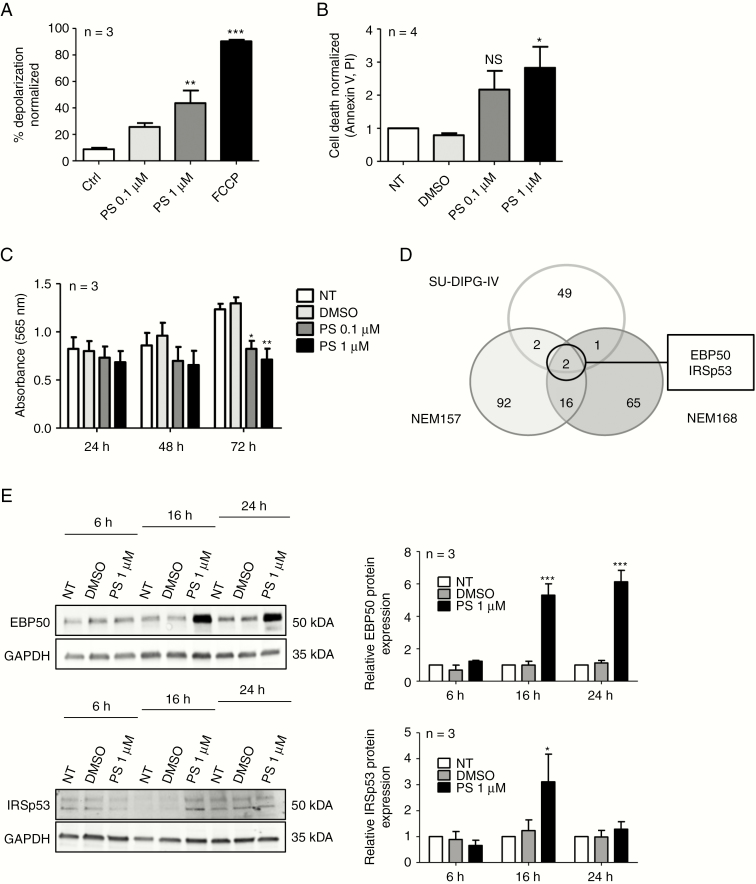
To gain insight into the way PS exerts its antitumor activity,3 we analyzed DMG cells by BH3 profiling after PS exposure. BH3 profiling provides a powerful method of early detection of cell commitment to apoptosis based on quantification of the degree of depolarization of the mitochondrial membrane.6 Even short-term exposure (16 hours) to PS engaged DMG cells to cell death. At 1 µM, we could evidence a significant depolarization of NEM157 cells compared with controls (Fig. 1A). Similar results were obtained in a second DMG cell line, NEM168 (Supplementary Fig. 1A). Completion of apoptosis was then confirmed using annexin-V and PI staining by FACS analysis after 72 hours of treatment at the same drug concentrations (Fig. 1B). Apoptosis was paralleled by inhibition of proliferation even at 0.1 µM of PS compared with solvent. Treatment of NEM157 cells with PS at 0.1 or 1 µM reduced cell proliferation after 72 hours of treatment compared with controls (Fig. 1C). Comparable results were also obtained in NEM168 and immortalized NEM157-i cells (Supplementary Fig. 1B, 1C).
Fig. 1.
Panobinostat treatment effects in DMG cell lines. (A) BH3 profiling of NEM157 cells at 90 minutes. Mitochondrial depolarization was measured with fluorescent dye JC-1, 16 hours after DMSO (solvent) or PS treatment. Data were normalized to DMSO. Ctrl: DMSO; FCCP (carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone) : positive control. (B) NEM157 cell death was measured by FACS using annexin-V–fluorescein isothiocyanate /PI staining 72 hours after treatment. Data were normalized to not-treated cells. (C) NEM157 cell growth was measured by absorbance at different timepoints using a sulforhodamine B assay. (D) Venn diagram resuming number of deregulated proteins (P < 0.05; fold change >1.5 or <0.7) after PS treatment on indicated cell lines. (E) Western blot validation of EBP50 and IRSp53 protein expression at different timepoints in NEM157 cells. Graphs were normalized to glyceraldehyde 3-phosphate dehydrogenase. NT: not treated.
EBP50 and IRSp53 Induction After Panobinostat Treatment
A proteomic approach was chosen to identify proteins deregulated by PS treatment using DMG cell lines SU-DIPG-IV, NEM157, and NEM168. We focused on proteins that are deregulated after 16 hours of PS treatment compared with controls. Following criteria of selection (0.7 <ratio >1.5, P < 0.05), we found a total of 227 proteins (Fig. 1D and Supplementary Table 1) deregulated in either of the cell lines, but only 2 proteins were upregulated in all 3 cell lines (Fig. 1D). EBP50 protein was found to be induced 2–3 times in treated DIPG cell lines and IRSp53 protein was induced 2–4 times, dependent on the cell line (Supplementary Table 1).
EBP50 and IRSp53 protein induction after PS treatment was validated by western blots in NEM157 cells (Fig. 1E) and NEM168, SU-DIPG-IV, and SU-DIPG-VI cells (Supplementary Fig. 2A, 2B). PS induced EBP50 and IRSp53 protein in a dose-dependent manner, with first visible induction at concentrations around 0.05 µM (Supplementary Fig. 2C).
Artificial Upregulation of EBP50 Using a Lentiviral System or mRNA Injection
To evaluate if high EBP50 levels have cytotoxic or growth promoting effects in cells or embryos, EBP50 was stably overexpressed by lentivirus infection, but no effects on cell growth were observed. Injection of mRNA in xenopus embryos causes no developmental defects (Supplementary Fig. 4 and Supplementary Results).
Localization Studies of EBP50 in DMG Cells
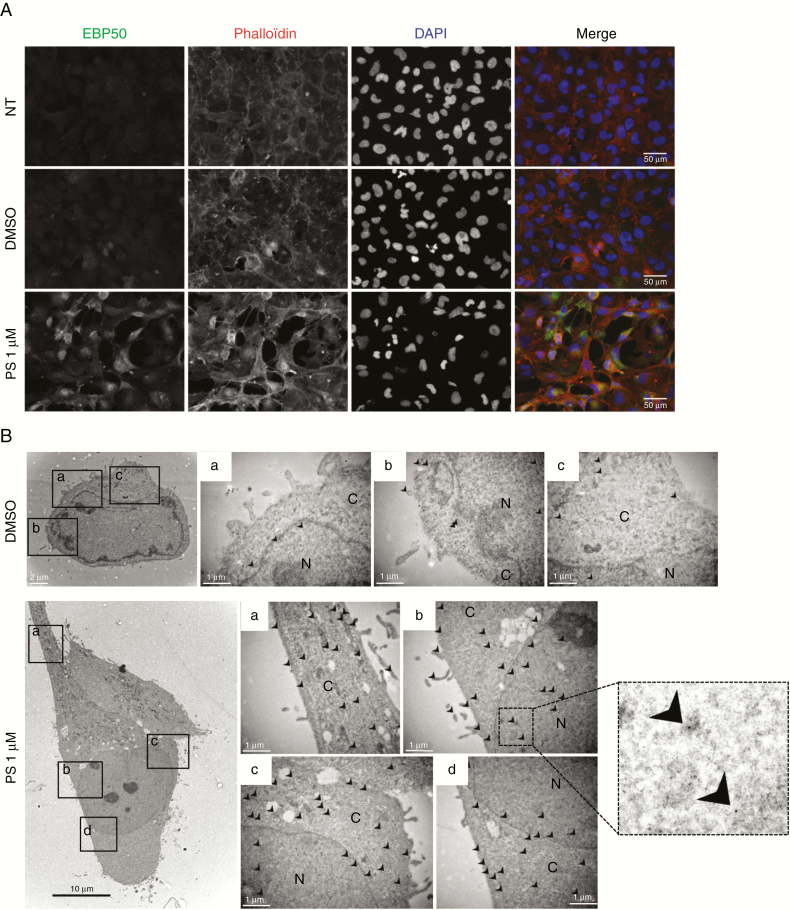
Cytoplasmic and nuclear expressions of EBP50 are associated with an oncogenic role of the protein.9 Using immunofluorescence or electron microscopy, we detected EBP50 in all cellular structures in untreated or control conditions. PS treatment did not delocalize EBP50 to a specific compartment but increased its expression in the nucleus and cytoplasm compared with controls (Fig. 2A, 2B). Similar results were also obtained in other DMG cell lines (Supplementary Fig. 3A, 3B). These localization data suggest an oncogenic function of EBP50 in DMG pathology.
Fig. 2.
EBP50 immunocytochemistry and immunogold labeling. NEM157-i cells were treated with DMSO or PS during 24 hours before fixation. (A) Immunofluorescence with EBP50 antibody (green), phalloïdin (red), and nuclei (blue) staining. Objective x40. (B) Immunogold electron microscopy localization of EBP50 proteins. Arrows indicate the localization of EBP50 detected by 10 nm gold particles in different cellular compartments. Box on the right represents a zoom to visualize gold particles. N: nuclei, C: cytoplasm. Scale bars are indicated on pictures.
Downregulation of EBP50 or IRSp53 Protein Level Affects Cellular Growth and Motility of DMG Cell Lines
siRNA silencing
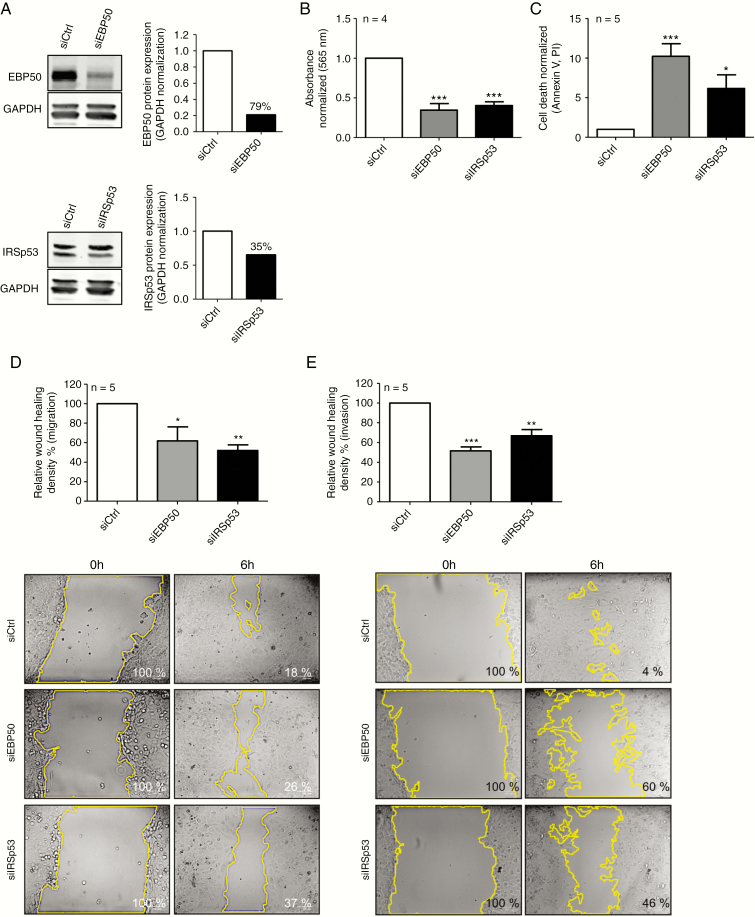
To assess the effect of EBP50 or IRSp53 downregulation in DMG cell lines, we transfected cells with siRNAs targeting SLC9A3R1 or BAIAP2, validated by Western blots (Fig. 3A). EBP50 and IRSp53 siRNA knock-down significantly reduced proliferation compared with controls (Fig. 3B). Low levels of the two proteins were associated with a strong increase in cell death after 72 hours of transfection (Fig. 3C). Similar results were obtained using the NEM168 cell line (Supplementary Fig. 5A–C).
Fig. 3.
Biological effects of EBP50 and IRSp53 downregulation by siRNAs (NEM157-i line). (A) Western blot validation of downregulation by siRNAs. Percent downregulation is indicated on graphs. (B) Cell proliferation assays 96 hours after transfection measured by sulforhodamine B assay. (C) Cell death was measured by FACS with annexinV–fluorescein isothiocyanate/PI staining 72 hours after transfection. (D) Migration 48 hours after transfection was measured using wound healing assay. Cells were incubated 6 hours after injury. (E) Invasion was measured 48 hours after transfection by wound healing assay coated with Matrigel. Cells were incubated 48 hours after injury. Representative photos of one experiment used for quantification are shown. All graphs were normalized to siCtrl.
EBP50 and IRSp53 are scaffolding proteins implicated in the interaction between membrane proteins and the cytoskeleton. We therefore investigated whether EBP50 or IRSp53 affects cell migration or invasion of NEM157-i cells. Wound closure 6 hours after scratching was significantly delayed by EBP50 or IRSp53 siRNAs (Fig. 3D). Invasion was also reduced after 48 hours in siRNA conditions (Fig. 3E). EBP50 or IRSp53 downregulation also significantly reduced cell motility in a second DMG cell line, NEM168 (Supplementary Fig. 5D, 5E).
Combination of EBP50 or IRSp53 inhibition with PS treatment in vitro
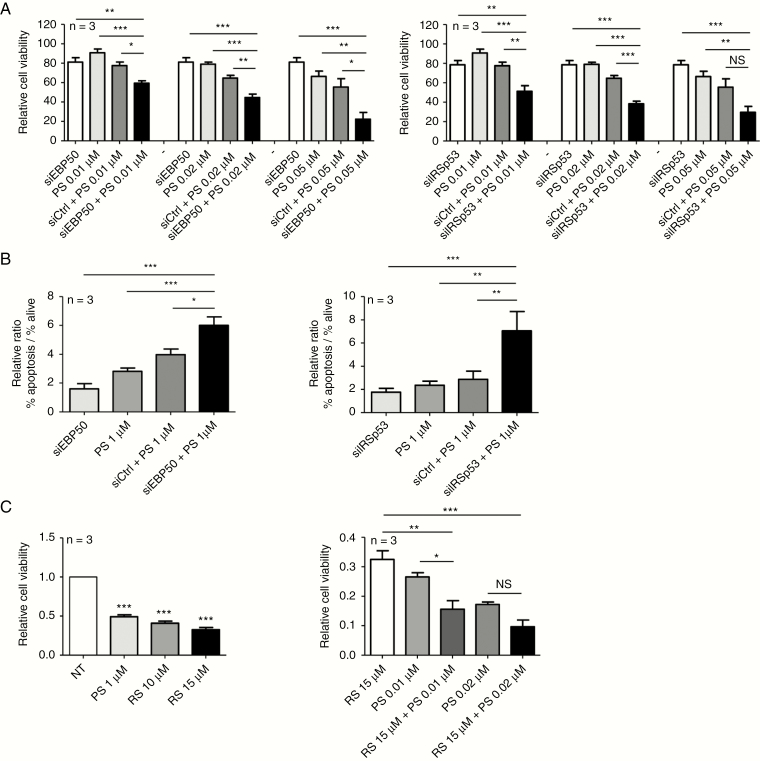
Given the fact that both proteins induced by PS play roles in cell growth and survival, we reasoned that combined inhibition could increase efficacy of PS. Cells were first transfected with siRNA and 24 hours after, cells were treated with PS at different concentration during 48 hours. Proliferation was significantly decreased in cells treated with siEBP50 in combination with PS from 0.01 µM to 0.05 µM compared with controls (siCtrl/PS), resulting in 20–50% greater inhibition (Fig. 4A). Similar results have been observed for siIRSp53, except at the highest dose of PS tested, where differences were observed but not significant (Fig. 4A). This reduction of cell growth in the combination siRNA/PS was accompanied by a 30% (siEBP50) or 50% (siIRSp53) increase in apoptotic cell number compared with controls (Fig. 4B).
Fig. 4.
Combinatory effects of PS treatment and EBP50 or IRSp53 inhibition (NEM157-i line). (A) Cells were transfected with indicated siRNAs in combination with different PS doses (0.01, 0.02, or 0.05 µM). Cell viability was measured by absorbance using sulforhodamine B assay. Graph was normalized to siCtrl. (B) Apoptosis was measured by flow cytometry (annexin-V–fluorescein isothiocyanate/PI) and graphs were normalized to nontransfected cells. (C) Cells were treated with EBP50 inhibitor RS5517 (RS) at different doses. Cell viability was measured by absorbance using sulforhodamine B assay. Graphs were normalized to not-treated cells.
RS5517 is a new PDZ1 (postsynaptic density protein, Drosophila disc large tumor suppressor, zonula occludens 1 protein) domain antagonist of EBP50. In colorectal cancer cells, this inhibitor has growth inhibitory effects.13 Viability of NEM157-i cells was decreased about 50% at all doses of RS5517 tested (1 µM to 15µM) compared with solvent control (Fig. 4C). Importantly, treatment with a low dose of PS (0.01 µM) in association with RS5517 at 15 µM significantly reduced tumor cell growth compared with single treatments alone. This synergy was lost at a higher dose of PS (Fig. 4C). These results further underline benefits of the combinatorial EBP50/PS inhibition strategy.
Antibody Array
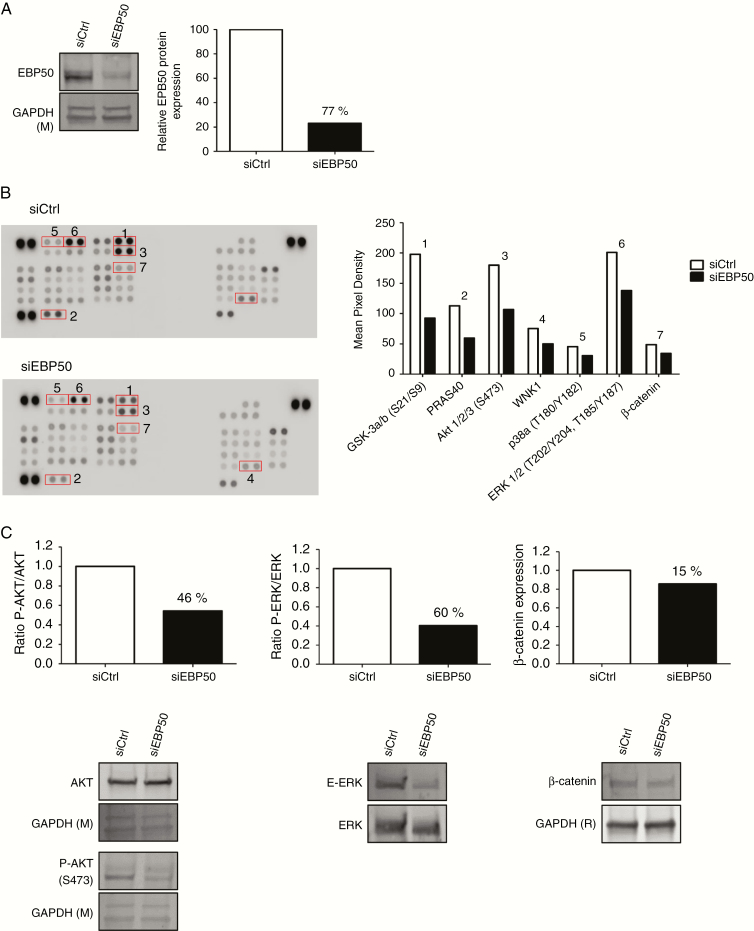
In order to shed light on the molecular alterations occurring in cells after siEBP50 treatment, we performed a phosphokinase assay. We obtained 77% reduction of EBP50 protein 48 hours after siRNA transfection (Fig. 5A). Proteins from these cells were spotted on the kinase array. Out of 43 human kinases presented on the array, only 7 were affected by reduction of EBP50 protein (Fig. 5B). We validated 46% reduction of phosphorylation at S473 of AKT, 60% reduction of phospho‒extracellular signal-regulated kinase (ERK), and 15% decrease of β-catenin (Fig. 5C).
Fig. 5.
Knock-down effects of siEBP50 on phosphorylation of protein kinases. (A) Validation of siEBP50 efficacy expression by western blot in NEM168-i cells, percent downregulation is indicated on graph. (B) Relative levels of phosphorylation of 43 kinase phosphorylation sites were compared between siCtrl and siEBP50 after 48 hours of transfection. Graphs represent mean pixel density of proteins that are deregulated with a ratio <0.7-fold. Proteins are classified by numbers 1–7. (C) Western blot validation of phospho-AKT (3), phospho-ERK (6), and β-catenin (7) in NEM168-i cells. Graphs illustrate percent downregulation of the blots below. Glyceraldehyde 3-phosphate dehydrogenase (R) or (M): rabbit or mouse antibody.
Development of an In Vivo DMG Model in Chick Embryos
To access the role of EBP50 in an in vivo setting, we developed a new DIPG tumor model on the CAM, adapted from our experimental glioma model.19 The initial DIPG cell line NEM157-i did not implant reliably on the CAM due to slow growth and insufficient angiogenic response. We therefore generated a NEM157-i cell line stably overexpressing VEGF165 (NEM157-i-VEGF), a growth factor which favors implantation of human tumor cells in this model.19
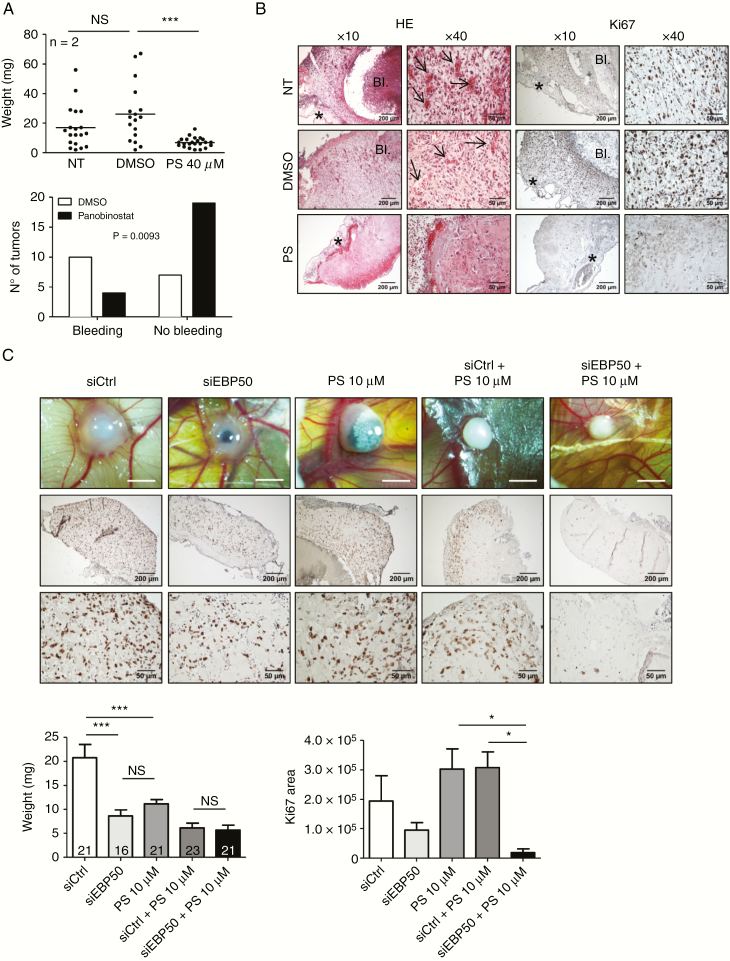
To validate the model and to evaluate the inhibitory effect of PS, tumor cell mixture (1:1 NEM157-i-VEGF with NEM157-i cells) was implanted on the CAM in Matrigel containing PS at 40 µM. Seven days after implantation, PS-treated tumors had significant 4- to 5-fold reduction of tumor growth compared with controls (Fig. 6A, upper graph). Control tumors often show bleeding associated with tumor growth, a phenomenon that was significantly less observed in PS-treated tumors (Fig. 6A, lower graph). Histological analysis of tumor sections confirmed the presence of chick blood vessels inside the tumor graft in control, but not in PS-treated tumors (Fig. 6B, arrows). Ki-67 immunostaining was strongly reduced in PS tumors compared with controls (Fig. 6B, right panels). This model confirmed the growth inhibitory effects of PS on DMG development and suggests anti-angiogenic effects of the drug in this pathology.
Fig. 6.
Development of a DMG CAM model. NEM157-i-VEGF and NEM157-i cells were mixed at 1:1 ratio embedded in Matrigel containing PS or solvent and implanted on CAMs. (A) Tumor weights 7 days after implantation and resection (upper graph) and classification of tumors presenting bleeding or not (lower graph). (B) Tumor sections stained by hematoxylin & eosin (HE) or Ki-67 immunostaining, 7 days after implantation. Arrows show chick blood vessels present in tumors. BI: bleeding; *CAM. (C) Tumor monitoring by stereomicroscopy and tumor sections immunostained for Ki-67, six days after implantation. White scale bar represents 2 mm. (D) Six days post-implantation, tumor weights and total Ki-67 area were measured. Numbers of CAMs per group are indicated on graph for tumor weights. Four CAMs were used to quantify Ki-67 area. NT: not treated.
In Vivo Effects of EBP50/PS Inhibition (CAM Assay)
Given the encouraging in vitro results showing an improved antitumor activity of a combined PS/anti-EBP50 approach, we sought to validate this effect in the DMG-CAM model. Tumors were transfected with siCtrl or siEBP50 in association or not with PS at 10 µM before implantation on CAM. Tumors were photographed by stereomicroscopy and analyzed by standard histology and Ki-67 staining (Fig. 6C). As expected, treatment with siEBP50 or PS alone did decrease tumor weight by approximately 50%. SiCtrl plus PS and siEBP50 plus PS even decreased tumor weight slightly more. There was no significant difference between these 2 groups (Fig. 6D). Immunostaining with Ki-67 revealed lower proliferation indexes in siEBP50 tumors compared with siCtrl. However, cell proliferation was nearly abolished in siEBP50/PS-treated tumors compared with PS alone or siCtrl/PS treatments (Fig. 6C, 6E). These results suggest a beneficial effect of combined anti-EBP50/PS treatment and are in line with the in vitro results.
Discussion
In this study, we analyzed biological effects of the HDAC inhibitor panobinostat on cell lines derived from pediatric glioma patients. Using the BH3 profiling technique, we could evidence that engagement to apoptosis occurs as early as 16 hours after exposure to the drug. Proteomic analysis was carried out to determine drug action on cells.
Only 2 proteins, IRSp53 and EBP50, were commonly induced by the treatment in 3 different DMG cell lines; we therefore studied their role in diffuse midline glioma pathology.
IRSp53 encoded by the BAIAP2 gene is a scaffolding protein that bundles actin filaments and interacts with the small GTPase Rac.20 It plays a role in filopodium dynamics by coupling actin elongation to membrane protrusions and is an important effector of CDC42 that orchestrates events required for filopodium formation.21 Little is known about the role of IRSp53 in cancer, but its pivotal role in the formation of lamellipodia via Rac activation suggests participation in the invasive behavior of cancer cells.22 Our in vitro results strongly confirm an important function of IRSp53 in the invasion but also proliferation of pediatric glioma cell lines.
We have chosen to study EBP50 protein more in detail. Depending on the subcellular location of the protein, EBP50 binding partners change and therefore the net outcome of the interaction for cell behavior. It is generally accepted that membrane-bound EBP50 has tumor suppressive functions, whereas cytoplasmic and nuclear localization is associated with oncogenic growth-promoting behavior.9 Our results clearly show that in DMG cells, EBP50 is expressed in the cytoplasm, nucleus, and membrane as evidenced by immunocytochemistry and immunogold electron microscopy. Nuclear presence of β-catenin protein seems not to be common in DMG tumors23; it is therefore possible that the oncogenic potential of EBP50 in DMG cells is mediated by other nuclear factors, such as Yes associated protein 1, which are known to interact with EBP5024 and are known to be overexpressed in central nervous system tumors, including pediatric glioma.25 In DMG, depletion of EBP50 leads to blockage of cell proliferation and increase of apoptosis, as reflected by a decrease of ERK and AKT phosphorylation. Further studies need to address the question of which proteins EBP50 specifically interacts with in DMG cells.
In adult glioma, EBP50 is associated with an invasive phenotype and is highly expressed in cells at the invading front of tumors. Reducing its expression inhibited glioma cell migration and sensitized tumor cells to temozolomide treatment.26
Induction of NHERF1/EBP50 after HDACi seems to be a conserved mechanism, since several microarray studies carried out after short-term treatment in normal human cell lines such as mesenchymal stromal cells and osteoblasts found consistent upregulation.27,28 Interestingly, in the list of genes highly induced by HDACi published by Dudakovic et al,29 we also found BAIAP2 encoding IRSp53. It is tempting to speculate that these 2 proteins participate in a signaling network initiated by HDACi. Even more intriguing, Garbett et al have shown that EBP50 and the T-isoform of IRSp53 physically interact in a human choriocarcinoma cell line.30 The very strong binding occurs via the C-terminal PDZ-binding domain of IRSp53-T directly to the PDZ1 domain of EBP50. Using an isoform-specific antibody against IRSp53-T, we could evidence expression of this isoform in NEM157 or SU-DIPG-IV glioma cell lines (Supplementary Fig. 6A), suggesting that physical interaction of these 2 proteins occurs in DMG cells. However, this isoform is not induced by PS treatment (Supplementary Fig. 6B).
Our data suggest that EBP50 protein has oncogenic functions in pediatric glioma progression. Several studies also show tumor suppressor roles for this protein. It is therefore important to determine for each cancer type for which HDACi will be proposed, where EBP50 is localized and which biological role it plays.
Inhibition of protein-protein interaction via small molecules targeting PDZ domains is a promising novel therapeutic approach.31 This might result in novel anticancer drugs which block specific binding of oncogenic signaling partners of EBP50. Saponaro et al have designed RS5517, which achieves cytotoxicity by blocking protein interactions with the PDZ1 domain of EBP50.13 In addition, efforts to restore EBP50 to its physiological antitumoral plasma membrane location deserve thorough consideration as a novel strategy for cancer treatment.32
Toxicity of PS remains a major concern.2 Our study using the xenopus embryo model revealed unexpected developmental defects in the central nervous system, leading to complete absence of eye development at high doses (Supplementary Fig. 7). Even though it is difficult to translate this finding into clinical application of the drug, this information underlines the necessity to evaluate alternative delivery methods such as convection-enhanced delivery,33 thereby limiting exposure to normal tissue.
Taken together, our results have several implications for the treatment of DMG and for anticancer therapy using HDACi in general. First, even though augmentation of EBP50 protein does not affect tumor cell viability, reduction significantly affects important protumoral features of cancer cells. EBP50 expression is required for DMG cell growth and HDACi provides cells with higher EBP50 levels. PS treatment prevents diminution of a protein important for DMG cell proliferation which indirectly could impact PS efficacy on cancer cells. Second, inhibition of EBP50 protein induced by HDACi significantly improves cytotoxicity of HDACi at lower doses, both in vitro and in vivo. This novel combinatory therapy concept warrants further investigation in other solid tumors with poor prognosis.
Funding
This study was supported by grants from the Agence Nationale de la Recherche (ANR) in the frame of the “Investments for the future” program, IdEx program Univ Bordeaux (reference ANR-10-IDEX-03-02), and the following charity associations: Eva Pour La Vie, Aidons Marina, Fondation Flavien, E.S.C.A.P.E., Les Amis de Marius, Kaëna et les lapinours foundation, the Cassandra Contre la Leucémie association, Les Motards Solidaires and the Sphères foundantion. CC was supported by “Fondation Groupama pour la santé” and Eva Pour La Vie.
Supplementary Material
Acknowledgments
The authors thank M. Monje, Stanford University and J. Grill, Institut Gustave Roussy, Paris, for providing DMG cell lines. E. Richard, INSERM U1035, Univ Bordeaux and D. Trono, EPF, Lausanne provided material and help for immortalization of DMG cells; N. D. Mazarakis, University College London and L. Naldini, Univ Milan provided plasmids for construction of VEGF overexpressing cells, and H. J. Kreienkamp, Univ Hamburg provided BAIAP2 and NHERF1 plasmid constructions. EBP50 inhibitor RS5517 was a gift from Prof R. Silvestri (Sapienza University of Rome, Italy). Imaging was performed at the Bordeaux Imaging Center, member of the FranceBioImaging national infrastructure (ANR-10-INBS-04). A. Bibeyran and V. Guyonnet-Duperat (Vect’UB, INSERM US005–CNRS UMS3427–TBM-Core, Univ Bordeaux) provided help for lentivirus production, C. Pain (INSERM U1035) helped with histological sections and staining, V. Pitard helped with flow cytometry and J. W. Dupuy with proteomic analysis. F. Sagliocco performed cloning of VEGF, BAIAP2, and NHERF lentivirus constructions and M. Ménard helped with cell culture and proliferation assays.
Conflict of interest statement. The authors declare no conflict of interest.
Authorship statement. Cell culture, histology, and CAM experiments: CC, AD, and JC. Western blots, antibody array, functional test (apoptosis, proliferation, and migration): CC. Cloning, preparation of lentiviral constructs: FS. Xenopus experiments: PT, NT, and SF. Bioinformatics analyses: KBH. Lentivirus production: VGD. Electron microscopy: MP. Proteomic analysis: AAR and JWD. Prepared figures, contributed to writing: CC. Supervised experiments: CFG and MH. Designed study, wrote the paper: MH.
References
- 1. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016; 131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 2. Hennika T, Hu G, Olaciregui NG, et al. Pre-clinical study of panobinostat in xenograft and genetically engineered murine diffuse intrinsic pontine glioma models. PLoS One. 2017; 12(1):e0169485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Grasso CS, Tang Y, Truffaux N, et al. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat Med. 2015; 21(6):555–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clinical Trials Website, 2019: https://clinicaltrials.gov/ct2/results?cond=DIPG&term=LBH-589.
- 5. Montero J, Sarosiek KA, DeAngelo JD, et al. Drug-induced death signaling strategy rapidly predicts cancer response to chemotherapy. Cell. 2015; 160(5):977–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Deng J, Carlson N, Takeyama K, Dal Cin P, Shipp M, Letai A. BH3 profiling identifies three distinct classes of apoptotic blocks to predict response to ABT-737 and conventional chemotherapeutic agents. Cancer Cell. 2007; 12(2):171–185. [DOI] [PubMed] [Google Scholar]
- 7. Truffaux N, Philippe C, Paulsson J, et al. Preclinical evaluation of dasatinib alone and in combination with cabozantinib for the treatment of diffuse intrinsic pontine glioma. Neuro Oncol. 2015; 17(7):953–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reczek D, Berryman M, Bretscher A. Identification of EBP50: A PDZ-containing phosphoprotein that associates with members of the ezrin-radixin-moesin family. J Cell Biol. 1997; 139(1):169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vaquero J, Nguyen Ho-Bouldoires TH, Clapéron A, Fouassier L. Role of the PDZ-scaffold protein NHERF1/EBP50 in cancer biology: from signaling regulation to clinical relevance. Oncogene. 2017; 36(22):3067–3079. [DOI] [PubMed] [Google Scholar]
- 10. Shiratsuchi T, Futamura M, Oda K, Nishimori H, Nakamura Y, Tokino T. Cloning and characterization of BAI-associated protein 1: a PDZ domain-containing protein that interacts with BAI1. Biochem Biophys Res Commun. 1998; 247(3):597–604. [DOI] [PubMed] [Google Scholar]
- 11. Scita G, Confalonieri S, Lappalainen P, Suetsugu S. IRSp53: crossing the road of membrane and actin dynamics in the formation of membrane protrusions. Trends Cell Biol. 2008; 18(2):52–60. [DOI] [PubMed] [Google Scholar]
- 12. Ahmed S, Goh WI, Bu W. I-BAR domains, IRSp53 and filopodium formation. Semin Cell Dev Biol. 2010; 21(4):350–356. [DOI] [PubMed] [Google Scholar]
- 13. Saponaro C, Sergio S, Coluccia A, et al. β-catenin knockdown promotes NHERF1-mediated survival of colorectal cancer cells: implications for a double-targeted therapy. Oncogene. 2018; 37(24):3301–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Salmon P, Oberholzer J, Occhiodoro T, Morel P, Lou J, Trono D. Reversible immortalization of human primary cells by lentivector-mediated transfer of specific genes. Mol Ther. 2000; 2(4):404–414. [DOI] [PubMed] [Google Scholar]
- 15. Richard E, Mendez M, Mazurier F, et al. Gene therapy of a mouse model of protoporphyria with a self-inactivating erythroid-specific lentiviral vector without preselection. Mol Ther. 2001; 4(4):331–338. [DOI] [PubMed] [Google Scholar]
- 16. Dull T, Zufferey R, Kelly M, et al. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998; 72(11):8463–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mélard P, Idrissi Y, Andrique L, et al. Molecular alterations and tumor suppressive function of the DUSP22 (dual specificity phosphatase 22) gene in peripheral T-cell lymphoma subtypes. Oncotarget. 2016; 7(42):68734–68748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Henriet E, Abou Hammoud A, Dupuy JW, et al. Argininosuccinate synthase 1 (ASS1): A marker of unclassified hepatocellular adenoma and high bleeding risk. Hepatology. 2017; 66(6):2016–2028. [DOI] [PubMed] [Google Scholar]
- 19. Hagedorn M, Javerzat S, Gilges D, et al. Accessing key steps of human tumor progression in vivo by using an avian embryo model. Proc Natl Acad Sci U S A. 2005; 102(5):1643–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Millard TH, Dawson J, Machesky LM. Characterisation of IRTKS, a novel IRSp53/MIM family actin regulator with distinct filament bundling properties. J Cell Sci. 2007; 120(Pt 9):1663–1672. [DOI] [PubMed] [Google Scholar]
- 21. Goh WI, Lim KB, Sudhaharan T, et al. mDia1 and WAVE2 proteins interact directly with IRSp53 in filopodia and are involved in filopodium formation. J Biol Chem. 2012; 287(7):4702–4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Funato Y, Terabayashi T, Suenaga N, Seiki M, Takenawa T, Miki H. IRSp53/Eps8 complex is important for positive regulation of Rac and cancer cell motility/invasiveness. Cancer Res. 2004; 64(15):5237–5244. [DOI] [PubMed] [Google Scholar]
- 23. Ballester LY, Wang Z, Shandilya S, et al. Morphologic characteristics and immunohistochemical profile of diffuse intrinsic pontine gliomas. Am J Surg Pathol. 2013; 37(9):1357–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mohler PJ, Kreda SM, Boucher RC, Sudol M, Stutts MJ, Milgram SL. Yes-associated protein 65 localizes p62(c-Yes) to the apical compartment of airway epithelia by association with EBP50. J Cell Biol. 1999; 147(4):879–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Orr BA, Bai H, Odia Y, Jain D, Anders RA, Eberhart CG. Yes-associated protein 1 is widely expressed in human brain tumors and promotes glioblastoma growth. J Neuropathol Exp Neurol. 2011; 70(7):568–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kislin KL, McDonough WS, Eschbacher JM, Armstrong BA, Berens ME. NHERF-1: modulator of glioblastoma cell migration and invasion. Neoplasia. 2009; 11(4):377–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schroeder TM, Nair AK, Staggs R, Lamblin AF, Westendorf JJ. Gene profile analysis of osteoblast genes differentially regulated by histone deacetylase inhibitors. BMC Genomics. 2007; 8:362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dudakovic A, Camilleri ET, Lewallen EA, et al. Histone deacetylase inhibition destabilizes the multi-potent state of uncommitted adipose-derived mesenchymal stromal cells. J Cell Physiol. 2015; 230(1):52–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dudakovic A, Evans JM, Li Y, et al. Histone deacetylase inhibition promotes osteoblast maturation by altering the histone H4 epigenome and reduces Akt phosphorylation. J Biol Chem. 2013; 288(40):28783–28791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Garbett D, Sauvanet C, Viswanatha R, Bretscher A. The tails of apical scaffolding proteins EBP50 and E3KARP regulate their localization and dynamics. Mol Biol Cell. 2013; 24(21):3381–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Grillo-Bosch D, Choquet D, Sainlos M. Inhibition of PDZ domain-mediated interactions. Drug Discov Today Technol. 2013; 10(4):e531–e540. [DOI] [PubMed] [Google Scholar]
- 32. Hayashi Y, Molina JR, Hamilton SR, Georgescu MM. NHERF1/EBP50 is a new marker in colorectal cancer. Neoplasia. 2010; 12(12):1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou Z, Singh R, Souweidane MM. Convection-enhanced delivery for diffuse intrinsic pontine glioma treatment. Curr Neuropharmacol. 2017; 15(1):116–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.