Abstract
Background
Early detection of increased aggressiveness of brain tumors is a major challenge in the field of neuro-oncology because of the inability of traditional imaging to uncover it. Isocitrate dehydrogenase (IDH)-mutant gliomas represent an ideal model system to study the molecular mechanisms associated with tumorigenicity because they appear indolent and non-glycolytic initially, but eventually a subset progresses toward secondary glioblastoma with a Warburg-like phenotype. The mechanisms and molecular features associated with this transformation are poorly understood.
Methods
We employed model systems for IDH1 mutant (IDH1mut) gliomas with different growth and proliferation rates in vivo and in vitro. We described the metabolome, transcriptome, and epigenome of these models in order to understand the link between their metabolism and the tumor biology. To verify whether this metabolic reprogramming occurs in the clinic, we analyzed data from The Cancer Genome Atlas.
Results
We reveal that the aggressive glioma models have lost DNA methylation in the promoters of glycolytic enzymes, especially lactate dehydrogenase A (LDHA), and have increased mRNA and metabolite levels compared with the indolent model. We find that the acquisition of the high glycolytic phenotype occurs at the glioma cytosine-phosphate-guanine island methylator phenotype (G-CIMP)-high molecular subtype in patients and is associated with the worst outcome.
Conclusion
We propose very early monitoring of lactate levels as a biomarker of metabolic reprogramming and tumor aggressiveness.
Keywords: IDH1-mutant, glioma, epigenetics, 13C-hyperpolarized MRSI, metabolism
Key Points.
Loss of methylation in glycolytic genes, especially LDHA, is associated with a more aggressive phenotype in IDH1mut gliomas.
High glycolytic metabolism occurs in the G-CIMP-high molecular subtype.
Lactate production is associated with increased aggressiveness in clinical samples and IDH1mut models.
Importance of the Study.
DNA methylation shapes the metabolism of tumors by silencing metabolic genes; however, this can be remodeled providing these tumors with new metabolic capabilities. While IDH1mut gliomas have been considered low glycolytic tumors, this work reveals how the remodeling of their epigenome is associated with a high glycolytic phenotype, and accordingly, patient’s outcome. Herein, we establish the feasibility of using the lactate:pyruvate ratio in preclinical models for monitoring tumor aggressiveness and metabolic reprogramming in IDH1mut gliomas. This biomarker could be readily applied to human clinical trials due to its non-invasive nature and has the potential to identify increased aggressiveness very early.
Rewiring of metabolism has been highlighted as a hallmark of cancer1,2 since nearly a century ago by Otto Warburg. He first described the upregulation of lactate production from glucose in cancer cells under aerobic conditions known as “aerobic glycolysis” or the “Warburg effect.” 3–6
Metabolic reprogramming through mutation of key metabolic enzymes can by itself lead to tumor formation. Most notably, gain-of-function mutations in isocitrate dehydrogenase 1 and 2 (IDH1/2mut) that produce D-2-hydroxyglutarate (D-2HG) are initiator events responsible for 80% of lower grade gliomas (LGGs, World Health Organization grades II–III) and close to 10% of glioblastomas (GBMs, grade IV).7–13 The accumulation of D-2HG leads to gliomagenesis through reshaping of the epigenome via the inhibition of α-ketoglutarate–dependent demethylases.14–16 IDH mutations are sufficient to produce the glioma cytosine-phosphate-guanine (CpG) island methylator phenotype (G-CIMP) in gliomas, which is a unique, genome-wide hypermethylated signature at CpG sites frequent in IDH1mut 1p/19q intact LGG and IDHmut secondary GBM. The G-CIMP signature can be further categorized according to the degree of methylation: prognostically favorable G-CIMP-high and less favorable G-CIMP-low.17–22
Despite their markedly better prognosis, LGGs that are IDH mutated often transform into higher grades and develop resistance to therapy.23–25 Therefore, IDH1mut gliomas that progress toward more aggressive phenotypes offer a unique opportunity to study metabolic reprogramming. They appear initially indolent and are metabolically defective—characterized by lower glycolytic rates and lower energy production—and yet evolve to meet the metabolic demands of fast-growing aggressive tumor phenotypes. However, few studies have profiled the metabolic reprogramming within the LGGs that are IDH1 mutated.25–27 Current hypotheses regarding disease progression within the IDH1mut subtypes are related to the global loss of DNA methylation from G-CIMP-high to G-CIMP-low, loss of IDH1 mutant allele, or the accumulation of multiple mutations.22,28–32
In this study, we used 3 patient-derived cell lines as model systems that recapitulate indolent and aggressive phenotypes in order to study the metabolic reprogramming associated with increasing aggressiveness of only IDH1mut LGGs. To determine the molecular mechanisms by which these models become aggressive, we compared their epigenomic, transcriptomic, and metabolomic profiles with those of a non-aggressive IDH1mut cell line in vitro, and in vivo via hyperpolarized (HP)-MRI. Our results show that the patient-derived IDH1mut aggressive cell lines have acquired a high glycolytic phenotype by the specific loss of promoter methylation in glycolytic genes, especially lactate dehydrogenase A (LDHA). We further showed that a high glycolytic signature in patient samples from The Cancer Genome Atlas (TCGA) is associated with the worst survival. The glycolytic signature splits the G-CIMP-high molecular group into 2 subgroups, which we have defined as “G-CIMP-high high-glycolytic” and “G-CIMP-high low-glycolytic.” We propose that the specific loss of promoter methylation of glycolytic genes, especially LDHA, is associated with aggressiveness and the non-invasive, in vivo monitoring of the pyruvate-to-lactate ratio as a predictive biomarker of aggressiveness of LGGs that are IDH1mut.
Methods
Cell Models and Culture
TS603 (grade III oligodendroglioma), BT142 (grade III oligoastrocytoma), and NCH1681 (grade III astrocytoma)33 were grown in Dulbecco’s modified Eagle’s medium/F12 medium supplemented with 1% N2 growth supplement, heparin sulfate, penicillin-streptomycin, epidermal growth factor, and fibroblast growth factor. Cell lines were cultured and utilized for experiments immediately after expanding in vitro in order to avoid the loss of heterozygosity previously reported.34 Proliferation rate was assessed through daily monitoring of cell growth using a Vi-Cell automatic counter. For 13C-tracing experiments, cells were grown in the same media described above but lacking glucose, which was added as 13C-U-glucose in the same concentrations as the original media. Cells were grown in this labeled media for 72 hours prior to collection for liquid chromatography–mass spectrometry (LC-MS) analysis.
Mice Bearing Tumors
Intracranial orthotopic mouse models with IDH1mut glioma cell lines TS603, NCH1681, BT142, and BT142 overexpressing LDHA were established according to approved animal study proposal NOB-008 by the National Cancer Institute‒Animal Use and Care Committee. Briefly, cells were harvested, washed with phosphate buffered saline and counted. The resulting pellet was resuspended in Hank’s Balanced Salt Solution, and 5 μL of the cell suspension was injected stereotactically into the striatum of female severe combined immunodeficient (SCID) mice 6–8 weeks old (Charles River Frederick Research Model Facility) using a stereotactic device. Tumor growth was monitored for neurological symptoms daily. For comparison of survival curves, the log-rank (Mantel–Cox) test has been used. The BT142 cell line overexpressing LDHA was generated as described in the Supplementary Methods and the corresponding mouse model following the same procedure described above.
Hyperpolarized 13C-MRI of Pyruvate Metabolism
Samples of [1-13C]-pyruvate acid (30 µL) containing 15 mM of OX063 and 2.5 mM of the gadolinium chelate ProHance (Bracco Diagnostics) were polarized in the Hypersense dynamic nuclear polarizer (Oxford Instruments). After 40–60 min, the hyperpolarized sample was rapidly dissolved in 4.5 mL of a superheated Tris based alkaline buffer. NaOH was added to the dissolution buffer to be pH 7.4 after mixture with [1-13C]-pyruvate. Hyperpolarized [1-13C]-pyruvate solution (96 mM) was intravenously injected through a catheter placed in the tail vein of the mouse (12 µL/g body weight). A more detailed description of animal handling can be found in the Supplementary Methods. 13C two-dimensional spectroscopic images were acquired 30 seconds after the start of the pyruvate injection, with a 28 × 28 mm2 field of view in an 8 mm slice through the tumor, a matrix size of 14 × 14, spectral width of 3330 Hz, repetition time of 85 ms, and Gaussian excitation pulse with a flip angle of 10°.
Magnetic Resonance Spectroscopic Data Analysis
13C chemical shift images and 1H anatomical images were merged using MatLab software v9.2. Displayed representative spectra were derived from spectra in tumor regions. Heatmap of the [1-13C] pyruvate, [1-13C] lactate, and [1-13C] lactate to [1-13C] pyruvate ratio was calculated in each pixel of chemical shift images and the resolution was digitally enhanced from matrix size of 14 × 14 to 32 × 32. The median value of the lactate:pyruvate ratio was calculated from chemical shift images in the tumor region. All magnetic resonance spectroscopic (MRS) data analysis except the merging process was performed using ImageJ software v1.51.
LC-MS Metabolomics
Analysis for cell extracts was performed on the Agilent Quadrupole Time-of-Flight Mass Spectrometer coupled with an Infinity II 1290 Liquid Chromatography Ultra-High-Pressure unit. LC-MS data acquisition was conducted through 3 experiments consisting of 3 gradients and 2 columns to enhance coverage and resolution of amino acid and central carbon metabolites. Global profiling of polar metabolites and relative quantification for steady state, time-dependent 13C-label flux of polar metabolites was conducted on both the AdvanceBio Glycan Map 2.1 x 150 mm 2.7 µm column (Agilent Technologies) and Acquity UPLC BEH Amide 2.1 x 100 mm, 1.7 µm column (Waters Corporation). A more detailed description of the methods and analysis of LC-MS experiments can be found in the Supplementary Methods.
DNA Methylation of Promoter Region in Genes Involved in the Glycolytic Pathway
For the DNA methylation probe selection, we got information of gene genomic location from the Gencode v2835 for the 9 genes involved in the glycolysis pathway. Next, we retrieved the promoter region of each gene by using the function “promoters” from the GenomicRanges package,36 considering 2000 bp upstream and downstream the transcription start site. We identified 159 probes from the EPIC array that overlapped with the promoter regions of the 9 genes of interest. We then selected only probes that were hypomethylated in the TS603 cell line compared with BT142 (mean methylation levels greater than 0.2). A total of 19 probes were selected (Fig. 5).
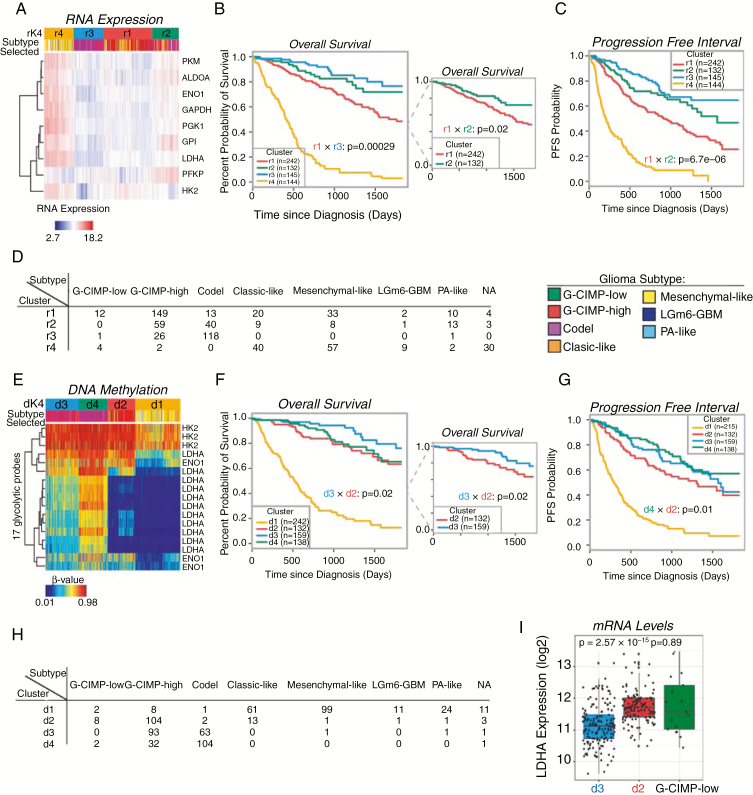
Fig. 5.
Glycolytic genes are prognostic markers for survival in patients. (A) Unsupervised hierarchical clustering of RNA transcripts from nine glycolytic genes and 672 samples from TCGA database. The 4 clusters are shown in orange (r4), light blue (r3), red (r1), and green (r2). (B, C) Overall survival (OS) and progression-free interval (PFI) are displayed for the clusters using the same color coding. The most significant P-value for the clusters containing IDH1mut samples is shown. The insert in (B) only displays clusters r1 and r2 for clarity purposes. (D) mRNA expression cluster composition is shown with the number of patients from each molecular subtype represented. (E) Heatmap for the unsupervised hierarchical clustering of DNA promoter methylation using TCGA database, 648 samples included. The clusters are shown in orange (d1), red (d2), light blue (d3), and green (d4). (F, G) OS and PFI are displayed for the clusters using the same color coding. The most significant P-values for the clusters containing IDH1mut samples are shown. The inset in (F) only displays clusters d2 and d3 for clarity purposes. (H) DNA methylation cluster composition is shown with the number of patients from each molecular subtype represented. (I) mRNA expression of LDHA from clusters d2 and d3 and G-CIMP-low molecular subtype.
TCGA Data Analysis
For methylation, a total of 656 glioma samples were downloaded from TCGA Data Portal using TCGAbiolinks functions “GDCquery” and “GDCdownload” importing into R (http://www.r-project.org) for further analysis.37,38 The raw IDAT files were processed according to the description above. For RNA expression data, TCGA RNA sequence normalized data were downloaded from the GDC Data Portal using TCGAbiolinks functions “GDCquery,” “GDCdownload,” and “GDCprepare” 38,39 importing into R (http://www.r-project.org) for further analysis. Glioma patient RNA sequencing, clinical, and molecular annotation data generated from TCGA Research Network (https://cancergenome.nih.gov/) were downloaded and processed using the R40 package TCGAWorkflow37 for the analysis of the 26 selected genes.
For a more detailed description of all the methods, see the Supplementary Material. RNA-seq and methylation data were deposited in the Gene Expression Omnibus database under accession number GSE138873 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE138873).
Results
Patients with Secondary GBM IDH1mut Have Increased mRNA Levels of Glycolytic Enzymes and Shorter Survival Compared with IDH1mut LGG
We compared mRNA levels of glycolytic enzymes of IDH1mut secondary GBM and IDH1mut LGGs using TCGA data revealing an upregulation of these enzymes in the progressed state of the disease (Fig. 1A). The fold change (FC) in mRNA varied from 1.1 to 4, with LDHA and phosphoglycerate mutase 2 experiencing the most significant changes (FC = 3.1 and FC = 3.7, respectively) (Supplementary Table 1). In addition, mRNA levels of LDHB, normally upregulated in the IDH1mut LGGs compared with IDH1 wild-type (IDH1wt), were lower in higher-grade tumors (Fig. 1A). As expected, the overall survival (OS) of patients with secondary GBM was significantly lower than that of patients with LGG despite their IDH1mut status (Fig. 1B, red and blue lines, respectively). These results, correlating metabolic pathway alterations and patients’ outcome, further suggest that glycolysis is an upregulated pathway between the lower- and higher-grade gliomas harboring IDH1 mutations.
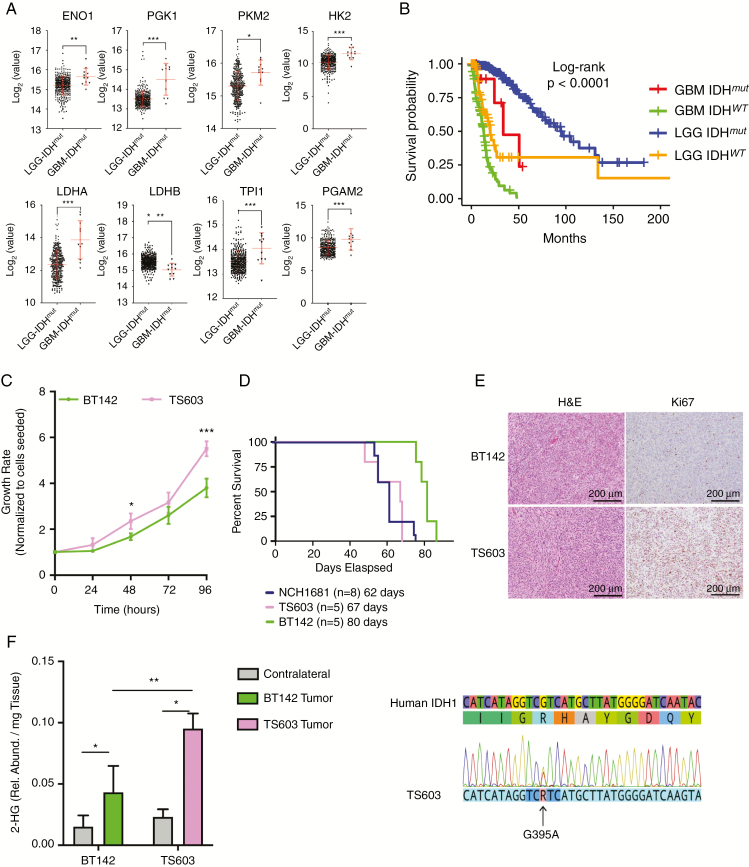
Fig. 1.
Identification of model systems that reflect patient’s LGG and secondary GBM glycolytic phenotype. (A) mRNA levels comparison between LGGs (circles, n = 369) and secondary GBM (triangles, n = 11) containing an IDHmut. The y-axes are displayed as log2 from the mRNA counts, including the mean and SD in red. FDR corrected P-values obtained from a t-test, *P < 0.05; **P < 0.005; ***P < 0.001. A detailed version of the fold change and p-values is located in the Supplementary Table. (B) Kaplan–Meier survival plot generated from TCGA data stratified according to the grade and IDH mutational status. Blue: IDHmut LGG patients. Red: secondary IDHmut GBM patients. Orange: IDHwt LGG patients. Green: IDHwt GBM. (C) Growth rate of TS603 ND BT142 over time. (D) Kaplan–Meier survival plot for mice inoculated with IDH1mut cell lines. (E) 2HG levels in both the tumor and contralateral tissue obtained from mice inoculated with IDH1mut cell lines. (F) Immunostaining of tissue collected from our IDH1mut mouse models displaying both hematoxylin and eosin and Ki67 as a marker of proliferation. Scale bars and percentage of staining are provided within the images. Significance based on t-test for 2HG levels and growth rate. *P < 0.05; **P < 0.005; ***P < 0.001.
IDH1mut Cell Lines with Aggressive Phenotypes as Model Systems to Study Metabolic Reprogramming
To identify cell lines that represent useful model systems for studying metabolic reprogramming and aggressiveness, we classified patient-derived IDH1mut cell lines according to their growth rate and proliferation patterns in vitro and in vivo (Fig. 1C–E). BT142 displayed slower growth in vitro, increased survival of mice, and decreased rate of proliferation in vivo assessed by Ki67 immunostaining. We therefore considered the TS603 cell line as aggressive, while BT142 was defined as indolent. In addition, we found NCH1681 to be the most aggressive in vivo based on the survival of mice harboring this cell line. Total 2-hydroxyglutarate (2HG) levels were also analyzed in vivo for both BT142 and TS603 mouse models (Fig. 1F) revealing higher levels for this metabolite in the tumor tissue compared with those of the contralateral region. Further confirmation of IDH1 mutant status for TS603 was carried out through DNA sequencing (Fig. 1G) to show the presence of both wild-type and mutant alleles with the same intensity at the DNA level. Using methylation analyses, BT142 and NCH1681 cell lines were classified as G-CIMP-low, while TS603 as 1p/19q codeleted (Supplementary Fig. 1A–F and Supplementary Fig. 2). The mRNA levels of IDH1mut were higher in the TS603 compared with the BT142 cell line (Supplementary Fig. 1G). Among the genes with significantly lower expression of mRNA in TS603 (RNA-seq data) were alpha thalassemia/mental retardation syndrome X-linked, adenomatous polyposis coli, ataxia telangiectasia mutated, Notch1, and MET, while higher mRNA expression in the TS603 cell line was displayed by Abelson murine leukemia viral oncogene homolog 1, Fms-like tyrosine kinase 3, O6-methylguanine-DNA methyltransferase, kinase insert domain receptor, tumor protein 53, Von Hippel–Lindau Tumor Suppressor, AKT1, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha, and phosphatase and tensin homolog (Supplementary Fig. 1H). Next, we set out to determine the metabolic, transcriptomic, and epigenomic differences that lead to such aggressive phenotype by comparing those cell lines in vitro and in vivo.
Aggressive Cell Lines Exhibit Decreased Promoter Methylation in the Glycolytic Genes
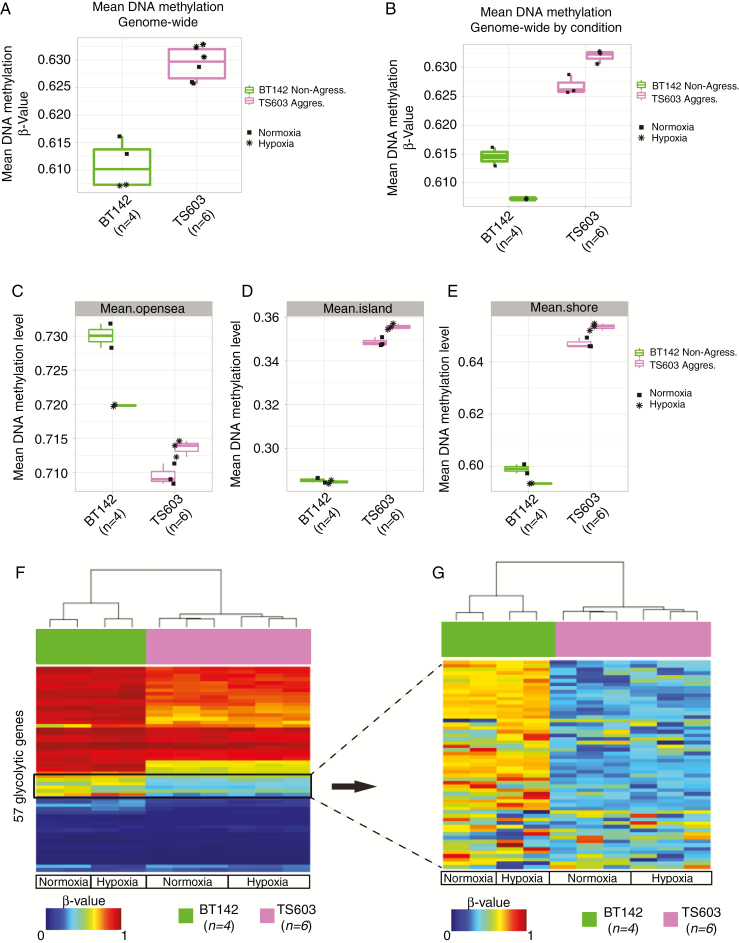
We examined whether global loss of DNA methylation is responsible for the aggressiveness of the IDH1mut cell lines (TS603 and NCH1681) by profiling their DNA methylation status using the Illumina Infinium MethylationEPIC approach.41 In order to dismiss the adaptation to the microenvironment42 as the cause of the metabolomic and transcriptomic profiles, we cultured cells in both normoxic and hypoxic conditions. The TS603 cell line had lower DNA methylation levels in the so-called “open sea” region of the genome (regions more than 2 Kbps from a known CpG island) but maintained a higher genome-wide methylation as well as NCH1681 (Supplementary Fig. 2C). We considered whether oxygen levels during growth affected the methylation subtype and applied a similar algorithm used to validate the subtypes as did Ceccarelli et al.22 We found that under controlled culture conditions (normoxia or hypoxia) all cell lines maintained their original epigenomic subtype (Fig. 2A–E and Supplementary Fig. 2C–F). We next compared the methylation status of the glycolytic and pentose phosphate pathway (PPP) genes and uncovered loss of promoter methylation of these genes in the aggressive cell lines (Fig. 2F–G) when cultured in both normoxia and hypoxia.
Fig. 2.
Aggressive IDH1mut cell line maintains higher genome-wide methylation but displays lower methylation in the promoter region of the glycolytic genes. (A, B) Boxplot of the genome-wide DNA methylation levels between 2 different IDH1mut cell lines under normoxia and hypoxia in vitro. The y-axis indicates mean DNA methylation beta-values. Key is provided to indicate cell type and condition. (C, D, E) Boxplots of the DNA methylation levels for the different regions where the probes are located. (F) Heatmap of DNA methylation. Column-wise represents IDH1mut glioma cell line samples, row-wise represents 57 probes from the glycolytic genes. (G) Zoom in of the heatmap of DNA methylation for the IDH1mut cell lines for the region where the differential methylated probes are located within the glycolytic gene set.
Increased Glycolytic mRNA and Metabolite Levels in Aggressive Cell Lines
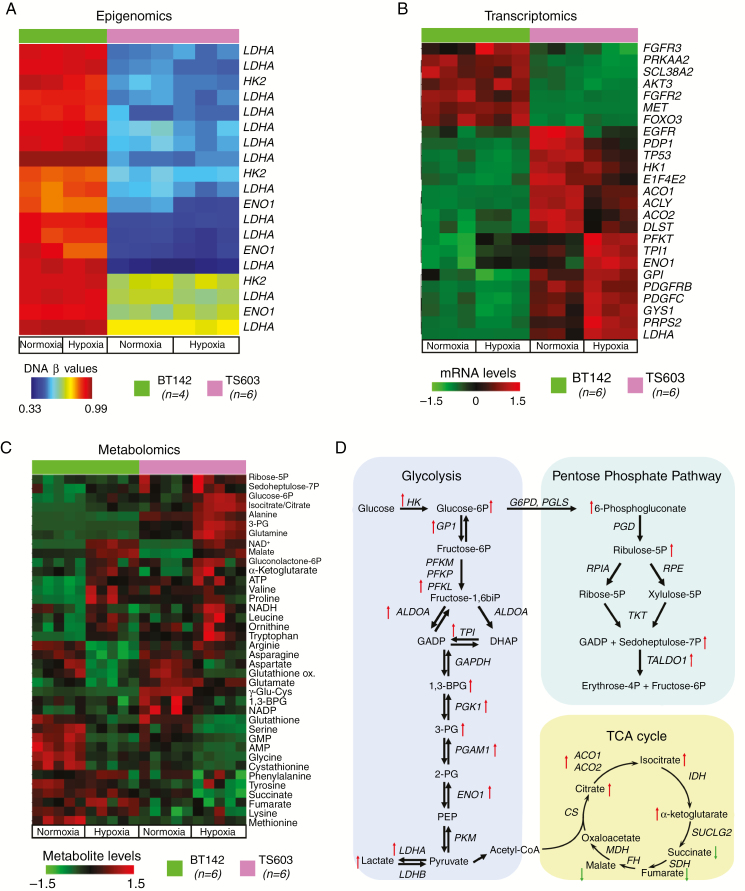
Our targeted epigenetic analysis of IDH1mut cell lines revealed decreased promoter methylation for metabolic enzymes in the aggressive cell lines (TS603 and NCH1681), including LDHA (Fig. 3A and Supplementary Fig. 3A–C). Using transcriptomic and metabolomic analyses in cultured cells, we verified that the loss of promoter methylation in glycolytic genes is translated into higher mRNA and metabolite levels for glycolysis and the PPP. Quantification of 180 mRNA transcripts revealed that the BT142 (indolent) and TS603 (aggressive) cell lines were clearly distinguishable in the principal component analysis biplot (Supplementary Fig. 3E). A significant increase in mRNA levels in the aggressive cell lines was observed when compared with the indolent one, regardless of the oxygen conditions. The differences in over 25 transcripts exhibited statistical significance (false discovery rate [FDR] <5%) (Fig. 3B). Seven out of the 25 most significantly altered transcripts are related to the glycolysis pathway: hexokinase 1, triosephosphate isomerase, glucose-6-phosphate isomerase, aldolase, LDHA, α-subunit of enolase (ENO1), and phosphofructokinase. Other glycolytic genes were upregulated as well, albeit with smaller fold changes (Supplementary Fig. 3D). The heatmap also revealed significantly higher expression levels of aconitase 1 and 2, both of which catalyze the conversion of citrate to isocitrate in either mitochondria or cytosol, in addition to PRPS2 (phosphoribosyl pyrophosphate synthetase 2), which is involved in the synthesis of purines and pyrimidines. Under hypoxic conditions, the differences in the transcriptome of these cell lines were even more pronounced.
Fig. 3.
Multi-omics approach to identify the mechanisms associated with aggressive phenotype in IDH1mut glioma cells. (A) Epigenomic analysis showing differentially methylated promoter regions for the glycolysis and PPP pathways between the aggressive (pink) and non-aggressive (green) IDH1mut cell lines. (B) Transcriptomics approach to identify differentially regulated mRNA transcripts out of 180 probes. Similar color coding as (A). Top 25 mRNAs as selected by their FDR-corrected P-values are displayed. (C) Metabolomics analysis for the relative quantification of metabolites from glycolysis, PPP, amino acids, energy, and TCA cycle. (D) Schematic representation of the enzymatic reactions from glycolysis, PPP, and TCA cycle highlighting the changes at the mRNA and metabolite levels between the 2 cell lines.
The transcriptomic changes reflect the metabolic reprogramming of the aggressive phenotype. The TS603 aggressive cell line displayed increased levels of metabolites from glycolysis, the PPP, and increased metabolites related to energy production, amino acids, and the tricarboxylic acid (TCA) cycle (Fig. 3C–D). Similarly, decreased DNA methylation for glycolytic genes, followed by an increase in mRNA, and higher levels of metabolites were observed for NCH1681 (Supplementary Fig. 4A–E). Additionally, analysis of (pyruvate-free) cell media collected from cells grown under hypoxic conditions revealed that BT142 has the lowest lactate:pyruvate ratio among the cell lines investigated (Supplementary Fig. 4F). This export of lactate into the media further demonstrates the acquisition of high glycolytic phenotype routinely observed in IDH1wt cell lines.
High Glycolytic Phenotype Occurs In Vivo in IDH1mut Mice
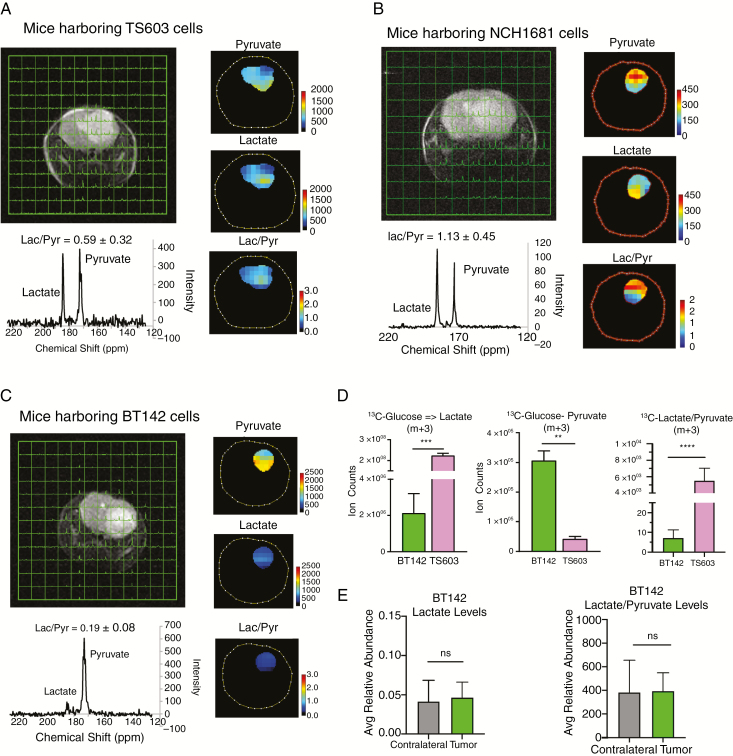
We next examined whether the aggressive IDH1mut cell lines displayed elevated glycolysis in vivo and in vitro. Magnetic resonance spectroscopic imaging (MRSI) on SCID mice implanted with TS603 and NCH1681 cells showed a higher lactate:pyruvate ratio when hyperpolarized [13C-1]-pyruvate was administered intravenously, validating the acquisition of a high glycolytic phenotype in vivo (Fig. 4A, B). In contrast, mice harboring BT142 displayed a lower lactate:pyruvate ratio, similar to previously reported studies (Fig. 4C).25 In vitro experiments involving 13C-U-glucose tracing also revealed a higher glycolytic activity in TS603, as observed by the formation of lactate and pyruvate m+3 (Fig. 4D). Additionally, LC-MS analysis of tissue extracts from the contralateral region of BT142 xenograft showed a low glycolytic flux, similar to that encountered in the tumor tissue (Fig. 4E). The results from HP-MRSI, the in vitro 13C analysis, and the growth and proliferation rates demonstrate that the high glycolytic phenotype is correlated with the aggressive phenotype in IDH1mut gliomas.
Fig. 4.
Validation of high glycolytic phenotype in vivo via HP-MRSI. (A, B) HP-MRI for mice harboring aggressive TS603 and NCH1681 cells respectively. (C) HP-MRI for mice harboring indolent BT142 cells. (D) Analysis of 13C incorporation into pyruvate and lactate from uniformly labeled 13C glucose, and lactate:pyruvate ratios in indolent and aggressive cell lines. (E) Total lactate levels and the ratio of lactate to pyruvate from mice tissue obtained from the BT142 xenograft.
High Glycolytic Phenotype Is Associated with Worse Prognosis in Patients
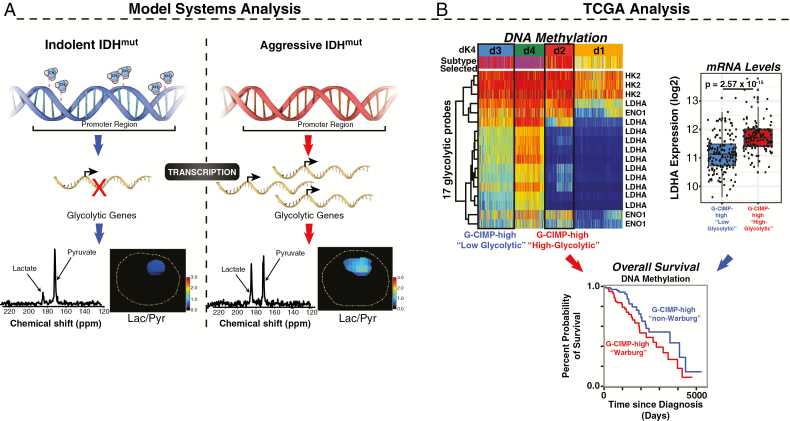
In order to evaluate the potential of glycolytic genes to predict a patient’s outcome, we undertook a detailed transcriptomic and epigenomic analysis using the database of TCGA. We identified 9 genes that are only glycolytic, and only the isozymes present in the brain to describe the high glycolytic phenotype.43–47 First, applying unsupervised hierarchical clustering to mRNA expression data, we identified 4 groups (r1-r4) (Fig. 5A, Supplementary Fig. 5). Both OS and progression-free interval (PFI) displayed good separation, revealing a trend characterized by high expression of glycolytic genes associated with aggressiveness (Figures 5B, C); indeed, the majority of OS clustering exhibited statistical significance (Fig. 5B). Including 26 genes from PPP did not modify the trend previously observed (Supplementary Fig. 6). The clusters are mixed in terms of molecular subtypes with r4, the most aggressive cluster, comprising mostly IDHwt gliomas (classic and mesenchymal), while r3, the least aggressive cluster, is predominantly formed by IDH1mut codeleted samples (n = 118) and few of the G-CIMP-high (n = 26) (Fig. 5D), which are included mainly in r1 and r2. These 2 groups show a significant difference (P = 0.02) for OS, which is even larger for PFI (P < 0.001). Due to the heterogeneity of r1 and r2 regarding the molecular subtypes, interpretation of the differences encountered herein present some limitations. Next, we mapped the promoter methylation of the 9 glycolytic genes in the cell lines in order to identify those probes that are hypomethylated in the aggressive cell line. The same exact probes that were hypomethylated in the aggressive cell line were matched in the 648 patient sample cohort from TCGA. Four clusters with different survival were also identified from the DNA methylation data (Fig. 5E, Supplementary Fig. 7). Out of the 9 genes, which in total present 46 probes at the promoter region, only 17 probes were hypomethylated in the cell lines. These probes correspond to 3 genes: LDHA, hexokinase 2, and ENO1. Interestingly, LDHA had the greatest number of methylation probes, being the main contributors to the 4 observed clusters. DNA methylation analysis also delivered 4 stable clusters (d1–d4) showing that the G-CIMP-high molecular subgroup of patients is further split by the addition of the high glycolytic signature (Fig. 5F). The unsupervised clustering strategy was able to group patients efficiently by their epigenetic or transcriptomic profiles, revealing a strong association between these biological profiles and patients’ OS and PFI (Figures 5B–H). Interestingly, mRNA levels of LDHA inversely correlated with the epigenome for the clusters d3 (defined as “G-CIMP-high low-glycolytic”) and d2 (defined as “G-CIMP-high high-glycolytic”), and there was no significant change in the mRNA levels of LDHA between “G-CIMP-high high-glycolytic” and G-CIMP-low (Fig. 5I). The association between the epigenome and the metabolome can be summarized in the diagram displayed in Fig. 6A. This classification of our model systems based on their metabolism and aggressiveness reflects the outcomes obtained from our analysis of TCGA data. G-CIMP-high molecular subtypes could be further stratified into 2 subgroups according to their glycolytic activity, which shows a correlation with the OS of these patients (Fig. 6B).
Fig. 6.
Proposed model for the aggressive secondary GBM with IDH1 mutation. (A) Loss of methylation at the promoter region of glycolytic genes could lead to increased mRNA and metabolite levels leading to the acquisition of high glycolytic phenotype. (B) TCGA data analysis can predict the molecular subtype in high glycolytic clinical samples. The addition of metabolic information provides an additional stratification by splitting the G-CIMP-high molecular subgroup into 2 distinctive subtypes by both clinical and molecular features. Comparison of mRNA expression levels of LDHA between d2 and d3 clusters suggests significant differences in the expression of this gene. Interestingly, there is no significant difference in the levels of mRNA between cluster d2 and G-CIMP-low molecular subtype.
Overexpression of LDHA in the Indolent Model System Increases Tumor Aggressiveness
The LDHA gene was the main glycolytic gene that contributed to the DNA methylation clustering in TCGA data analysis. Therefore, we wondered whether overexpressing this gene in our indolent model system might affect the rate of proliferation and mice survival. Indeed, we noticed a decreased survival of mice and increased proliferation rate in vivo (Supplementary Fig. 8) in those mouse xenografts generated by injecting (intracranially) the BT142 cell line overexpressing the LDHA gene.
Discussion and Conclusions
To gain insights into the molecular mechanisms underlying glioma evolution and to identify metabolic markers of aggressiveness, we undertook detailed multi-omics analyses on preclinical model systems that closely recapitulate patient IDH1mut LGG metabolic phenotypes. Epigenetically, our cell lines were classified into G-CIMP-low and 1p/19q codeleted subtypes. We identified 2 G-CIMP-low IDH1mut cell lines which differ in aggressiveness despite their global epigenetic similarities. The methylation status of all the cell lines corresponded with the epigenomic subtypes of those patients they belong to, increasing confidence in their use as models of the human tumor.
A smaller study examined 4 specific cases of grades II–III IDH1mut astrocytomas that unexpectedly and rapidly progressed to GBM. This study reported that progression is correlated with hypermutation, which increases genomic instability and amplification of genes that are frequently mutated in GBM.32 The results of this investigation show inconsistencies in the degree to which global or specific epigenetic changes contribute to glioma progression.
Our DNA methylation analyses indicate that the aggressive phenotype has decreased methylation levels of those genes involved in glycolysis and the PPP. Distinct from previous reports, the loss of DNA promoter methylation in our models is specific to the glycolytic genes and not a global event. In fact, the indolent BT142 and the aggressive NCH1681 have similar methylation classification (G-CIMP-low); however, NCH1681 displays higher global methylation compared with BT142. The aggressive TS603 (1p/19q codeletion) has the highest methylation level among the 3. This is expected given the known epigenomic differences between IDH1mut codeletion and IDH1mut non-codeletion (G-CIMP-low and -high).22 We therefore propose that there is a correlation between the increasing aggressiveness of the tumor and the loss of DNA methylation at very specific loci and does not require global demethylation.
Furthermore, we demonstrate that the specific epigenomic modifications are translated at the level of the transcriptome and metabolome, and we validate the use of 13C-pyruvate to monitor this transformation non-invasively. Integrating the current data, we propose a mechanism of increasing aggressiveness that involves the loss of promoter methylation in the glycolytic genes (Fig. 6, left panel). In our preclinical models, the loss of these specific epigenetic markers leads to increased expression of glycolytic and PPP-related mRNAs. We validated the presence of high glycolytic activity in the aggressive models in vivo using hyperpolarized 13C-pyruvate MRSI. This validation demonstrates that the acquisition of a high glycolytic phenotype is not due to cell culture adaptation, and that it is in fact supported by the brain microenvironment. The lactate:pyruvate ratio obtained through HP-MRSI can serve as a non-invasive marker of aggressiveness for clinical applications.48 Combining 2HG detection via MRS and the proposed biomarkers could result in the improvement of disease management by anticipating the increased aggressiveness of the tumor. Although there is interference from the lactate in the normal brain,49,50 monitoring lactate:pyruvate ratio in longitudinal studies in combination with other imaging modalities will still inform about the disease progression toward a potential more aggressive phenotype.
Our study also demonstrates that metabolic profiling of patients adds an extra dimension of complexity and enhances the potential of finding novel therapeutic targets that are more tailored to a distinct subset of patients. The finding that we can further stratify the G-CIMP-high molecular subgroup into 2 distinct metabolic subgroups suggests that metabolomic profiling complements the epigenomic and transcriptomic profiles. In addition, our study opens up the hypothesis that the shift from IDH1mut driven to high glycolytic metabolism, necessary for increasing the tumor aggressiveness, occurs at the G-CIMP-high level, and before the global loss of methylation from G-CIMP-high to G-CIMP-low. This relatively early metabolic reprogramming provides a good rationale to screen newly identified patients for their glycolytic activity in order to assess the aggressiveness of the tumor. Therefore, the identification of both epigenetic and metabolic biomarkers such as the ones coming from glycolysis presented here, or others elsewhere, have the potential to contribute to improved diagnosis, prognosis, and personalized therapy in glioma.51 Indeed, clinical trials investigating the correlation between the conversion rate of hyperpolarized [1-13C]-pyruvate to lactate and Ki67 levels as a marker of aggressiveness are under way (ClinicalTrials.gov: NCT03830151).
Limitations of the Study
Interestingly, TCGA analysis revealed that metabolic reprogramming toward higher glycolytic phenotypes occurs at the G-CIMP-high molecular subtype, contrary to our preclinical models, which are classified as either G-CIMP-low or 1p/19q codeleted. Since patient-derived cell lines are very difficult to grow and might adapt in cell culture conditions, there is a possibility that matched tumor samples might be G-CIMP-high and this methylation is not retained in the cell culture for these cell lines, therefore appear as G-CIMP-low. Future comparisons between matched patient samples and their derived cell lines will reveal the extent to which the global methylation profile is retained in cell culture.
Funding
This work was supported by the Intramural Research Program, Center for Cancer Research, National Cancer Institute, National Institutes of Health (ML, MG, MKC), with internal grants from the Henry Ford Health System and the US Department of Defense (TMM, TSS, HN).
Conflict of interest statement. The authors declare no competing interests.
Authorship statement. VRR, TMM, HN, and ML carried out the experimental work and design, interpreted the results, and wrote the paper. TM and MKC did the hyperpolarized MRI experiments and interpreted the results. TMM and HN did the epigenetics analysis and subtype classification parts into the 7 epigenomic subtypes as well as interpretation of the epigenetic data. TMM, AL, TD, ACS, OC, AL, WZ, HS, DD, SL, JBN, TSS, JM, HN, CHM, MRG, MKC, CZY, YL, and SH were involved in experimental interpretation and writing.
Supplementary Material
References
- 1. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. [DOI] [PubMed] [Google Scholar]
- 2. Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23(1):27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol. 1927;8(6):519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. [DOI] [PubMed] [Google Scholar]
- 5. Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11(5):325–337. [DOI] [PubMed] [Google Scholar]
- 6. Liberti MV, Locasale JW. The Warburg effect: how does it benefit cancer cells? Trends Biochem Sci. 2016;41(3):211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Khasraw M, Ameratunga MS, Grant R, Wheeler H, Pavlakis N. Antiangiogenic therapy for high-grade glioma. Cochrane Database Syst Rev. 2014(9):CD008218. [DOI] [PubMed] [Google Scholar]
- 8. Khasraw M, Lassman AB. Advances in the treatment of malignant gliomas. Curr Oncol Rep. 2010;12(1):26–33. [DOI] [PubMed] [Google Scholar]
- 9. Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60(3):166–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 11. Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174(4):1149–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nobusawa S, Watanabe T, Kleihues P, Ohgaki H. IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin Cancer Res. 2009;15(19):6002–6007. [DOI] [PubMed] [Google Scholar]
- 14. Xu W, Yang H, Liu Y, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19(1):17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chowdhury R, Yeoh KK, Tian YM, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12(5):463–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502(7472):472–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brat DJ, Verhaak RG, Aldape KD, et al. ; Cancer Genome Atlas Research Network Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu XY, Gerges N, Korshunov A, et al. Frequent ATRX mutations and loss of expression in adult diffuse astrocytic tumors carrying IDH1/IDH2 and TP53 mutations. Acta Neuropathol. 2012;124(5):615–625. [DOI] [PubMed] [Google Scholar]
- 19. Noushmehr H, Weisenberger DJ, Diefes K, et al. ; Cancer Genome Atlas Research Network Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shinawi T, Hill VK, Krex D, et al. DNA methylation profiles of long- and short-term glioblastoma survivors. Epigenetics. 2013;8(2):149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Duncan CG, Barwick BG, Jin G, et al. A heterozygous IDH1R132H/WT mutation induces genome-wide alterations in DNA methylation. Genome Res. 2012;22(12):2339–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ceccarelli M, Barthel FP, Malta TM, et al. ; TCGA Research Network Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Houillier C, Wang X, Kaloshi G, et al. IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology. 2010;75(17):1560–1566. [DOI] [PubMed] [Google Scholar]
- 24. SongTao Q, Lei Y, Si G, et al. IDH mutations predict longer survival and response to temozolomide in secondary glioblastoma. Cancer Sci. 2012;103(2):269–273. [DOI] [PubMed] [Google Scholar]
- 25. Chaumeil MM, Radoul M, Najac C, et al. Hyperpolarized (13)C MR imaging detects no lactate production in mutant IDH1 gliomas: implications for diagnosis and response monitoring. Neuroimage Clin. 2016;12:180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jalbert LE, Elkhaled A, Phillips JJ, et al. Metabolic profiling of IDH mutation and malignant progression in infiltrating glioma. Sci Rep. 2017;7:44792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chesnelong C, Chaumeil MM, Blough MD, et al. Lactate dehydrogenase A silencing in IDH mutant gliomas. Neuro Oncol. 2014;16(5):686–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mazor T, Pankov A, Johnson BE, et al. DNA methylation and somatic mutations converge on the cell cycle and define similar evolutionary histories in brain tumors. Cancer Cell. 2015;28(3):307–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bai H, Harmancı AS, Erson-Omay EZ, et al. Integrated genomic characterization of IDH1-mutant glioma malignant progression. Nat Genet. 2016;48(1):59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. de Souza CF, Sabedot TS, Malta TM, et al. A distinct DNA methylation shift in a subset of glioma CpG island methylator phenotypes during tumor recurrence. Cell Rep. 2018;23(2):637–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mazor T, Chesnelong C, Pankov A, et al. Clonal expansion and epigenetic reprogramming following deletion or amplification of mutant IDH1. Proc Natl Acad Sci U S A. 2017;114(40):10743–10748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Richardson TE, Snuderl M, Serrano J, et al. Rapid progression to glioblastoma in a subset of IDH-mutated astrocytomas: a genome-wide analysis. J Neurooncol. 2017;133(1):183–192. [DOI] [PubMed] [Google Scholar]
- 33. Dettling S, Stamova S, Warta R, et al. Identification of CRKII, CFL1, CNTN1, NME2, and TKT as novel and frequent T-cell targets in human IDH-mutant glioma. Clin Cancer Res. 2018;24(12):2951–2962. [DOI] [PubMed] [Google Scholar]
- 34. Luchman HA, Chesnelong C, Cairncross JG, Weiss S. Spontaneous loss of heterozygosity leading to homozygous R132H in a patient-derived IDH1 mutant cell line. Neuro Oncol. 2013;15(8):979–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Harrow J, Frankish A, Gonzalez JM, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22(9):1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lawrence M, Huber W, Pagès H, et al. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9(8):e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Silva TC, Colaprico A, Olsen C, et al. TCGA workflow: analyze cancer genomics and epigenomics data using Bioconductor packages. F1000Res. 2016;5:1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Silva T, Colaprico A, Olsen C, et al. TCGAbiolinksGUI: a graphical user interface to analyze cancer molecular and clinical data. F1000Research. 2018;7(439). [Google Scholar]
- 39. Colaprico A, Silva TC, Olsen C, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44(8):e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huber W, Carey VJ, Gentleman R, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12(2):115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fortin JP, Triche TJ Jr, Hansen KD. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics. 2017;33(4):558–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Venneti S, Thompson CB. Metabolic reprogramming in brain tumors. Annu Rev Pathol. 2017;12:515–545. [DOI] [PubMed] [Google Scholar]
- 43. Wolf A, Agnihotri S, Micallef J, et al. Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. J Exp Med. 2011;208(2):313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Muller FL, Colla S, Aquilanti E, et al. Passenger deletions generate therapeutic vulnerabilities in cancer. Nature. 2012;488(7411):337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yang W, Lu Z. Pyruvate kinase M2 at a glance. J Cell Sci. 2015;128(9):1655–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang W, Zheng Y, Xia Y, et al. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol. 2012;14(12):1295–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liang J, Cao R, Zhang Y, et al. PKM2 dephosphorylation by Cdc25A promotes the Warburg effect and tumorigenesis. Nat Commun. 2016;7:12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Golman K, Zandt RI, Lerche M, Pehrson R, Ardenkjaer-Larsen JH. Metabolic imaging by hyperpolarized 13C magnetic resonance imaging for in vivo tumor diagnosis. Cancer Res. 2006;66(22):10855–10860. [DOI] [PubMed] [Google Scholar]
- 49. Grist JT, McLean MA, Riemer F, et al. Quantifying normal human brain metabolism using hyperpolarized [1-13C]pyruvate and magnetic resonance imaging. Neuroimage. 2019;189:171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Park I, Larson PEZ, Gordon JW, et al. Development of methods and feasibility of using hyperpolarized carbon-13 imaging data for evaluating brain metabolism in patient studies. Magn Reson Med. 2018;80(3):864–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Klughammer J, Kiesel B, Roetzer T, et al. The DNA methylation landscape of glioblastoma disease progression shows extensive heterogeneity in time and space. Nat Med. 2018;24(10):1611–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.