Enteric viruses are transmitted through the fecal-oral route, but how enteric viruses survive in the environment is unclear. Previously, we found that bacterial polysaccharides enhance poliovirus stability against heat or bleach inactivation, but the specific molecular requirements have been unknown. Here, we showed that certain short-chain oligosaccharides can bind to poliovirus but do not increase virion stability. Long-chain polysaccharides bind and may bridge adjacent sites on the viral surface, thus increasing capsid rigidity and stability. This work defines the unique interactions of poliovirus and glycans, which provides insight into virion environmental stability and transmission.
KEYWORDS: enteric virus, glycovirology, poliovirus, virion stability
ABSTRACT
Enteric viruses infect the gastrointestinal tract, and bacteria can promote replication and transmission of several enteric viruses. Viruses can be inactivated by exposure to heat or bleach, but poliovirus, coxsackievirus B3, and reovirus can be stabilized by bacteria or bacterial polysaccharides, limiting inactivation and aiding transmission. We previously demonstrated that certain N-acetylglucosamine (GlcNAc)-containing polysaccharides can stabilize poliovirus. However, the detailed virus-glycan binding specificity and glycan chain length requirements, and thus the mechanism of virion stabilization, have been unclear. A previous limitation was our lack of defined-length glycans to probe mechanisms and consequences of virus-glycan interactions. Here, we generated a panel of polysaccharides and oligosaccharides to determine the properties required for binding and stabilization of poliovirus. Poliovirus virions are nonenveloped icosahedral 30-nm particles with 60 copies of each of four capsid proteins, VP1 to VP4. VP1 surrounds the 5-fold axis, and our past work indicates that this region likely contains the glycan binding site. We found that relatively short GlcNAc oligosaccharides, such as a six-unit GlcNAc oligomer, can bind poliovirus but fail to enhance virion stability. Virion stabilization required binding of long GlcNAc polymers of greater than 20 units. Our data suggest a model where GlcNAc polymers of greater than 20 units bind and bridge adjacent 5-fold axes, thus aiding capsid rigidity and stability. This study provides a deeper understanding of enteric virus-bacterial glycan interactions, which are important for virion environmental stability and transmission.
IMPORTANCE Enteric viruses are transmitted through the fecal-oral route, but how enteric viruses survive in the environment is unclear. Previously, we found that bacterial polysaccharides enhance poliovirus stability against heat or bleach inactivation, but the specific molecular requirements have been unknown. Here, we showed that certain short-chain oligosaccharides can bind to poliovirus but do not increase virion stability. Long-chain polysaccharides bind and may bridge adjacent sites on the viral surface, thus increasing capsid rigidity and stability. This work defines the unique interactions of poliovirus and glycans, which provides insight into virion environmental stability and transmission.
INTRODUCTION
Enteric viruses infect and replicate primarily in the gastrointestinal tract of the host and are transmitted by the fecal-oral route. Over 100 types of pathogenic enteric viruses are excreted in human and animal feces (1). Infections are associated primarily with diarrhea and vomiting in humans and may also cause respiratory infections, hepatitis, diseases that have high mortality rates (such as encephalitis), and paralysis (2). The picornavirus poliovirus is spread by the fecal-oral route but can rarely disseminate to the central nervous system, causing paralysis and death (3).
In the intestine, enteric viruses encounter huge numbers of microbes, collectively known as the microbiota. Intestinal microbiota can promote infection of enteric viruses such as poliovirus, coxsackievirus B3 (CVB3), reovirus, rotavirus, mouse mammary tumor virus, and norovirus through direct effects on virion stability/attachment or indirect effects on host immune responses (4–11). For example, using poliovirus as a model enteric virus, we found that poliovirus binds to bacterial surface lipopolysaccharide (LPS) (5, 8). LPS binding reduced premature genomic RNA release and increased virion stability and host cell attachment. We found that long N-acetylglucosamine (GlcNAc)-containing polysaccharides can stabilize poliovirus but that a six-unit GlcNAc oligosaccharide, GlcNAc6, could not. Recent comparison studies of poliovirus with other members of the Picornaviridae, such as coxsackievirus B3, Aichi virus, and mengovirus, revealed shared and distinct effects of bacteria on virion stability (12).
A poliovirus mutant with reduced LPS binding has shed light on potential binding sites on the virion as well as consequences of polysaccharide binding. Poliovirus capsids are icosahedral structures composed of 60 copies of each of four proteins, VP1 to VP4. We previously showed that a VP1 T99K mutant poliovirus has reduced binding to LPS and is not stabilized by LPS at physiological temperature (8). Deletion of the T99 residue did not influence LPS binding, and the T99K mutant phenotype was conditional, with no LPS-mediated stabilization at 37°C but with wild-type levels of LPS-mediated stabilization at 42°C. These results suggest that VP1 T99 is not directly involved in LPS binding but is likely close to the binding site. Importantly, the T99K mutant virus with low LPS binding had a transmission defect in mice when virion instability was a selective pressure (8).
Our previous glycan binding studies with poliovirus were performed using bacterial LPS, which is composed of polysaccharide and lipid A (5, 8). Whether one or both parts of LPS contribute to binding and stabilization of poliovirus is not clear. The polysaccharides of LPS are very diverse in structure, and not all of them contain GlcNAc residues, indicating that binding specificity may be not limited to GlcNAc-containing glycans. The relatively high molecular weight and heterogeneity of LPS add complexity and hinder further characterization of glycan binding avidity and binding sites. Moreover, the glycan chain length requirements for binding/stabilization of poliovirus are unclear.
In this study, we used poliovirus as a model enteric virus and examined the properties of glycans required for binding and stabilization of viral particles. A limitation of prior work was the absence of key reagents to examine effects of glycan chain length on virion binding and stabilization. While our past studies examined virion stability in the presence of dozens of glycans, we lacked glycans of defined lengths in a key range (6- to 30-unit-long oligomers), and our direct binding assays were limited to commercially available biotinylated LPS (5, 8). Here, we generated a library of different polysaccharides and purified oligosaccharides of different chain lengths, and we found that acetylated polysaccharides bind to and stabilize poliovirus. Short-chain GlcNAc oligomers could bind but did not stabilize poliovirus. Viral stabilization required GlcNAc oligomers longer than 20 units, likely through bridging of multiple binding sites on the virion surface. In conclusion, this work defines the unique interactions of poliovirus and bacterial glycans, which may provide insight into virion environmental stability and transmission.
(This article was submitted to an online preprint archive [13].)
RESULTS
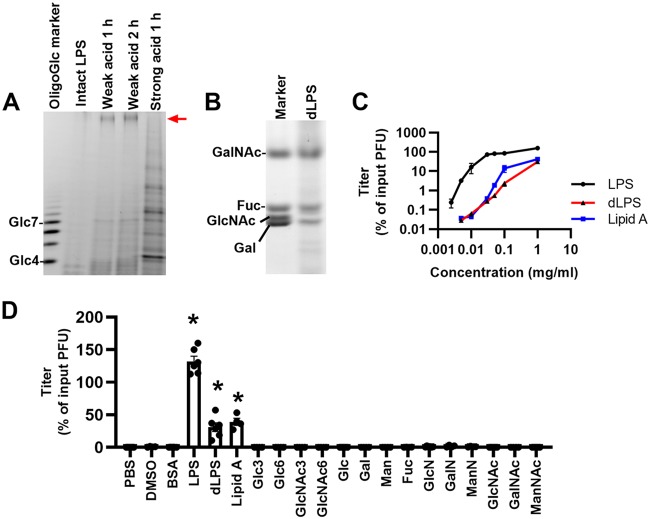
Previously, we found that bacterial LPS can stabilize poliovirus against heat- or bleach-induced inactivation (5, 8). LPS contains a hydrophobic phosphoglycolipid moiety (lipid A) and a polysaccharide moiety (detoxified LPS), but whether one or both contribute to poliovirus stabilization is unclear. To investigate this, we used LPS from Escherichia coli O127:B8 and performed weak-acid hydrolysis (Fig. 1A), which was sufficient to release polysaccharide from lipid A without polysaccharide degradation, as shown by fluorophore-assisted carbohydrate electrophoresis (FACE). FACE is a rapid and sensitive gel electrophoresis method to examine monosaccharide composition and to profile oligosaccharide length. In FACE, a fluorophore is linked to the reducing termini of glycans by reductive amination, followed by gel electrophoresis and detection using UV light. The identity of each band can be deduced by comparing the migration rate with a proper standard. We analyzed the monosaccharide composition of LPS from E. coli O127:B8 and confirmed that it contains mainly N-acetylgalactosamine (GalNAc), galactose, and fucose (Fig. 1B), consistent with previous reports (14). We purified detoxified LPS (dLPS) and lipid A by phase separation and tested their abilities to stabilize poliovirus using an in vitro thermal inactivation assay. Poliovirus (106 PFU) was incubated with compounds at 45°C for 5 h, followed by plaque assay on Vero cells to determine the number of remaining infectious particles. We found that LPS very efficiently stabilized poliovirus over a wide concentration range (0.01 to 1 mg/ml) (Fig. 1C), in accordance with previous results (5, 8). Detoxified LPS, which is a polysaccharide usually consisting of 50 to 100 monosaccharide residues, stabilized poliovirus but was about 10- to 100-fold less effective than intact LPS. Lipid A from E. coli stabilized poliovirus similarly to dLPS. In contrast, 10 different monosaccharides and 4 oligosaccharides did not stabilize poliovirus, even at high concentrations (20 mg/ml) (Fig. 1D). Overall, these data indicate that both the polysaccharide and lipid A moieties of LPS contribute to poliovirus stabilization. Polysaccharides, but not monosaccharides or short oligosaccharides, can stabilize poliovirus against heat inactivation.
FIG 1.
Components of LPS that contribute to poliovirus stabilization. (A) LPS from E. coli O127:B8 was hydrolyzed by acid and analyzed by FACE. Intact LPS cannot be labeled with fluorophore and therefore does not generate a band in the FACE gel. Weak-acid treatment of LPS (2% acetic acid, 100°C) is sufficient to release intact polysaccharide (detoxified LPS), as indicated by the red arrow. Strong-acid treatment of LPS (0.1 M HCl, 100°C) generated multiple degraded fragments of the polysaccharide. Glucose oligomers (Glc4 to Glc7) were used to roughly indicate glycan sizes. (B) FACE analysis of the monosaccharide composition of the detoxified LPS (dLPS) from E. coli O127:B8 following strong-acid hydrolysis. N-Acetylgalactosamine, fucose, and galactose were detected by comparison with monosaccharide standards. (C) Detoxified LPS from E. coli O127:B8 stabilizes poliovirus but at a lower efficiency than intact LPS. Poliovirus (106 PFU) was incubated with compounds at 45°C for 5 h. Remaining titers after incubation were calculated by plaque assays and normalized to the result for the untreated control. (D) Polysaccharide (dLPS) and lipid components of LPS contribute to poliovirus stabilization, but monosaccharides or short oligosaccharides do not. Concentrations of compounds: 1 mg/ml for dimethyl sulfoxide (DMSO), BSA, LPS, dLPS, and lipid A; 20 mg/ml for monosaccharides; 5 mg/ml for oligosaccharides. Poliovirus (106 PFU) was incubated with compounds at 45°C for 5 h. Remaining titers after incubation were calculated by plaque assays and normalized to the result for the untreated control. The saccharides used were d-glucose (Glc), d-galactose (Gal), d-mannose (Man), l-fucose (Fuc), d-glucosamine (GlcN), d-galactosamine (GalN), d-mannosamine (ManN), N-acetyl-d-glucosamine (GlcNAc), N-acetyl-d-galactosamine (GalNAc), N-acetyl-d-mannosamine (ManNAc), maltotriose (Glc3), maltohexaose (Glc6), tri-N-acetylchitotriose (GlcNAc3), and hexa-N-acetylchitohexaose (GlcNAc6). Results represent mean ± standard error of the mean (SEM) (n = 4 to 8). *, P < 0.05 by Kruskal-Wallis test.
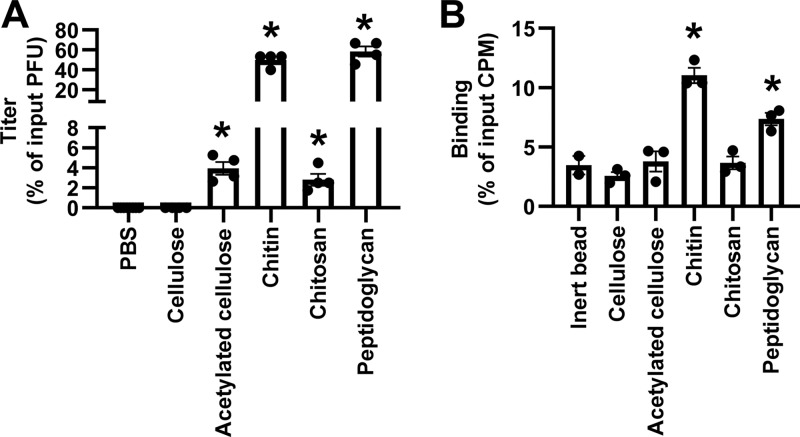
Since the detoxified LPS from E. coli O127:B8 contains GalNAc but not GlcNAc, we hypothesized that acetyl groups in polysaccharides are important for binding and stabilization of poliovirus. To test this, we examined the effects of a group of polysaccharides on viral particle thermal stability (Fig. 2A). We found that chitin (a GlcNAc homopolymer) and peptidoglycan (a GlcNAc-containing polysaccharide) stabilized more than 50% of input viruses against heat inactivation. The acetyl groups in chitin can be removed to convert chitin into chitosan. We found that chitosan (85% deacetylated) stabilized only about 5% of input viruses, and more than 99.99% of input viruses were inactivated when coincubated with phosphate-buffered saline (PBS) or cellulose (a glucose homopolymer). Randomly introducing O-acetyl groups into cellulose (acetylated cellulose) protected 5% of input viruses against heat inactivation. Thus, acetyl groups in polysaccharide contribute to poliovirus stabilization. We also examined binding of poliovirus with insoluble polysaccharides using 35S-labeled virus in a pulldown assay (Fig. 2B). 35S-labeled poliovirus (105 PFU) was incubated with insoluble polysaccharides at 37°C for 3 h. Unbound viruses were washed away with PBS. Polysaccharide-associated viruses were quantified by scintillation counting. Cellulose bound 2.5% of input viruses, comparable to the background binding of the inert bead control. The acetylated glycans peptidoglycan and chitin had higher binding capacity (7.5 to 11%). Similarly, Erickson et al. found that Lactobacillus johnsonii bacteria with O-acetylated exopolysaccharides (EPS) had more efficient poliovirus binding than isogenic mutant strains that do not produce EPS (15). Although chitosan and acetylated cellulose had marginal activity at stabilizing poliovirus, we did not detect any statistically significant binding compared to that for cellulose in our pulldown assay. Overall, these data suggest that acetyl groups in polysaccharides contribute to binding and stabilization of poliovirus but that other factors, such as carbohydrate conformation and linkage of the acetyl group, may also impact binding.
FIG 2.
Poliovirus binds to and is stabilized by acetylated polysaccharides. (A) Acetylated polysaccharides stabilize poliovirus against in vitro thermal inactivation. Poliovirus (106 PFU) was incubated with 1 mg/ml of each compound at 45°C for 5 h. Remaining titers after incubation were quantified by plaque assays and normalized to the result for the untreated control. (B) Acetylated polysaccharides bind poliovirus in a direct pulldown assay. 35S-labeled poliovirus (105 PFU) was incubated with 500 μg insoluble polysaccharides. After incubation, samples were centrifuged and washed, and polysaccharide-associated virus in the pellet was measured by scintillation counting. Results represent mean ± SEM (n = 2 to 4). *, P < 0.05 by one-way ANOVA.
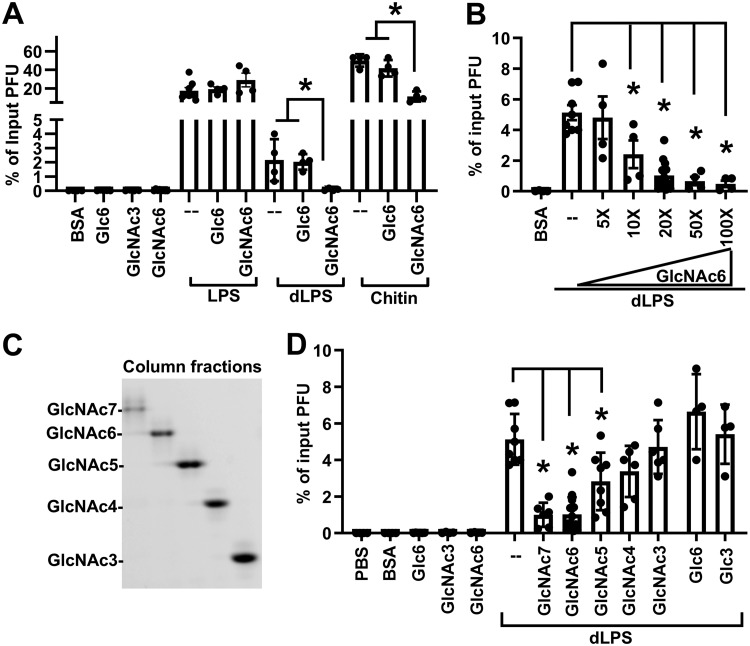
Short oligosaccharides do not stabilize poliovirus, but whether they bind to the virion is unknown. Chitin is a GlcNAc homopolymer, and we used it to generate low-molecular-weight GlcNAc oligosaccharides by acid hydrolysis and then fractionated them by size exclusion chromatography. We obtained GlcNAc oligomers containing 6 to 17 units, as shown by FACE (Fig. 3A). To examine their virion binding abilities, we used lectin-cross-linked agarose beads to immobilize oligosaccharides on the bead surface followed by pulldown assay (Fig. 3B). Wheat germ agglutinin (WGA) specifically binds GlcNAc-containing glycans. A previous study has shown that GlcNAc oligomers longer than 3 units bind WGA efficiently with a dissociation constant (Kd) of around 45 μM (16). Therefore, we used WGA-agarose beads to bind GlcNAc oligomers. As a negative control, concanavalin A (ConA) binds glucose-containing glycans. Nonspecific binding sites were blocked with bovine serum albumin (BSA). We used a high molar ratio of glycans to ensure that the glycan binding sites on the bead surface were saturated. Excess unbound glycans were washed away with PBS. We coincubated 105 PFU of 35S-labeled polioviruses with oligosaccharide-coated beads at 37°C for 3 h and then measured bead-associated viruses by liquid scintillation counting. We found GlcNAc3-coated WGA beads did not pull down virus, similar to the case for the PBS control. GlcNAc6-coated WGA beads bound 4% of input viruses, while GlcNAc12 to -14 bound 50% of input viruses, indicating that the pulldown efficiency of WGA beads is correlated with the chain length of the coated GlcNAc oligosaccharides. Because WGA binds one to two GlcNAc residues within its glycan binding sites, GlcNAc3-coated WGA beads likely have only 1 to 2 units exposed on the bead surface, which likely explains the deficiency in the pulldown assay. Although the detoxified LPS of E. coli O127:B8 can stabilize poliovirus, it did not pull down poliovirus here due to a lack of GlcNAc residues required for WGA bead binding. The negative control, glucose oligosaccharide-coated ConA beads, also did not bind poliovirus. These data suggest that short GlcNAc oligosaccharides can bind to poliovirus but fail to stabilize the virus.
FIG 3.
Poliovirus binds low-molecular-weight GlcNAc oligosaccharides. (A) Preparation of low-molecular-weight GlcNAc oligosaccharides. Chitin was hydrolyzed by acid to generate water-soluble oligosaccharides. GlcNAc oligomers were fractionated with a Bio-Gel P-4 size exclusion column and analyzed by FACE. Left lane, oligo(GlcNAc) standard. (B) Poliovirus binds to GlcNAc oligomer-coated WGA-agarose beads. 35S-labeled poliovirus (105 PFU) was incubated with glycan-coated lectin-agarose beads. After incubation, bead-associated virus was measured by scintillation counting. WGA lectin binds GlcNAc-containing glycans; ConA binds glucose-containing glycans. Results represent mean ± SEM (n = 2 to 5). *, P < 0.05 by one-way ANOVA.
GlcNAc6 binds but does not stabilize poliovirus, suggesting that it may act as a competitive inhibitor of polysaccharide-mediated virion stabilization. To test this, we coincubated poliovirus with polysaccharides in the presence or absence of excess GlcNAc6 in our in vitro thermal inactivation assay (Fig. 4A). We found that coincubation of 1 mg/ml GlcNAc6 with 0.05 mg/ml detoxified LPS reduced the recovered titer 10- to 100-fold compared to that with detoxified LPS alone. Similarly, 1 mg/ml GlcNAc6 inhibited stabilization of poliovirus mediated by 0.5 mg/ml chitin, suggesting that GlcNAc6 competes with long polysaccharides for the limited glycan binding sites on the virion surface. However, we did not observe any titer decrease when poliovirus was coincubated with 0.01 mg/ml LPS in the presence of 1 mg/ml GlcNAc6. The lack of competition of GlcNAc6 for intact LPS may be due to the virion-stabilizing effects of lipid A. Control Glc6 had no effect on polysaccharide-mediated stabilization of poliovirus. The effects of GlcNAc6 were concentration dependent, with inhibition requiring at least 10-fold excess GlcNAc6 by weight (Fig. 4B). Since excess GlcNAc6 decreased dLPS-mediated virion stabilization, we wondered whether shorter GlcNAc oligomers would have activity. We performed acid hydrolysis of chitin, purified short GlcNAc oligomers using size exclusion chromatography, and analyzed them by FACE (Fig. 4C). Poliovirus was exposed to 0.05 mg/ml dLPS in the presence or absence of 1 mg/ml GlcNAc oligomers 3, 4, 5, 6, or 7 units long in our in vitro thermal inactivation assay (Fig. 4D). We found that inhibition of dLPS-mediated stabilization became stronger as the glycan chain length increased from 3 to 6 GlcNAc residues. GlcNAc7 did not cause further titer reduction. Overall, these data indicate that excess short GlcNAc oligosaccharides inhibit polysaccharide-mediated stabilization of poliovirus, suggesting that GlcNAc oligomers as short as 5 units may bind poliovirus but fail to stabilize.
FIG 4.
Excess GlcNAc6 inhibits polysaccharide-mediated stabilization of poliovirus. (A) Excess GlcNAc6 inhibits detoxified LPS (dLPS) and chitin-mediated stabilization of poliovirus. Poliovirus (106 PFU) was incubated with compounds at 45°C for 5 h. Remaining titers after incubation were quantified by plaque assay and normalized to the result for the untreated control. Concentrations of compounds: 0.01 mg/ml for LPS; 0.05 mg/ml for dLPS; 0.5 mg/ml for chitin; 1 mg/ml for BSA, GlcNAc3, GlcNAc6 (20-fold of dLPS), Glc3, and Glc6. Results represent mean ± SEM (n = 4 to 8). *, P < 0.05 by one-way ANOVA. (B) The inhibition effect of GlcNAc6 is concentration dependent. Experiments were performed as for panel A, but different concentrations of GlcNAc6 were used. Concentrations of compounds: 0.05 mg/ml for dLPS, 1 mg/ml for BSA, 0.25 mg/ml (5×) to 5 mg/ml (100×) for GlcNAc6. Results represent mean ± SEM (n = 4 to 14). *, P < 0.05 by one-way ANOVA. (C) FACE analysis of purified glycans containing 3 to 7 GlcNAc units. Chitin was hydrolyzed by acid to generate water-soluble oligosaccharides. GlcNAc oligomers were fractionated with a Bio-Gel P-4 size exclusion column and analyzed by FACE. (D) The chain lengths of GlcNAc oligomers affect their inhibition efficiency. Experiments were performed as for panel A. Concentrations of compounds: 0.05 mg/ml for dLPS; 1 mg/ml for BSA, GlcNAc3, GlcNAc4, GlcNAc5, GlcNAc6, GlcNAc7, Glc3, and Glc6. Results represent mean ± SEM (n = 4 to 14). *, P < 0.05 by one-way ANOVA.
To define the minimum glycan length requirement for stabilization, we examined the activity of GlcNAc oligomers of a variety of lengths in titer-based virion stability assays and in a virion RNA release assay. We performed acid hydrolysis of chitin, purified high-molecular-weight GlcNAc oligomers using size exclusion chromatography, and analyzed them by FACE. Due to limitations of FACE gel resolution, glycans longer than 20 GlcNAc residues appear as a smear in the top of the gel, whereas glycans containing fewer than 20 GlcNAc residues are well separated (Fig. 5A). We performed in vitro thermal inactivation experiments by incubating 106 PFU of poliovirus with 0.5 mg/ml of each column fraction at 45°C for 5 h, followed by viral titer assay. We found that fractions 1 and 2, which contained long-chain oligosaccharides (>20 GlcNAc residues), were the most effective at stabilizing poliovirus, comparable to the results with intact chitin (Fig. 5B). Fractions 3 and 4 were mixture of short and long glycans (14 to >20 residues), and they also stabilized poliovirus at lower efficiency. Fractions 5 to 10, containing glycans with fewer than 20 GlcNAc residues, did not stabilize poliovirus. We directly measured poliovirus stability using a technique independent from the plaque assay: the particle stability thermal release assay (PaSTRY) (17) (Fig. 5C and D). PaSTRY quantifies virion stability by monitoring the temperature at which virion RNA is released. Poliovirus was mixed with SYBR green II dye and heated in a real-time instrument on a stepwise temperature gradient with fluorescence monitoring. We found that the viral RNA release temperature in the presence of PBS was about 45°C, in line with previous results (17). Preexposure of poliovirus with fraction 1 or fraction 2, which contain GlcNAc oligomers >20 units, at 37°C for 1 h increased the RNA release temperature by 2°C. Coincubation with fraction 5, which contains GlcNAc oligomers of ∼13 to 18 units, before PaSTRY did not change the RNA release temperature, which was similar to that for the PBS control. Collectively, these results indicated that glycans containing more than 20 GlcNAc residues can stabilize poliovirus.
FIG 5.
High-molecular-weight GlcNAc oligosaccharides (>20 units) stabilize poliovirus. (A) FACE analysis of Bio-Gel P-10 column-fractionated high-molecular-weight GlcNAc oligosaccharides. (B) GlcNAc oligomers longer than 20 units stabilize poliovirus against heat inactivation. Poliovirus (106 PFU) was incubated with 0.5 mg/ml of each compound at 45°C for 5 h. The remaining titer after incubation was quantified by plaque assay and normalized to the result for the untreated control. Results represent mean ± SEM (n = 2 to 6). *, P < 0.05 by one-way ANOVA. (C) Representative profiles of poliovirus particle stability thermorelease assay (PaSTRy). Poliovirus was preincubated with 0.5 mg/ml compound for 1 h at 37°C. Subsequently, viruses were mixed with SYBR green and heated in a real-time machine over a stepwise temperature gradient. The intensity of SYBR green fluorescence was plotted over increasing temperature. The virion RNA release temperature (TR) represents the temperature causing the highest instantaneous rate of fluorescence change and is indicated by an arrow. AU, arbitrary units. (D) Quantification of RNA release temperatures measured by PaSTRy experiments. Results represent mean ± SEM (n = 3). *, P < 0.05 by one-way ANOVA.
DISCUSSION
Enteric viruses are in close contact with bacteria in the gastrointestinal tract, and previous studies found that bacteria promote enteric virus replication and transmission (18–20). Using poliovirus as a model system, we previously demonstrated that binding of bacteria or bacterial polysaccharides can prevent premature release of viral RNA, which is important for the stability of the virus in the environment and may aid transmission to the next host. For example, a viral mutant with reduced LPS binding, VP1 T99K, had a transmission defect in mice due to virion instability in feces (8). Despite the importance of bacterium-virus interactions, the binding specificity and how glycans bind and stabilize enteric viruses are unclear. The major goal of this study was to determine the glycan requirements for binding and stabilization of poliovirus.
We found that both polysaccharide and lipid components of LPS contribute to poliovirus stabilization (Fig. 1). Poliovirus capsid protein VP1 has a hydrophobic pocket which is occupied by a fatty acid-like molecule, the so-called “pocket factor” (21). Antiviral agents can replace the pocket factor and bind to the hydrophobic pocket, which can stabilize the virus and prevent uncoating (22). A previous study reported increased poliovirus stability in the presence of fatty acids (23). Unlike the diverse structures of bacterial LPS glycan moieties, the lipid A structure in general is highly conserved and contains 6 acyl chains linked to the disaccharide backbone (24). It is possible that one of the acyl chains of lipid A can insert into the hydrophobic pocket and stabilize poliovirus.
The glycan binding specificity of poliovirus is restricted not to GlcNAc-containing polysaccharides but rather to acetylated glycans. Acetylated glycans are frequently found in bacterial surface polysaccharides and mammalian intestinal mucin glycans. Glycans can be acetylated through an amide bond (N-acetylation) or an ester bond (O-acetylation). Glycan acetylation can alter biological properties of bacterium-host interactions, such as bacterial virulence, disease pathogenesis, and the host innate and adaptive immune responses (25). Moreover, acetylation is a reversible modification. Some bacteria encode deacetylating enzymes which can tune the acetylation level of surface polysaccharides according to environmental stress (26). Interactions of poliovirus with both N-acetylated polysaccharides (such as chitin and peptidoglycan) and O-acetylated polysaccharides (such as Lactobacillus johnsonii EPS) suggest that modulation of the acetylation level of bacterial polysaccharides may impact poliovirus.
We examined binding of poliovirus with insoluble polysaccharides via direct pulldown assays (Fig. 2) and purified GlcNAc oligosaccharides via lectin bead pulldown assays (Fig. 3). We found that long, insoluble GlcNAc-containing polysaccharides such as chitin and peptidoglycan bound poliovirus. Furthermore, short GlcNAc oligomers immobilized on lectin-agarose beads bound poliovirus, whereas glucose-containing glycans did not. For the GlcNAc oligomers, viral binding efficiency was proportional to glycan length, with 7- to 8-unit-long oligomers binding weakly but 14- to 17-unit-long oligomers binding efficiently (Fig. 3B). Interestingly, GlcNAc6 had minimal viral binding in the lectin bead pulldown assay (Fig. 3B) but was able to competitively inhibit dLPS-mediated viral stabilization (Fig. 4A, B, and D). These results suggest that our direct binding assays may be stringent and may underestimate binding of short glycans. Regardless, cumulatively, our data show that relatively short GlcNAc oligomers can bind to poliovirus.
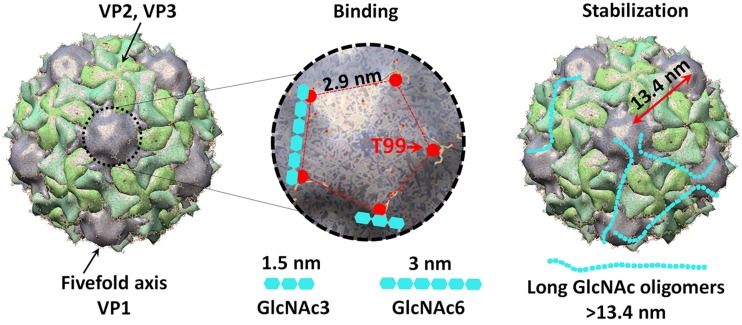
Although short GlcNAc glycans could bind to poliovirus, we found that virion stabilization required long GlcNAc polymers (>20 units). Why do short glycans bind virions but not stabilize them? Poliovirus capsids contain 60 copies of each capsid protein, VP1 to VP4. VP1 surrounds the 5-fold axis, and defective LPS binding of the VP1 T99K mutant suggests that this region contains the glycan binding site (8). We measured the distance of T99 residues on the virion surface, using Chimera software, based on a poliovirus structure at 3.0-Å resolution (27, 28). We found that the distance between the closest T99 residues within a 5-fold axis is 2.9 nm, which approximately matches the length of GlcNAc6 (3 nm) (29) (Fig. 6). Conversely, the distance between a T99 residue at one 5-fold axis and a T99 residue at an adjacent 5-fold axis is 13.4 nm, which approximately matches the length of GlcNAc27 (13.5 nm) (Fig. 6). Thus, we propose a model where short oligomers such as GlcNAc6 are long enough to bind two adjacent sites within a 5-fold axis, but this is insufficient to stabilize the virus; conversely, long oligomers (>13.4 nm) bind to and bridge two adjacent 5-fold axes, thus aiding the structural rigidity and stability of the capsid. This model would explain why excess GlcNAc6 inhibits stabilization by long glycans. We propose that long glycans form chain-like structures on the virion surface and restrain heat-induced virion conformation changes that precede premature RNA release. It is also possible that long glycans bind two or more virions, inducing viral aggregates. Previous work has shown that virion aggregates are more resistant to heat inactivation (30, 31), so it is possible that glycan-induced aggregation also induces stability. Future studies will test this possibility.
FIG 6.
Model of glycan binding and stabilization of poliovirus. The poliovirus capsid contains 60 copies each of VP1, VP2, VP3, and VP4. VP4 is internal and not surface exposed. VP1 surrounds the 5-fold symmetry axis, whereas VP2 and VP3 alternate around the 3-fold axis. Based on the number of capsid proteins present, each virion may have 60 glycan binding sites. The exposed T99 residue of VP1 is important for polysaccharide binding and is likely near the glycan binding site. The distance between two nearby T99 residues is 2.9 nm, corresponding with the length of GlcNAc6 (3 nm). It is possible that GlcNAc6 can bind two adjacent glycan binding sites within a 5-fold axis, resulting in increased avidity compared with that of monovalent nonbinding GlcNAc3 (1.5 nm). Long-chain polysaccharides (>13.4 nm) are long enough to potentially bridge nearby 5-fold axes, form cage-like structures on the virion surface, and stabilize poliovirus against heat-induced conformational change causing premature RNA release.
The glycan binding specificity of poliovirus may not be conserved among other enteric viruses. Human rotavirus and norovirus interact with polymorphic human histo-blood group antigens (HBGAs), and this interaction is thought to be important for infection (32). LPS stabilizes coxsackievirus B3 (CVB3) in vitro, but CVB3 is more sensitive to microbiota perturbation than poliovirus (11, 12), indicating that it may have distinct glycan binding specificity. Bacterial LPS and peptidoglycan also increase reovirus thermostability (33), but the detailed glycan binding specificity is unclear.
Proviral effects of microbiota have been observed for viruses in four different families (18). Antibiotic treatment causes microbiota imbalances and can cause antibiotic resistance (34), making perturbation of the microbiota unviable for treatment of enteric virus infection. As an alternative, targeting the binding of enteric virus to bacterial glycans may provide a promising approach for control of enteric virus infection. Short GlcNAc oligosaccharides can inhibit polysaccharide-mediated poliovirus stabilization. The concentration of bacterial polysaccharide present in the gastrointestinal tract is estimated to be in the milligram/milliliter range (35). Due to such a high concentration, oral administration of short GlcNAc oligosaccharides is not likely to have any antiviral effects. However, it is useful to characterize glycan-virion binding, which may aid the design of high-binding small molecules as novel antiviral drugs. Future structural biology studies may illuminate virus-glycan interactions in more detail. Overall, our work has provided new insight into glycan-virus interactions that impact viral stability.
MATERIALS AND METHODS
Virus.
Poliovirus work was performed in biosafety level 2+ (BSL2+) areas in accordance with practices recommended by the World Health Organization. Poliovirus (serotype 1 virulent Mahoney) cell culture infections and plaque assays were performed using HeLa and Vero cells, respectively (5).
Acid hydrolysis of LPS.
In our previous work (5), we tested LPS from Escherichia coli, Salmonella enterica, and Klebsiella pneumoniae. We found that each of these LPS types can stabilize poliovirus. In this study, we chose LPS from E. coli for detailed study. LPS from E. coli O127:B8 (L3129; Sigma-Aldrich) was dissolved in 2% acetic acid at 5 mg/ml. After incubation at 100°C for 2 h with occasional shaking, chloroform and methanol were added to yield a final chloroform/methanol/water ratio of 2:1:3. The mixture was vortexed and centrifuged at 5,000 × g for 10 min. The resulting upper phase containing detoxified LPS and the lower phase containing lipid A were collected separately and dried in a Speed-Vac. For monosaccharide analysis, detoxified LPS was dissolved in 1 M HCl and heated at 100°C for 30 min. Samples were dried in a Speed-Vac.
FACE analysis.
Detoxified LPS, LPS monosaccharides, and chitin oligosaccharides were analyzed by FACE (36, 37). Briefly, monosaccharides were labeled with 2-aminoacridone (06627; Sigma-Aldrich), and oligosaccharides were labeled with 7-amino-1,3-naphthalenedisulfonic acid (81529; AnaSpec) prior to electrophoresis on 20% polyacrylamide gels. Gel images were acquired with a UVP Chemidoc-It II scanner (Analytik Jena, Jena, Germany).
Preparation and purification of GlcNAc oligomers.
Chitin oligomers were prepared via chitin hydrolysis in concentrated hydrochloric acid followed by acetone precipitation (38). Two grams of chitin (C9752, Sigma) was dissolved in 80 ml concentrated HCl and incubated at 40°C for 1 h. Samples were added to 1,100 ml acetone and stirred overnight at 4°C. The glycan pellet was collected by centrifugation at 5,000 × g for 10 min. The pellet was washed twice with acetone and dried at 43°C in an oven, and then 200 ml water was added and stirred overnight at 4°C. Insoluble polysaccharides were removed by centrifugation at 12,000 × g for 10 min. The supernatant containing soluble chitin oligosaccharides was loaded on a Dowex AG 50W-X8 hydrogen-form cation-exchange column to eliminate any deacetylated products. The flowthrough fraction was concentrated in a Speed-Vac. The concentrations of oligosaccharides were measured by absorbance at 210 nm using GlcNAc6 as a standard.
For purification of low-molecular-weight GlcNAc oligomers, a hand-packed Bio-Gel P-4 size exclusion column (1.0 by 50 cm) (1504128; Bio-Rad) was used for glycan separation. Samples were eluted in water at 0.1 ml/min and fractionated for 5 min/tube. For purification of high-molecular-weight GlcNAc oligomers, a hand-packed Bio-Gel P-10 size exclusion column (1.5 by 50 cm) (1504144; Bio-Rad) was used. Samples were eluted in 20 mM acetic acid at 0.33 ml/min and fractionated for 10 min/tube. Glycans in each fraction were quantified by measuring absorbance at 210 nm and analyzed by FACE.
35S-labeled poliovirus pulldown assay.
35S-labeled polioviruses were generated by growing virus in the presence of 35S-labeled methionine-cysteine and purifying by CsCl gradient ultracentrifugation (5). For the insoluble-polysaccharide pulldown assay, 5,000 cpm of 35S-labeled polioviruses (105 PFU) was mixed with 500 μg polysaccharides in 0.5 ml PBS and incubated at 37°C for 3 h. Polysaccharides were pelleted at 1,500 × g for 2 min and washed with 1 ml PBS three times. Polysaccharide-associated virus was measured by scintillation counting. Peptidoglycan (77140; Sigma) was further purified by proteinase K digestion and PBS washing. All other insoluble polysaccharides were from Sigma without further purification. Inert beads (Dynabeads 65305; Thermo Fisher) were used as a background binding control.
For binding of low-molecular-weight GlcNAc oligomers, agarose bead-cross-linked lectin (AL-1023; Vector Laboratories) was used to immobilize oligosaccharides on the bead surface. Ten microliters of bead slurry was incubated overnight with 20 μg glycans in PBS containing 4% BSA. Beads were washed three times to remove unbound glycans and mixed with 35S-labeled poliovirus following the same protocol as for insoluble polysaccharides. ConA-agarose beads (AL-1003; Vector Laboratories) were used as a negative control.
Poliovirus in vitro thermal stability assay.
CsCl gradient-purified polioviruses were used for thermal stability experiments. For in vitro thermal inactivation, 106 PFU of poliovirus was mixed with compounds at the indicated concentrations and incubated at 45°C for 5 h. After incubation, plaque assays were performed in Vero cells, and the remaining PFU was compared with the input to calculate the percentage of input PFU. d-Glucose (Glc), d-galactose (Gal), d-mannose (Man), l-fucose (Fuc), d-glucosamine (GlcN), d-galactosamine (GalN), d-mannosamine (ManN), N-acetyl-d-glucosamine (GlcNAc), N-acetyl-d-galactosamine (GalNAc), N-acetyl-d-mannosamine (ManNAc), maltotriose (Glc3), and maltohexaose (Glc6) were purchased from Sigma. Tri-N-acetylchitotriose (GlcNAc3) and hexa-N-acetylchitohexaose (GlcNAc6) were from Caymanchem. The identities of these glycans were confirmed by FACE.
For PaSTRY, 1 μg of poliovirus was preincubated with compounds for 1 h at 37°C to promote binding. After incubation, a 10× final concentration of SYBR green II and buffer (10 mM HEPES, 200 mM NaCl, pH 8.0) were added to a final volume of 30 μl (17). Samples were heated in an ABI 7500 real-time instrument from 25°C to 99°C on a 1% stepwise gradient with fluorescence monitoring.
Statistics.
Either ordinary one-way analysis of variance (ANOVA) or the Kruskal-Wallis test was used for comparing multiple independent samples. A P value of <0.05 was considered statistically significant.
ACKNOWLEDGMENTS
This work was supported by a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Diseases award, a Howard Hughes Medical Institute Faculty Scholar Award, and grant R01 AI74668 to J.K.P. and by grant R01 GM038545 to M.A.L.
REFERENCES
- 1.Hurst CJ, Gerba CP. 1989. Fate of viruses during wastewater sludge treatment processes. Crit Rev Environ Control 18:317–343. doi: 10.1080/10643388909388352. [DOI] [Google Scholar]
- 2.Kocwa-Haluch R. 2001. Waterborne enteroviruses as a hazard for human health. Pol J Environ Stud 10:485–487. [Google Scholar]
- 3.Racaniello VR. 2006. One hundred years of poliovirus pathogenesis. Virology 344:9–16. doi: 10.1016/j.virol.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 4.Kane M, Case LK, Kopaskie K, Kozlova A, MacDearmid C, Chervonsky AV, Golovkina TV. 2011. Successful transmission of a retrovirus depends on the commensal microbiota. Science 334:245–249. doi: 10.1126/science.1210718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuss SK, Best GT, Etheredge CA, Pruijssers AJ, Frierson JM, Hooper LV, Dermody TS, Pfeiffer JK. 2011. Intestinal microbiota promote enteric virus replication, and systemic pathogenesis. Science 334:249–252. doi: 10.1126/science.1211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones MK, Watanabe M, Zhu S, Graves CL, Keyes LR, Grau KR, Gonzalez-Hernandez MB, Iovine NM, Wobus CE, Vinje J, Tibbetts SA, Wallet SM, Karst SM. 2014. Enteric bacteria promote human and mouse norovirus infection of B cells. Science 346:755–759. doi: 10.1126/science.1257147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kernbauer E, Ding Y, Cadwell K. 2014. An enteric virus can replace the beneficial function of commensal bacteria. Nature 516:94–98. doi: 10.1038/nature13960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson CM, Jesudhasan PR, Pfeiffer JK. 2014. Bacterial lipopolysaccharide binding enhances virion stability and promotes environmental fitness of an enteric virus. Cell Host Microbe 15:36–46. doi: 10.1016/j.chom.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uchiyama R, Chassaing B, Zhang B, Gewirtz AT. 2014. Antibiotic treatment suppresses rotavirus infection and enhances specific humoral immunity. J Infect Dis 210:171–182. doi: 10.1093/infdis/jiu037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baldridge MT, Nice TJ, McCune BT, Yokoyama CC, Kambal A, Wheadon M, Diamond MS, Ivanova Y, Artyomov M, Virgin HW. 2015. Commensal microbes and interferon-lambda determine persistence of enteric murine norovirus infection. Science 347:266–269. doi: 10.1126/science.1258025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson CM, Acevedo W, McCune B, Pfeiffer JK. 2019. Related enteric viruses have different requirements for host microbiota in mice. J Virol 93:e01339-19. doi: 10.1128/JVI.01339-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aguilera ER, Nguyen Y, Sasaki J, Pfeiffer JK, Aguilera ER, Nguyen Y, Sasaki J, Pfeiffer JK. 2019. Bacterial stabilization of a panel of picornaviruses. mSphere 4:e00183-19. doi: 10.1128/mSphere.00183-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu H, Lehrman MA, Pfeiffer JK. 2019. Use of a glycan library reveals a new model for enteric virus oligosaccharide binding and virion stabilization. bioRxiv 10.1101/834101. [DOI] [PMC free article] [PubMed]
- 14.Stenutz R, Weintraub A, Widmalm G. 2006. The structures of Escherichia coli O-polysaccharide antigens. FEMS Microbiol Rev 30:382–403. doi: 10.1111/j.1574-6976.2006.00016.x. [DOI] [PubMed] [Google Scholar]
- 15.Erickson AK, Jesudhasan PR, Mayer MJ, Narbad A, Winter SE, Pfeiffer JK. 2018. Bacteria facilitate enteric virus co-infection of mammalian cells and promote genetic recombination. Cell Host Microbe 23:77–88. doi: 10.1016/j.chom.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lienemann M, Paananen A, Boer H, de la Fuente JM, García I, Penadés S, Koivula A. 2009. Characterization of the wheat germ agglutinin binding to self-assembled monolayers of neoglycoconjugates by AFM and SPR. Glycobiology 19:633–643. doi: 10.1093/glycob/cwp030. [DOI] [PubMed] [Google Scholar]
- 17.Walter TS, Ren J, Tuthill TJ, Rowlands DJ, Stuart DI, Fry EE. 2012. A plate-based high-throughput assay for virus stability and vaccine formulation. J Virol Methods 185:166–170. doi: 10.1016/j.jviromet.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfeiffer JK, Virgin HW, Pfeiffer JK, Virgin HW. 2016. Viral immunity. Transkingdom control of viral infection and immunity in the mammalian intestine. Science 351:aad5872. doi: 10.1126/science.aad5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson CM. 2019. Enteric viruses exploit the microbiota to promote infection. Curr Opin Virol 37:58–62. doi: 10.1016/j.coviro.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roth AN, Grau KR, Karst SM, Roth AN, Grau KR, Karst SM. 2019. Diverse mechanisms underlie enhancement of enteric viruses by the mammalian intestinal microbiota. Viruses 11:760. doi: 10.3390/v11080760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Filman DJ, Syed R, Chow M, Macadam AJ, Minor PD, Hogle JM. 1989. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J 8:1567–1579. doi: 10.1002/j.1460-2075.1989.tb03541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fox MP, Otto MJ, McKinlay MA. 1986. Prevention of rhinovirus and poliovirus uncoating by WIN 51711, a new antiviral drug. Antimicrob Agents Chemother 30:110–116. doi: 10.1128/aac.30.1.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorval BL, Chow M, Klibanov AM. 1989. Stabilization of poliovirus against heat inactivation. Biochem Biophys Res Commun 159:1177–1183. doi: 10.1016/0006-291x(89)92234-1. [DOI] [PubMed] [Google Scholar]
- 24.Steimle A, Autenrieth IB, Frick JS. 2016. Structure and function: lipid A modifications in commensals and pathogens. Int J Med Microbiol 306:290–301. doi: 10.1016/j.ijmm.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Berti F, De Ricco R, Rappuoli R, Berti F, De Ricco R, Rappuoli R. 2018. Role of O-acetylation in the immunogenicity of bacterial polysaccharide vaccines. Molecules 23:1340. doi: 10.3390/molecules23061340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benachour A, Ladjouzi R, Le Jeune A, Hebert L, Thorpe S, Courtin P, Chapot-Chartier MP, Prajsnar TK, Foster SJ, Mesnage S. 2012. The lysozyme-induced peptidoglycan N-acetylglucosamine deacetylase PgdA (EF1843) is required for Enterococcus faecalis virulence. J Bacteriol 194:6066–6073. doi: 10.1128/JB.00981-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller ST, Hogle JM, Filman DJ. 2001. Ab initio phasing of high-symmetry macromolecular complexes: successful phasing of authentic poliovirus data to 3.0 A resolution. J Mol Biol 307:499–512. doi: 10.1006/jmbi.2001.4485. [DOI] [PubMed] [Google Scholar]
- 28.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 29.Nakamura A, Okazaki KI, Furuta T, Sakurai M, Iino R. 2018. Processive chitinase is Brownian monorail operated by fast catalysis after peeling rail from crystalline chitin. Nat Commun 9:3814. doi: 10.1038/s41467-018-06362-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Young DC, Sharp DG. 1977. Poliovirus aggregates and their survival in water. Appl Environ Microbiol 33:168–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mattle MJ, Crouzy B, Brennecke M, Wigginton KR, Perona P, Kohn T. 2011. Impact of virus aggregation on inactivation by peracetic acid and implications for other disinfectants. Environ Sci Technol 45:7710–7717. doi: 10.1021/es201633s. [DOI] [PubMed] [Google Scholar]
- 32.Tan M, Jiang X. 2014. Histo-blood group antigens: a common niche for norovirus and rotavirus. Expert Rev Mol Med 16:e5. doi: 10.1017/erm.2014.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berger AK, Yi H, Kearns DB, Mainou BA. 2017. Bacteria and bacterial envelope components enhance mammalian reovirus thermostability. PLoS Pathog 13:e1006768. doi: 10.1371/journal.ppat.1006768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Becattini S, Taur Y, Pamer EG. 2016. Antibiotic-induced changes in the intestinal microbiota and disease. Trends Mol Med 22:458–478. doi: 10.1016/j.molmed.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bates JM, Akerlund J, Mittge E, Guillemin K. 2007. Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe 2:371–382. doi: 10.1016/j.chom.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao N, Holmes J, Lehrman MA. 2013. Letter to the Glycoforum: improved protocols for preparing lipid-linked and related saccharides for fluorophore-assisted carbohydrate electrophoresis (FACE). Glycobiology 23:1111. doi: 10.1093/glycob/cwt067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao N, Lehrman MA. 2006. Non-radioactive analysis of lipid-linked oligosaccharide compositions by fluorophore-assisted carbohydrate electrophoresis. Methods Enzymol 415:3–20. doi: 10.1016/S0076-6879(06)15001-6. [DOI] [PubMed] [Google Scholar]
- 38.Kazami N, Sakaguchi M, Mizutani D, Masuda T, Wakita S, Oyama F, Kawakita M, Sugahara Y. 2015. A simple procedure for preparing chitin oligomers through acetone precipitation after hydrolysis in concentrated hydrochloric acid. Carbohydr Polym 132:304–310. doi: 10.1016/j.carbpol.2015.05.082. [DOI] [PubMed] [Google Scholar]