Cells use the editing activity of adenosine deaminase acting on RNA proteins (ADARs) to prevent autoimmune responses induced by self dsRNA, but viruses can exploit this process to their advantage. Borna disease virus (BoDV), a nuclear-replicating RNA virus, must escape nonself RNA sensing by the host to establish persistent infection in the nucleus. We evaluated whether BoDV utilizes ADARs to prevent innate immune induction. ADAR2 plays a key role throughout the BoDV life cycle. ADAR2 knockdown reduced A-to-I editing of BoDV genomic RNA, leading to the induction of a strong innate immune response. These data suggest that BoDV exploits ADAR2 to edit nonself genomic RNA to appear as self RNA for innate immune evasion and establishment of persistent infection.
KEYWORDS: nuclear-replicating RNA virus, A-to-I editing, self/nonself, innate immunity, persistent infection
ABSTRACT
Cells sense pathogen-derived double-stranded RNA (dsRNA) as nonself. To avoid autoimmune activation by self dsRNA, cells utilize A-to-I editing by adenosine deaminase acting on RNA 1 (ADAR1) to disrupt dsRNA structures. Considering that viruses have evolved to exploit host machinery, A-to-I editing could benefit innate immune evasion by viruses. Borna disease virus (BoDV), a nuclear-replicating RNA virus, may require escape from nonself RNA-sensing and immune responses to establish persistent infection in the nucleus; however, the strategy by which BoDV evades nonself recognition is unclear. Here, we evaluated the involvement of ADARs in BoDV infection. The infection efficiency of BoDV was markedly decreased in both ADAR1 and ADAR2 knockdown cells at the early phase of infection. Microarray analysis using ADAR2 knockdown cells revealed that ADAR2 reduces immune responses even in the absence of infection. Knockdown of ADAR2 but not ADAR1 significantly reduced the spread and titer of BoDV in infected cells. Furthermore, ADAR2 knockout decreased the infection efficiency of BoDV, and overexpression of ADAR2 rescued the reduced infectivity in ADAR2 knockdown cells. However, the growth of influenza A virus, which causes acute infection in the nucleus, was not affected by ADAR2 knockdown. Moreover, ADAR2 bound to BoDV genomic RNA and induced A-to-G mutations in the genomes of persistently infected cells. We finally demonstrated that BoDV produced in ADAR2 knockdown cells induces stronger innate immune responses than those produced in wild-type cells. Taken together, our results suggest that BoDV utilizes ADAR2 to edit its genome to appear as “self” RNA in order to maintain persistent infection in the nucleus.
IMPORTANCE Cells use the editing activity of adenosine deaminase acting on RNA proteins (ADARs) to prevent autoimmune responses induced by self dsRNA, but viruses can exploit this process to their advantage. Borna disease virus (BoDV), a nuclear-replicating RNA virus, must escape nonself RNA sensing by the host to establish persistent infection in the nucleus. We evaluated whether BoDV utilizes ADARs to prevent innate immune induction. ADAR2 plays a key role throughout the BoDV life cycle. ADAR2 knockdown reduced A-to-I editing of BoDV genomic RNA, leading to the induction of a strong innate immune response. These data suggest that BoDV exploits ADAR2 to edit nonself genomic RNA to appear as self RNA for innate immune evasion and establishment of persistent infection.
INTRODUCTION
Cells sense exogenous dsRNA as “nonself” and induce innate immune responses to eliminate the pathogen. This defense system critically influences the outcome of pathogen infection (1, 2). In addition, cells possess machinery to prevent autoimmune activation by “self” double-stranded RNA (dsRNA). To escape recognition by dsRNA sensors such as melanoma differentiation-associated protein 5 (MDA5), dsRNA-dependent protein kinase R (PKR), and 2′,5′-oligoadenylate synthetase (OAS), adenosine deaminase acting on RNA-1 (ADAR1) mediates A-to-I editing to induce a conformational change in the stem-loop structure at the 3′ untranslated region (UTR) of mRNAs and Alu-derived dsRNAs (3–7).
A-to-I editing is the process by which adenosine (A) nucleotides in dsRNA are converted to inosine (I) nucleotides, and this editing is catalyzed by ADARs (8–10). The ADAR family contains three members, ADAR1, ADAR2, and ADAR3. Although all 3 have conserved deaminase domains, only ADAR1 and ADAR2 catalyze A-to-I editing in mammals (11–13). ADAR1 is ubiquitously expressed and has two isoforms, the constitutive p110 isoform and the interferon-inducible p150 isoform. While ADAR1 p110 localizes to both the nucleus and cytoplasm, p150 is a cytoplasmic protein (14). ADAR2 is highly expressed in the nuclei of cells in the central nervous system (CNS) (12). ADAR1 and ADAR2 partially share substrates; however, ADAR1 primarily catalyzes repetitive sites, while ADAR2 prefers nonrepetitive coding regions (15, 16).
In addition to self/nonself discrimination, the consequences of A-to-I editing are diverse in cellular systems. Conformational changes in RNA by A-to-I editing play roles in the regulation of RNA-binding protein accessibility to mediate the stability and nuclear retention of RNA (17–19). Furthermore, A-to-I editing modulates RNA splicing by generating donor or acceptor sites or eliminating branch point sites (20–22). Direct editing of the coding region, mainly catalyzed by ADAR2 in the CNS, leads to alterations in protein structure and function through amino acid substitutions (16, 23). Some neurotransmitter receptors require A-to-I editing by ADAR2 to rewrite mRNA sequences and produce multiple variants, which are necessary for the normal development and function of the brain (24, 25). ADAR2 provides an additional layer of epigenetic regulation of the biological activity of genes; however, unlike for ADAR1, the roles of ADAR2 in the A-to-I editing of noncoding regions and in the discrimination of self/nonself RNA have not been demonstrated.
Viruses can utilize cellular RNA modification systems to evade recognition as nonself by dsRNA sensors. Measles virus (MV) produces defective interfering (DI) RNAs from the viral genome, which can form dsRNA (26). ADAR1 knockout reduces A-to-I editing in DI RNA of MV and suppresses viral growth depending on the innate immune response through PKR activation (27, 28). These observations suggest that MV employs A-to-I editing by ADAR1 to escape recognition as nonself. In addition to MV, hepatitis delta virus (HDV) is also known to utilize A-to-I editing by ADAR1 to accomplish its life cycle. HDV antigenomic RNA is edited by ADAR1 to produce two viral proteins from one transcript (29, 30). The hyperediting of transcripts of mouse polyomavirus at the overlapping region of the early and late genes is potentially mediated by ADARs and might be involved in the viral early-to-late switch (31–33). Although ADARs can impact infection by other RNA viruses, such as HIV-1 (34–38) and influenza A virus (IAV) (39–41), in an editing-dependent or editing-independent manner, the biological significance of ADARs and their mechanisms of action in viral infection are varied (42).
Borna disease virus (BoDV), a nonsegmented, negative-sense RNA virus, is a neurotropic virus that replicates in the cell nucleus. BoDV infection causes severe immune-mediated encephalitis in highly susceptible species, such as horses and sheep (43–45). However, in most mammals, BoDV infection does not trigger fatal symptoms and progresses to persistent infection without overt cytopathic effects (46, 47). In addition, unlike any other RNA virus, BoDV maintains its replication complex in the cell nucleus in an episomal form and acts as a part of the cell component through close interaction with host chromosomes (48). To achieve persistent infection in the nucleus, therefore, BoDV may need to prevent the recognition of its genomic RNA as nonself by RNA sensors. Previous studies revealed that BoDV trims triphosphate at the 5′ termini of its genomic RNA to monophosphate to avoid recognition by retinoic acid-inducible gene-I (RIG-I) (49, 50). However, considering that BoDV elongates the 3′ termini of genomic and antigenomic RNA through a back-priming process and that other single-stranded negative-sense RNA viruses generate dsRNA, BoDV RNAs can also form intra- or intermolecular dsRNAs during replication in the nucleus (49, 51). Despite these observations, the mechanisms by which BoDV escapes recognition by dsRNA sensors in persistently infected cells are unclear.
Here, to elucidate the mechanism by which the BoDV genome mimics self RNA in the nucleus, we evaluated the role of ADARs in BoDV infection. We demonstrated that A-to-I editing of the BoDV genome by ADAR2 but not by ADAR1 is involved in efficient viral replication in the nucleus, leading to the maintenance of persistent infection in the nucleus.
RESULTS
Knockdown of ADARs affects the initial infection of BoDV.
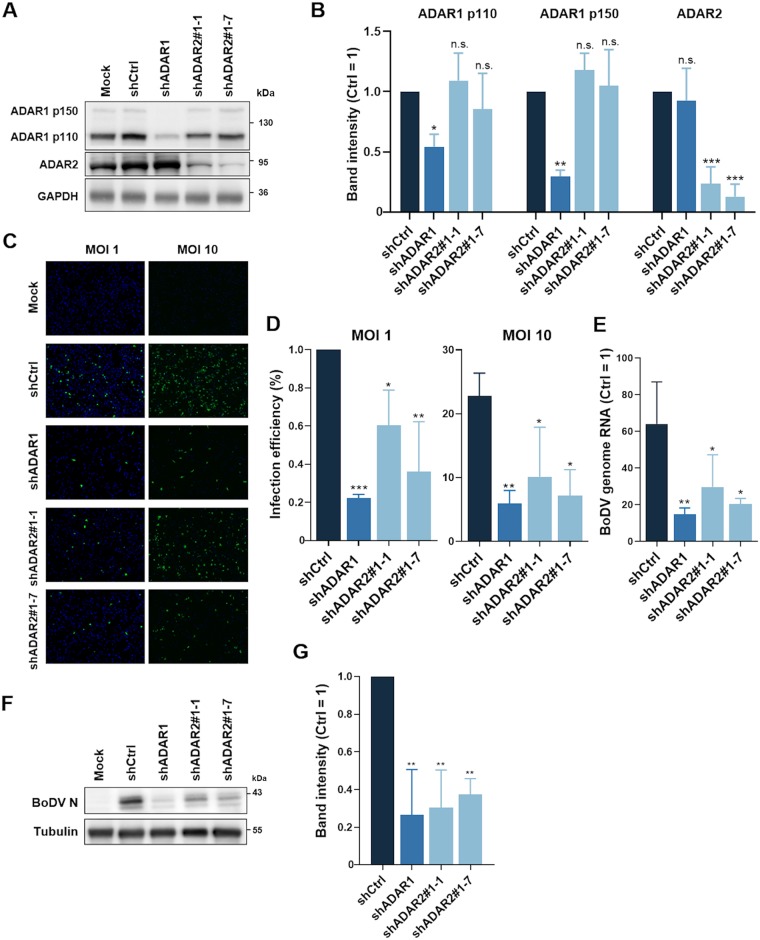
To assess the role of ADARs in BoDV infection, we first introduced short hairpin RNA (shRNA) against ADAR1 or ADAR2 into human oligodendroglial (OL) cells via a lentiviral vector system (Fig. 1A and B). Knockdown cells were infected with BoDV at a multiplicity of infection (MOI) of 1 or 10, and the infection efficiencies were then determined by an indirect immunofluorescence assay (IFA) at 4 days postinfection (dpi). As shown in Fig. 1C and D, knockdown of either ADAR1 or ADAR2 significantly decreased the efficiency of BoDV infection at the early phase compared with that in control cells. Consistent with these findings, the expression levels of BoDV genomic RNA and protein were decreased in ADAR knockdown cells (Fig. 1E to G). These results suggested that knockdown of ADARs reduces the BoDV replication efficiency at the early phase of infection.
FIG 1.
Knockdown of ADARs decreased the BoDV infection efficiency at the early phase. (A and B) Western blot analysis of ADAR1 and ADAR2 knockdown cells. OL cells were infected with lentivirus harboring shRNA targeting ADAR1 or ADAR2 or control (Ctrl) shRNA. Cells were cloned and used for subsequent experiments. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the loading control. The band intensities were normalized by the signal intensities of GAPDH. (C and D) Cell lines with knockdown of ADARs were infected with BoDV at an MOI of 1.0 or 10. At 4 dpi, an IFA was performed with anti-BoDV P antibody (green) and DAPI (blue). The infection efficiencies were calculated by dividing the number of infected cells by that of total cells. (E) RT-qPCR of viral genomic RNA at 4 dpi. (F and G) Western blot analysis of the viral proteins nucleoprotein (N) at 4 dpi. Alpha-tubulin was used as the loading control. The band intensities were normalized by the signal intensities of alpha-tubulin. All experiments were performed in three biologically independent replicates. The values are expressed as the means + standard error (SE) of the results from three independent experiments. Significance was analyzed by one-way ANOVA and Dunnett’s multiple-comparison test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., nonsignificant.
ADAR2 knockdown induces antiviral immune responses.
ADAR1 knockout or knockdown induces aberrant innate immune responses through autoimmune activation by nonedited mRNA (3, 4); however, the effect of ADAR2 knockdown on innate immune responses has not been determined. Therefore, we next performed a comprehensive analysis using a microarray system to assess whether antiviral genes are upregulated by ADAR2 knockdown. As shown in Fig. 2A, the expression levels of 512 genes were changed more than 2-fold by ADAR2 knockdown. Moreover, by the rank product method, 65 genes were extracted as differentially expressed genes (DEGs) and subjected to enrichment analysis to elucidate their biological importance. As shown in Fig. 2B, the DEGs were significantly enriched in inflammation- and immune response-related genes. These results indicated that ADAR2 reduces autoimmune responses even in the absence of infection and stimulation, suggesting that like ADAR1, ADAR2 is involved in the avoidance of immune activation (3, 4).
FIG 2.
ADAR2 knockdown induces an anti-BoDV state via the upregulation of Rab27b and CXCL1. (A) Scatter plot of DEGs in ADAR2 knockdown and control cells. (B) Summary of the GO terms enriched in the 65 DEGs identified by the rank product analysis, as analyzed by DAVID. Inflammation-related and immune response-related genes are shown in blue, and the others are shown in gray. (C) Infection efficiencies of BoDV in ADAR2 knockdown cells transduced with a series of siRNAs. The names of the target genes are listed in Table 2. siRNA no. 26, Rab27b; siRNA no. 49, CXCL1. (D) Confirmation experiment of panel C results using different siRNAs against Rab27b and CXCL1. All experiments were performed in 2 to 3 biologically independent replicates. The values are expressed as the means + SE. Significance was analyzed by one-way ANOVA and Dunnett’s multiple-comparison test. *, P < 0.05; **, P < 0.01; n.s., nonsignificant.
To identify the genes responsible for the reduced BoDV infection efficiency in ADAR2 knockdown cells, we next performed a small interfering RNA (siRNA) screen of the upregulated genes identified in the microarray analysis. Briefly, ADAR2 knockdown cells were transfected with a series of siRNAs and inoculated with BoDV at 24 h posttransfection. Infection efficiencies were determined by IFA at 3 dpi. As shown in Fig. 2C, siRNA against Rab27b and CXCL1 increased the infection efficiency of BoDV in ADAR2 knockdown cells to the same extent as in control cells. To further confirm this result, an infection experiment was carried out with the same system using different siRNAs against Rab27b and CXCL1. Consistent with the siRNA screening results, the infectivity of BoDV was rescued by knockdown of Rab27b and CXCL1 (Fig. 2D). Taken together, our results suggest that ADAR2 knockdown induces antiviral immune responses leading to the suppression of initial BoDV infection.
ADAR2 affects the intranuclear replication of BoDV.
We next evaluated whether knockdown of ADARs influences BoDV propagation in addition to initial infection. To this end, we monitored the growth rate of BoDV in ADAR knockdown cells. As shown in Fig. 3A, the growth kinetics of BoDV in ADAR1 knockdown cells were similar to those in control cells (P > 0.05 by two-way analysis of variance [ANOVA] and Dunnett’s post hoc test). In contrast, BoDV propagation was slower in ADAR2 knockdown cells than in control cells (P < 0.01 for both ADAR2 knockdown cell lines by two-way ANOVA and Dunnett’s post hoc test). Interestingly, the amount of genomic RNA in ADAR2 knockdown cells was significantly smaller than that in control cells even after the infection efficiency of BoDV reached almost 100% at 48 dpi (Fig. 3B). Furthermore, ADAR2 knockdown significantly decreased the expression level of BoDV nucleoprotein (BoDV-N) at 48 dpi (Fig. 3C and D). These data suggest that ADAR2 knockdown affects not only the initial infection but also the intranuclear replication of BoDV, while ADAR1 is involved only in the initial infection.
FIG 3.
ADAR2 knockdown impairs BoDV propagation. (A) Growth curves of BoDV in cells with knockdown of ADARs. Knockdown cell lines were infected with BoDV at an MOI of 1.0, and the infection efficiency was determined by IFA every 4 days after infection. (B) Relative amounts of BoDV genomic RNA in infected cells at 48 dpi. (C and D) Western blot analysis of the BoDV proteins nucleoprotein (N) at 48 dpi. Alpha-tubulin was used as the loading control. The band intensities were normalized by the signal intensities of alpha-tubulin. All experiments were performed in three biologically independent replicates. Significance was analyzed by one-way or two-way ANOVA and Dunnett’s multiple-comparison test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., nonsignificant.
To verify that ADAR2 is involved in the replication of BoDV in the nucleus, we next knocked down ADAR2 with two different shRNAs in OL cells persistently infected with BoDV (Fig. 4A and B). Thirty days later, the infection efficiencies and BoDV genomic RNA expression levels were analyzed. Although ADAR2 knockdown did not alter the infection efficiencies of BoDV in the cells (Fig. 4C), the level of viral genomic RNA was significantly reduced in the knockdown cells (Fig. 4D). Consistent with the decrease in the viral genomic RNA level, ADAR2 knockdown led to a significant reduction in the titer of virus collected from the cells (Fig. 4E). Considering that BoDV infection is maintained in persistently infected cultured cells mainly through cell division, not by reinfection (48), these observations strongly suggest that ADAR2 contributes to efficient viral replication within cell nuclei, which may be important for the maintenance of persistent BoDV infection.
FIG 4.
Effect of ADAR2 knockdown on persistent infection of BoDV. (A and B) Western blotting of BoDV-infected OL cells transduced with two shRNAs against ADAR2. GAPDH was used as the loading control in Western blotting. The band intensities were normalized by the signal intensity of GAPDH. (C) Infection efficiencies were determined by IFA using anti-BoDV-P antibody. (D) Expression levels of viral genomic RNA were measured by RT-qPCR. (E) Titers of BoDV were measured by IFA. Sonicated BoDV was prepared from ADAR2 knockdown cells infected with BoDV. OL cells were infected with sonicated BoDV, and IFA was performed at 3 dpi using anti-BoDV-P antibody. FFU, focus-forming units. (F) WSN virus was collected from ADAR2 knockdown cells and control cells. Viral titers were determined by a plaque assay. All experiments were performed in three biologically independent replicates. The values are expressed as the means + SE of the results from three independent experiments. Significance was analyzed by Student's t test or by one-way ANOVA and Dunnett’s multiple-comparison test. *, P < 0.05; **, P < 0.001; ***, P < 0.0001; n.s., nonsignificant.
In addition, we performed an ADAR2 knockdown experiment using influenza A/WSN/33 (H1N1) (WSN) virus, which, like BoDV, replicates in the nucleus. However, WSN virus growth was not inhibited in ADAR2 knockdown cells (Fig. 4F), suggesting that ADAR2 knockdown attenuates persistent BoDV infection but not acute IAV infection in the nucleus.
ADAR2 knockout reduces infection efficiency of BoDV.
To further confirm the significance of ADAR2, the infection experiments were carried out using ADAR2 knockout cells generated by the CRISPR-Cas9 system (Fig. 5A). Consistent with the results in ADAR2 knockdown cells, the infection efficiencies of BoDV in ADAR2 knockout cells were significantly decreased at 4 dpi compared with that in mock cells (Fig. 5B). In parallel, the expression levels of BoDV genomic RNA were also reduced by ADAR2 knockout (Fig. 5C). These results strongly support the significance of ADAR2 in BoDV infection.
FIG 5.
ADAR2 knockout reduces infection efficiency of BoDV. (A) Western blot analysis of ADAR2 knockout (KO) cells. OL cells were introduced with Cas9-gRNA ribonucleoprotein by electroporation and cloned (KO#1 and KO#2). GAPDH was used as the loading control. (B) ADAR2 knockout cells were infected with BoDV at an MOI of 1.0. The infection efficiencies were determined by IFA at 4 dpi. (C) RT-qPCR of viral genomic RNA at 4 dpi. All experiments were performed in three biologically independent replicates. The values are expressed as the means + SE of the results from three independent experiments. Significance was analyzed by one-way ANOVA and Dunnett’s multiple-comparison test. **, P < 0.01; ***, P < 0.001.
ADAR2 edits BoDV genomic RNA in persistently infected nuclei.
To assess whether the effect of ADAR2 knockdown on BoDV replication is dependent on A-to-I editing, we first evaluated the A-to-I editing activity of ADAR2 in uninfected OL cells. The A-to-I editing activity is generally determined by measuring the editing efficiency of the Q/R site in host glutamate receptor 2 (GluR2), which was edited by ADAR2 with almost 100% efficiency (Fig. 6A) (52). Since I is read as guanosine (G) in sequencing, an A-to-G substitution is observed at the Q/R site as the result of A-to-I editing. We amplified the GluR2 fragment containing the Q/R site by seminested PCR and sequenced it. As illustrated in Fig. 6B, only a G signal was detected at the Q/R site in uninfected OL cells, suggesting that the Q/R site was almost completely edited by ADAR2, as reported previously (52). However, only an A signal was detected in ADAR2 knockdown cells, indicating that the A-to-I editing activity in ADAR2 knockdown cells was below the detection limit.
FIG 6.
ADAR2 binds and edits BoDV genomic RNA. (A) Schematic of the editing assay. (B) Results of sequencing analysis at the Q/R site in control cells and ADAR2 knockdown cells (shADAR2#1-7). (C) ADAR2 knockdown cells (shADAR2#1-7) were transfected with a series of plasmids expressing ADAR2 WT, ADAR2 E/A (an editing activity-deficient mutant) or EGFP as a control. After 48 h, the cells were inoculated with BoDV at an MOI of 1.0 and subjected to IFA at 4 dpi. All experiments were performed in three biologically independent replicates. The values are expressed as the means + SE of the results from three independent experiments. (D) Left, schematic diagram of A-to-I editing sites in BoDV genomic RNA. The indicated numbers show the positions of A-to-G mutations. Right, summary of the A-to-G position and amino acid changes. (E) The interaction between BoDV genomic RNA and ADAR2 was examined by RNA immunoprecipitation. FLAG-tagged ADAR2 WT, ADAR2 EAA (an RNA binding-deficient mutant), BoDV-N, and EGFP were separately overexpressed in BoDV-infected OL cells. Coimmunoprecipitated proteins and RNAs were analyzed by Western blotting and RT-PCR, respectively. (F) Coimmunoprecipitated BoDV genomic RNA was quantified by RT-qPCR. All experiments were performed in three biologically independent replicates. The values are expressed as the means + SE of the results from three independent experiments. Significance was analyzed by one-way ANOVA and Dunnett’s multiple-comparison test. *, P < 0.05.
To assess whether A-to-I activity of ADAR2 is involved in BoDV infection, phenotypic rescue was performed by overexpressing ADAR2 in the knockdown cells. ADAR2 knockdown cells were transfected with a series of plasmids expressing wild-type (WT) ADAR2, an editing activity-deficient mutant (E/A) (53), or red fluorescent protein (RFP) as a control. At 48 h posttransfection, the cells were infected with BoDV at an MOI of 1, and the infection efficiencies were measured at 4 dpi. As shown in Fig. 6C, overexpression of ADAR2 WT recovered the infection efficiency of BoDV, but overexpression of ADAR2 E/A did not. These data suggest that the A-to-I editing activity of ADAR2 is important for BoDV infection.
We next investigated whether BoDV genomic RNA is edited by ADAR2. We performed sequence analysis on genomic RNA of BoDV collected from ADAR2 knockdown cells by Sanger sequencing. We identified six possible A-to-I editing sites, in which G residues existed as a minor population (10%), in BoDV genomic RNA collected from WT cells but not from ADAR2 knockdown cells (Fig. 6D). All six sites were in the coding regions. Among the A-to-G mutations, three were synonymous, and the others were nonsynonymous substitutions. ADAR2 has a triplet preference for deamination. Triplets are classified into type I (high preference), type II (moderate preference), and type III (low preference) depending on the deamination efficiency (26). All six sites appeared to belong to the higher-preference type I and II categories, suggesting that the G residues identified as the minor population could be generated by the A-to-I editing activity of ADAR2. These results showed that BoDV genomic RNA is edited by ADAR2 in persistently infected nuclei.
To confirm the editing of viral genomic RNA by ADAR2, we investigated the interaction between BoDV genomic RNA and ADAR2 by RNA immunoprecipitation analysis. Plasmids expressing FLAG-tagged ADAR2 WT, the ADAR2 RNA binding-deficient mutant (EAA) (54), enhanced green fluorescent protein (EGFP) (negative control), or BoDV-N (positive control) were transfected into cells persistently infected with BoDV. At 48 h posttransfection, cell extracts were subjected to immunoprecipitation using anti-FLAG antibody and were then analyzed by Western blotting and reverse transcription-quantitative PCR (RT-qPCR). As shown in Fig. 6E and F, BoDV genomic RNA was clearly coimmunoprecipitated with ADAR2 WT but not with ADAR2 EAA. These data show that BoDV genomic RNA interacts with ADAR2 through its dsRNA-binding domain, indicating that ADAR2 is involved in the A-to-I editing of BoDV genomic RNA in the nucleus.
BoDV utilizes the editing activity of ADAR2 to avoid recognition as nonself in infected nuclei.
Our observations imply that BoDV exploits the A-to-I editing activity of ADAR2 to avoid recognition as nonself, ensuring persistent infection in the nucleus. Therefore, we next investigated the innate immune response induced by BoDV particles recovered from ADAR2 knockdown cells. OL cells were inoculated with BoDV particles collected from ADAR2 knockdown cells infected with BoDV, and the induction of innate immune responses was evaluated. As shown in Fig. 7A, the proinflammatory cytokines interleukin 6 (IL-6) and CXCL10, which were shown to be upregulated in BoDV-infected cells (55, 56), were significantly upregulated by infection with nonedited BoDV collected from ADAR2 knockdown cells. However, the expression levels of these proinflammatory cytokines did not significantly differ between the HEPES solutions collected from noninfected ADAR2 knockdown cells and control cells (Fig. 7B). These data indicate that infection with BoDV recovered from ADAR2 knockdown cells can induce an enhanced innate immune response.
FIG 7.
BoDV suppresses the induction of innate immunity through the RNA-editing activity of ADAR2. (A) Cell-free BoDV particles were collected from ADAR2 knockdown (shADAR2#1-7) or control cells infected with BoDV, and the normalized amount of viral genomic RNA was determined by RT-qPCR. OL cells were inoculated with an equal amount of BoDV particles for 1 h at 37°C. After 8 h, total cell lysate was collected and subjected to RNA extraction, reverse transcription, and RT-qPCR. (B) The procedure described in panel A was performed, except that noninfected cells were used. (C) Phenotypic rescue experiments were performed. Cell-free BoDV particles were collected from ADAR2 knockdown cells expressing ADAR2 WT, ADAR2 E/A, or EGFP as a control. OL cells were inoculated with an equal amount of BoDV particles for 1 h at 37°C. After 8 h, total cell lysate was collected and subjected to RNA extraction, reverse transcription, and RT-qPCR. (D) The expression levels of CXCL10 in ADAR2 knockdown cells with persistent BoDV infection or mock infection were quantified by RT-qPCR. All experiments were performed in three biologically independent replicates. The values are expressed as the means + SE of the results from three independent experiments. Significance was analyzed by Student's t test or one-way ANOVA and Dunnett’s multiple-comparison test. *, P < 0.05; **, P < 0.001; ***, P < 0.0001; n.s., nonsignificant.
To verify whether the editing activity of ADAR2 is important for the enhanced immune response in the above-described experiments, we performed a rescue experiment using BoDV collected from ADAR2 knockdown cells overexpressing ADAR2 WT, ADAR2 E/A, or EGFP. As shown in Fig. 7C, compared to infection with BoDV from ADAR2 knockdown cells transfected with ADAR2 E/A, infection with BoDV from ADAR2 knockdown cells transfected with ADAR2 WT suppressed the expression level of IL-6. However, for CXCL10, only ADAR2 WT rescued the ADAR2 knockdown phenotype (Fig. 7C). These results suggest that BoDV manipulates the A-to-I editing activity of ADAR2 to avoid recognition as nonself by the host and prevent the induction of the innate immune response.
We finally assessed whether ADAR2 suppresses the expression of innate immune genes even in persistently infected cells. To this end, we knocked down ADAR2 in OL cells persistently infected with BoDV and investigated the expression level of CXCL10 mRNA. As shown in Fig. 7D, in uninfected cells, the expression level of CXCL10 mRNA did not differ between ADAR2 knockdown and control cells; in contrast, CXCL10 was significantly upregulated by ADAR2 knockdown in cells persistently infected with BoDV. Considering that ADAR2 is required for the efficient replication of BoDV in persistently infected cells (Fig. 4), these observations indicate that the genomic RNA in persistently infected nuclei must also undergo A-to-I editing for efficient replication. In turn, this requirement suggests that the mechanism for sensing nonself RNA is active in the nucleus and that BoDV utilizes an intranuclear host editing system, ADAR2, to evade this sensing mechanism.
DISCUSSION
This report is the first to demonstrate that ADAR2 knockdown upregulates immune responses and affects RNA virus infection. We showed that the genomic RNA of persistently infecting BoDV is recognized as nonself RNA and edited by ADAR2 in the nucleus. This finding demonstrates a novel viral strategy utilizing cellular mechanisms to prevent antiviral responses in the nucleus.
Viruses have evolved original strategies to evade the host innate immune system; however, the strategy used by a persistently infecting RNA virus to escape from nonself RNA sensors in the nucleus is poorly understood. Given that viruses hijack host machinery to survive, BoDV may conceivably utilize intranuclear machinery to avoid recognition of its RNA as nonself. In this study, we obtained the following experimental evidence that BoDV uses ADAR2 in the nucleus: (i) knockdown or knockout of ADAR2 reduces the replication efficiency of BoDV, and overexpression of ADAR2 WT rescues the reduced replication (Fig. 3 and 5), (ii) ADAR2 can edit BoDV genomic RNA in persistently infected nuclei (Fig. 6), (iii) ADAR2 binds to BoDV genomic RNA through the dsRNA-binding domain (Fig. 6), (iv) BoDV produced in ADAR2 knockdown cells induces an enhanced innate immune response (Fig. 6 and 7), and (v) the enhanced innate immune response induced by ADAR2 knockdown cell-derived BoDV is rescued by the expression of ADAR2 WT (Fig. 7C). Collectively, these findings strongly suggest that BoDV manipulates ADAR2 to edit viral RNA to appear as self RNA in order to prevent the induction of host innate immune responses in the nucleus. However, in this study, we cannot exclude the possibility that one or more host factors related to viral growth is edited by ADAR2 and affects BoDV infection and innate immune induction. Nevertheless, to our knowledge, this report is the first to describe the involvement of the RNA-editing enzyme in the intranuclear replication of RNA virus.
Our study also uncovers a previously undetermined function of ADAR2, which is suppression of the autoimmune response. Microarray analysis revealed that ADAR2 knockdown induced inflammatory and innate immune responses even in the absence of infection or stimulation (Fig. 2). Previous studies showed that ADAR2 mediates the A-to-I editing of mRNAs constitutively transcribed from host genomic DNA (16, 23–25). Our results indicated that, like ADAR1, ADAR2 might prevent the recognition of endogenous dsRNAs as nonself by RNA sensors such as MDA5 and PKR (3).
The involvement of ADAR2 in dsRNA recognition suggests the intriguing possibility that ADARs differentially recognize target RNAs in a manner dependent on the intracellular localization of the RNAs. Interferon-inducible ADAR1 p150 is a cytoplasmic protein, and p110 is constitutively expressed in both the nucleus and cytoplasm, while ADAR2 is localized in the nucleus. BoDV establishes persistent infection in the nucleus. During persistent infection, BoDV does not need to be outside the nucleus because its genomic RNA is associated with host chromosomes throughout the cell cycle (48). Considering this localization, ADAR2 is the most likely major partner of BoDV genomic RNA for editing in infected cells. Indeed, we demonstrated the interaction between ADAR2 and genomic RNA in infected cells (Fig. 6). Furthermore, ADAR1 did not affect the establishment of persistent BoDV infection (Fig. 3A). Conceivably, the selection of target RNA by the differential localization of ADAR1 and ADAR2 contributes to efficient RNA editing in cells. However, we did not find that ADAR2 knockdown affected the growth of IAV (Fig. 4F). IAV also replicates in the nucleus but causes acute infection. Although ADAR2 might be able to edit IAV genomic RNA in the nucleus, it would only mildly affect viral replication because IAV genomic RNA is localized in both the cytoplasm and nucleus during infection and has been shown to be recognized by RNA sensors, such as RIG-I, which results in the induction of strong immune responses (57–59). This recognition may mask the effect of ADAR2 in editing IAV genomic RNA in the nucleus. Alternatively, the target RNA specificity of ADARs may be determined by the preferential sites of A-to-I editing within the sequences. The A-to-I editing sites in BoDV genomic RNA are located in coding regions (Fig. 6D), consistent with the preference of ADAR2 for triplets, while ADAR1 shows a greater preference for repetitive sites (16). These data may explain why ADAR1 knockdown does not affect BoDV propagation (Fig. 3). Further analysis is necessary to elucidate the target RNA specificity of ADARs.
ADARs mediate hyperediting on RNAs of various viruses (60–62). In MV, A-to-I editing by ADAR1 may disrupt dsRNA structures within the DI RNA to avoid recognition by PKR and MDA5 (26–28). We found six editing positions in BoDV genomic RNA where A residues accounted for 90% and G residues for 10% of the residues in the read sequences (Fig. 6D). However, the G residue was totally eliminated in these six positions in the genomic RNA of virus produced in ADAR2 knockdown cells, indicating that the A-to-G mutation was introduced by ADAR2. In this study, we could not demonstrate the mechanism by which the A-to-G mutations in the genomic RNA reduce the induction of innate immune responses in cells. Although we could not find drastic changes in the predicted RNA secondary structures in the edited regions of BoDV genomic RNA (data not shown), small nucleotide changes in a long RNA molecule, such as viral genomic RNA, could conceivably change intramolecular interactions and influence structural stability or accessibility. Alternatively, during replication and transcription, BoDV RNA could form intermolecular dsRNA structures containing genomic and antigenomic RNAs and mRNAs. Since both intra- and intermolecular dsRNAs are substrates for ADARs (63), the edited sites in BoDV genomic RNA may prevent the generation of intermolecular dsRNA structures, resulting in the elimination of innate immune induction. Furthermore, of the six mutations, three contribute to nonsynonymous mutations in the P, M, and L genes (Fig. 6D). Thus, investigating the possibility that viral proteins harboring such mutations can directly suppress the innate immune response and contribute to efficient viral growth may be interesting.
We showed that ADAR2 knockdown significantly reduces the level of BoDV genomic RNA in persistently infected cells (Fig. 4D). Considering that only 10% of BoDV genomic RNA was edited by ADAR2 in the cells (Fig. 6D), such a small population of mutants in the nucleus could be critical for the maintenance of efficient viral replication. Another possibility is that BoDV genomic RNA is extensively edited at a low frequency that cannot be detected by Sanger sequencing, as reported by Li et al. (64), and the entire viral population is required to suppress the innate immune response, as is observed in poliovirus infection (65). Further analysis to detect whether or not BoDV generates minor mutations during the replication in the nucleus using RNA sequencing (RNA-seq) analysis would be of interest. On the other hand, we found no BoDV isolates with A-to-G mutations in the NCBI database, suggesting that although the edited mutants can reduce the innate immune response and maintain persistent infection as an overall viral population in infected cells, the presence of these mutations in the BoDV genome is lethal or deleterious for viral growth, and these mutants cannot remain a major population. Regulation of the ratio of mutant BoDV genomic RNA in the population may counterbalance innate immune activation and viral replication, which could be critical for the maintenance of persistent infection in the nucleus. Our findings may shed light on the new role of quasispecies among the viral population in persistently infected cells.
In summary, this study reveals an elaborate strategy by which RNA viruses manipulate host machinery to allow innate immune evasion and the establishment of persistent infection in the nucleus. Additionally, our findings provide new insight into the cellular mechanism by which intranuclear RNA editing allows the recognition of nonself signatures to maintain self functions.
MATERIALS AND METHODS
Cells.
OL cells derived from human embryonic oligodendroglial cells (66, 67), BoDV-infected OL cells, and ADAR2 knockout OL cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific) supplemented with 5% fetal calf serum (FCS; MP Biomedicals, CA, USA). OL cells expressing shRNA against ADAR1 or ADAR2 or a scrambled shRNA sequence were maintained in the same medium containing 2 μg/ml puromycin (Merck, Darmstadt, Germany). 293T LTV cells (Cell Biolabs, CA, USA) were grown in DMEM-low glucose (1.0%) (Nacalai Tesque, Kyoto, Japan) containing 10% FCS and 1% minimum essential medium (MEM) nonessential amino acid solution (Thermo Fisher Scientific). MDCK cells were cultured in MEM (Thermo Fisher Scientific) supplemented with 5% FCS, 7.5% NaHCO3 (Merck), a 1% MEM nonessential amino acid solution, a 1% MEM vitamin solution (Thermo Fisher Scientific), 1% l-glutamine (Merck), and a 1% penicillin-streptomycin solution (Wako, Osaka, Japan).
Plasmid construction.
To construct the ADAR2 expression plasmid (pcDNA3-3×FLAG-ADAR2), the ADAR2 fragment amplified by PCR with cDNA reverse transcribed from OL cells was cloned into the pcDNA3 vector, which contains three FLAG tags upstream of the cloning site. The plasmid expressing the ADAR2 editing-deficient mutant (pcDNA3-3×FLAG-ADAR2 E/A) was generated by PCR-based mutagenesis at the catalytic active site of ADAR2 (glutamic acid to alanine at position 396). The plasmid expressing ADAR2 EAA, an RNA binding-deficient mutant (pcDNA3-3×FLAG-ADAR2 EAA), was kindly provided by Kazuko Nishikura (The Wistar Institute, USA) (54). To obtain the EGFP expression plasmid (pcDNA3-3×FLAG-EGFP), EGFP cDNA was amplified using pEGFP-N1 (Addgene, MA, USA) as the template and inserted into the same vector. The BoDV-N expression plasmid was previously constructed (68).
shRNA expression.
To obtain shRNA expression plasmids, shRNA sequences against ADAR1 or ADAR2 or a scrambled shRNA sequence were cloned into pRSI-U6-(sh)-UbiC-RFP-2A-Puro (Cellecta, CA, USA). The sequence information of shRNAs used in this study was as follows: for shADAR1, 5′-CCAGCACAGCGGAGUGGUA-3′; for shADAR2#1, 5′-TACATGAGTGATCGTGGCC-3′; and for ADAR2#2, 5′-GATAGACACCCAAATCGTA-3′ (69). The plasmids were cotransfected with psPAX2 packaging plasmid (Cellecta) and pMD2.G (Cellecta) expressing vesicular stomatitis virus G protein into 293T LTV cells. After 72 h, supernatants containing lentiviral vectors were collected and filtered through a 0.45-μm filter. To establish the ADAR1 and ADAR2 knockdown cell lines, OL cells were infected with the lentiviral vectors and were then selected with 2 μg/ml puromycin. Puromycin-resistant cells were picked and cloned. The numbers of shRNAs and cell clones were 1 for shADAR2#1 and 7 for shADAR2#1.
Sonicated virus preparation.
Sonicated BoDV was prepared as previously described (70). Briefly, BoDV-infected OL cells were suspended in DMEM supplemented with 2% FCS and subjected to sonication. After centrifugation of sonicated cells at 1,200 × g for 25 min at 4°C, the supernatant was collected and stored at –80°C.
BoDV infection.
Cells were inoculated with sonicated BoDV or cell-free BoDV particles. After absorption for 1 h, cells were washed with phosphate-buffered saline (PBS) and maintained in DMEM containing 2% FCS. To assess viral propagation, the infected cells were passaged every 4 days.
Indirect IFA.
Cells were fixed with 4% paraformaldehyde (Nacalai Tesque) and permeabilized in PBS containing 0.1% Triton X-100 (Nacalai Tesque) and 2% FCS. After washing with PBS, cells were incubated with anti-BoDV P antibody and then with Alexa Fluor 488-conjugated secondary antibody (Thermo Fisher Scientific) and 4′,6-diamidino-2-phenylindole (DAPI; Merck) at room temperature for 1 h. BoDV-positive cells and DAPI-positive cells were counted using a BZ-X710 All-in-One fluorescence microscope (Keyence, Osaka, Japan). Infection efficiency was calculated by dividing the number of infected cells by the number of total cells.
RNA extraction.
Total RNA was extracted using a NucleoSpin RNA kit (Macherey-Nagel, Düren, Germany) or TRIzol LS (Thermo Fisher Scientific), according to the manufacturers’ protocols.
RT-qPCR.
Total RNA was reverse transcribed using a Verso cDNA synthesis kit (Thermo Fisher Scientific) using BoDV genome-specific, oligo(dT), or random hexamer primers. RT-qPCR assays were performed with a Rotor-Gene Q (Qiagen, Hilden, Germany) or a CFX Connect real-time system (Bio-Rad, CA, USA) using Thunderbird SYBR quantitative PCR (qPCR) mix (Toyobo, Osaka, Japan) or Thunderbird Probe qPCR mix (Toyobo). The primer sequences are available upon request.
Western blot analysis.
Cell lysates were prepared and subjected to SDS-PAGE and transferred to polyvinylidene difluoride membranes. Membranes were blocked with Blocking One solution (Nacalai Tesque) and were then incubated with the primary antibodies listed in Table 1 at room temperature for 1 h. After washing, membranes were probed with horseradish peroxidase (HRP)-conjugated secondary antibodies at room temperature for 1 h. Membranes were developed with ECL Plus Western blot detection reagents (GE Healthcare, IL, USA), according to the manufacturer’s instructions. Band intensities were measured using the Multi Gauge v3.2 software (Fujifilm, Tokyo, Japan).
TABLE 1.
Antibody list and characteristics
| Antibody | Assaya | Reference or source | Dilution | Catalog no. |
|---|---|---|---|---|
| Anti-human ADAR1 monoclonal antibody | WB | Santa Cruz Biotechnology | 1/1,000 | Sc-73408 |
| Anti-human ADAR2 monoclonal antibody | WB | Santa Cruz Biotechnology | 1/1,000 | Sc-73409 |
| Anti-human GAPDH monoclonal antibody | WB | Santa Cruz Biotechnology | 1/1,000 | Sc-47724 |
| Anti-human tubulin monoclonal antibody | WB | Sigma-Aldrich | 1/2,000 | T5168 |
| Anti-BoDV P polyclonal antibody | IFA | Hirai et al. (75) | 1/1,000 | |
| Anti-BoDV N monoclonal antibody | WB | Hirai et al. (75) | 1/4 | |
| Anti-FLAG M2 monoclonal antibody | WB, RIP | Sigma-Aldrich | 1/1,000 | F1804 |
| Alexa Fluor 488 goat anti-rabbit IgG | IFA | Thermo Fisher Scientific | 1/1,000 | A11034 |
| Peroxidase-conjugated AffiniPure donkey anti-mouse IgG | WB | Jackson ImmunoResearch | 1/1,000 | 715-035-150 |
| Peroxidase-conjugated AffiniPure donkey anti-rabbit IgG | WB | Jackson ImmunoResearch | 1/1,000 | 711-035-152 |
WB, Western blotting; IFA, indirect immunofluorescence assay; RIP, RNA immunoprecipitation.
Microarray analysis.
Microarray analysis was performed with a Human Clariom S array (Thermo Fisher Scientific), according to the manufacturer’s protocol. The results were analyzed using the Transcriptome Analysis Console (TAC) software (Thermo Fisher Scientific). DEGs were extracted by rank product analysis (false-discovery rate, <0.05) (71).
Gene Ontology term analysis.
Gene Ontology (GO) term analysis was conducted on the identified DEGs using the Database for Annotated Visualization, Integration and Discovery (DAVID) software (72, 73).
siRNA transfection for phenotypic rescue.
ADAR2 knockdown cells were transfected with a series of siRNAs using HiPerfect reagent (Qiagen), according to the manufacturer’s protocol. Twenty-four hours after transfection, cells were inoculated with BoDV at an MOI of 1 at 37°C for 1 h and subsequently subjected to IFA at 3 dpi. The siRNAs used in this study are listed in Table 2. The catalog numbers for the siRNAs are available upon request.
TABLE 2.
siRNAs used in Fig. 2C
| siRNA no. | Gene name |
|---|---|
| 1 | TMPRSS15 |
| 2 | CXCL5 |
| 3 | ANPEP |
| 4 | AOX1 |
| 5 | TMEM156 |
| 6 | CXorf57 |
| 7 | GPR85 |
| 8 | S100A16 |
| 9 | IL1B |
| 10 | GCA |
| 11 | QPRT |
| 12 | SERPINB7 |
| 13 | ZNF804A |
| 14 | EVI2A |
| 15 | TFPI |
| 16 | C3 |
| 17 | IL1RAPL1 |
| 18 | NAV3 |
| 19 | TMEM47 |
| 20 | CSMD3 |
| 21 | NCAM2 |
| 22 | C6orf99 |
| 23 | G0S2 |
| 24 | GLIPR1 |
| 25 | BIRC3 |
| 26 | RAB27B |
| 27 | AKR1C1 |
| 28 | EMP1 |
| 29 | GRPR |
| 30 | HDAC9 |
| 31 | ABI3BP |
| 32 | PTX3 |
| 33 | SYTL2 |
| 34 | SPP1 |
| 35 | TENM1 |
| 36 | SYT1 |
| 37 | PDCD1LG2 |
| 38 | CXCL8 |
| 39 | FGF5 |
| 40 | CDCP1 |
| 41 | PLAU |
| 42 | SNAI2 |
| 43 | PRTFDC1 |
| 44 | CDK15 |
| 45 | SRPX2 |
| 46 | CPED1 |
| 47 | HS3ST3A1 |
| 48 | ANTXR2 |
| 49 | CXCL1 |
| 50 | FGF1 |
| 51 | Nontargeting control |
| 52 | GAPDH |
Rescue of infection efficiency in ADAR2 knockdown cells.
ADAR2 knockdown cells were transfected with pcDNA3-3×FLAG-ADAR2, pcDNA3-3×FLAG-ADAR2 E/A, or pcDNA3-3×FLAG-EGFP. After 48 h, the cells were inoculated with BoDV at an MOI of 1 at 37°C for 1 h and subsequently subjected to IFA at 3 dpi.
IAV infection in ADAR2 knockdown cells.
WSN virus was kindly provided by Yoshihiro Kawaoka (The University of Tokyo, Japan). ADAR2 knockdown and control cells were inoculated with WSN virus at an MOI of 1 for 1 h. After washing with PBS, cells were maintained in the medium at 37°C. After 48 h, the supernatant was collected and centrifuged at 3,000 rpm for 10 min at 4°C to remove cell debris. Viral titers in the supernatants were determined by a plaque assay. MDCK cells in 12-well plates were inoculated with 10-fold serial dilutions of WSN virus for 1 h at 37°C, and agarose containing 0.2% tosylsulfonyl phenylalanyl chloromethyl ketone-treated trypsin (Merck) was then added. Plates were incubated at room temperature for 15 min and then at 37°C for 36 h. Then, 20% formalin was added to each well and incubated for 30 min at room temperature. After the formalin and agarose were removed, the wells were stained with 0.005% amido black at room temperature for 1 h. After the wells were washed, the number of plaques was counted.
Establishment of ADAR2 knockout cell lines with CRISPR-Cas9 system.
ADAR2 knockout cell lines were generated by the Alt-R CRISPR-Cas9 system (Integrated Device Technology, CA, USA), according to the manufacturer’s protocols. The target sequence was 5′-GCCATGCAGAAATAATATCTCGG-3′. Alt‐R CRISPR‐Cas9 CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA) were mixed and incubated for 5 min at 95°C. The genomic RNA (gRNA) was mixed with Alt-R S.p. HiFi Cas9 nuclease V3 at room temperature for 20 min. OL cells were electroporated with the Cas9-gRNA complex using 4D-Nucleofector and SF Cell Line 4D-Nucleofector X kit L (Lonza, Basel, Switzerland). The electroporated cells were cloned and used for subsequent experiments.
RNA editing detection assay.
The editing efficiency of the Q/R site in host GluR2, which was edited by ADAR2 with almost 100% efficiency, is commonly measured to assess the editing activity of ADAR2 (52). To this end, the GluR2 fragment containing the Q/R site was amplified by seminested PCR with PrimeSTAR GXL DNA polymerase (TaKaRa Bio, Shiga, Japan) and sequenced. The conditions for the first PCR were as follows: 35 cycles of 10 s at 98°C, 30 s at 60°C, and 30 s at 72°C. In the next reaction mixture, 1 μl of the first PCR product was used as the template. The conditions for the second reaction were as follows: 35 cycles of 10 s at 98°C, 30 s at 66°C, and 30 s at 68°C. The primer information is available upon request.
Direct sequencing of BoDV genomic RNA.
BoDV genomic RNA contained in viral particles collected from ADAR2 knockdown or control BoDV-infected cells was reverse transcribed with a Verso cDNA synthesis kit and amplified by PCR using KOD One PCR master mix -blue- (Toyobo). The PCR products were cloned into the pCR4Blunt-TOPO vector (Thermo Fisher Scientific), following the manufacturer’s guidelines. Ten colonies per PCR product were picked and sequenced. The sequences were analyzed using the Geneious software (Biomatters, Auckland, New Zealand). The primer sequences are available upon request.
RNA immunoprecipitation.
BoDV-infected OL cells were transfected with pcDNA3-3×FLAG-ADAR2 WT, pcDNA3-3×FLAG-ADAR2 EAA, pcDNA3-3×FLAG-EGFP, or pcDNA3-3×FLAG-BoDV-N using Lipofectamine 2000 (Thermo Fisher Scientific). At 48 h posttransfection, RNA immunoprecipitation (RIP) was performed using a RiboCluster Profiler/RIP-assay kit, according to the manufacturer’s instructions, except that Dynabeads protein G (Thermo Fisher Scientific) was used instead of the supplied agarose beads for immunoprecipitation. The coimmunoprecipitated sample was analyzed by Western blotting and RT-PCR. To quantitate BoDV genomic RNA, RT-qPCR was performed.
Cell-free BoDV preparation.
Cell-free BoDV particles were prepared as previously described (74). BoDV-infected cells were incubated with 20 mM HEPES buffer (pH 7.4) containing 250 mM MgCl2 and 1% FCS for 90 min at 37°C. Supernatants were filtered through 0.22-μm membrane filters and stored at –80°C.
Data availability.
We stored the microarray data in the NCBI Gene Expression Omnibus (accession number GSE138927).
ACKNOWLEDGMENTS
This work was supported in part by JSPS KAKENHI grants JP17H04083 (to K.T.), 18K05991 (to A.M.), 18J14718 (to M.Y.), and 17J07483 (to R.K.); MEXT KAKENHI grants JP16H06429, JP16K21723, and JP16H06430 (all to K.T.) and 19H04834 (to A.M.); JSPS Core-to-Core Program, AMED grant JP19fm0208014 (to K.T.); and the Joint Usage/Research Center Program on inFront, Kyoto University.
We thank Yoshihiro Kawaoka (The University of Tokyo, Japan) for the influenza A/WSN/33 (H1N1) virus and Kazuko Nishikura (The Wistar Institute, USA) for the expression plasmid of the ADAR2 RNA binding-deficient mutant.
REFERENCES
- 1.Kawai T, Akira S. 2006. Innate immune recognition of viral infection. Nat Immunol 7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 2.Kenney AD, Dowdle JA, Bozzacco L, McMichael TM, St. Gelais C, Panfil AR, Sun Y, Schlesinger LS, Anderson MZ, Green PL, López CB, Rosenberg BR, Wu L, Yount JS. 2017. Human genetic determinants of viral diseases. Annu Rev Genet 51:241–263. doi: 10.1146/annurev-genet-120116-023425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liddicoat BJ, Ramaswami G, Hartner JC, Li JB, Piskol R, Higuchi M, Chalk AM, Seeburg PH, Walkley CR. 2015. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 349:1115–1120. doi: 10.1126/science.aac7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung H, Calis JJA, Wu X, Sun T, Yu Y, Sarbanes SL, Dao Thi VL, Shilvock AR, Hoffmann HH, Rosenberg BR, Rice CM. 2018. Human ADAR1 prevents endogenous RNA from triggering translational shutdown. Cell 172:811–824.e14. doi: 10.1016/j.cell.2017.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao L, Jha BK, Wu A, Elliott R, Ziebuhr J, Gorbalenya AE, Silverman RH, Weiss SR. 2012. Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe 11:607–616. doi: 10.1016/j.chom.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Banerjee S, Goldstein SA, Dong B, Gaughan C, Rath S, Donovan J, Korennykh A, Silverman RH, Weiss SR. 2017. Ribonuclease l mediates the cell-lethal phenotype of double-stranded RNA editing enzyme ADAR1 deficiency in a human cell line. Elife 6:e25687. doi: 10.7554/eLife.25687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.George CX, Ramaswami G, Li JB, Samuel CE. 2016. Editing of cellular self-RNAs by adenosine deaminase ADAR1 suppresses innate immune stress responses. J Biol Chem 291:6158–6168. doi: 10.1074/jbc.M115.709014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bass BL, Weintraub H. 1987. A developmentally regulated activity that unwinds RNA duplexes. Cell 48:607–613. doi: 10.1016/0092-8674(87)90239-x. [DOI] [PubMed] [Google Scholar]
- 9.Bass BL, Weintraub H. 1988. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell 55:1089–1098. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- 10.Wagner RW, Smith JE, Cooperman BS, Nishikura K. 1989. A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc Natl Acad Sci U S A 86:2647–2651. doi: 10.1073/pnas.86.8.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. 1994. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc Natl Acad Sci U S A 91:11457–11461. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melcher T, Maas S, Herb A, Sprengel R, Seeburg PH, Higuchi M. 1996. A mammalian RNA editing enzyme. Nature 379:460–464. doi: 10.1038/379460a0. [DOI] [PubMed] [Google Scholar]
- 13.Chen CX, Cho DS, Wang Q, Lai F, Carter KC, Nishikura K. 2000. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA 6:755–767. doi: 10.1017/s1355838200000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patterson JB, Samuel CE. 1995. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol Cell Biol 15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehmann KA, Bass BL. 2000. Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry 39:12875–12884. doi: 10.1021/bi001383g. [DOI] [PubMed] [Google Scholar]
- 16.Tan MH, Li Q, Shanmugam R, Piskol R, Kohler J, Young AN, Liu KI, Zhang R, Ramaswami G, Ariyoshi K, Gupte A, Keegan LP, George CX, Ramu A, Huang N, Pollina EA, Leeman DS, Rustighi A, Goh YPS, GTEx Consortium, Chawla A, Del Sal G, Peltz G, Brunet A, Conrad DF, Samuel CE, O’Connell MA, Walkley CR, Nishikura K, Li JB. 2017. Dynamic landscape and regulation of RNA editing in mammals. Nature 550:249–254. doi: 10.1038/nature24041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anantharaman A, Tripathi V, Khan A, Yoon J-H, Singh DK, Gholamalamdari O, Guang S, Ohlson J, Wahlstedt H, Öhman M, Jantsch MF, Conrad NK, Ma J, Gorospe M, Prasanth SG, Prasanth KV. 2017. ADAR2 regulates RNA stability by modifying access of decay-promoting RNA-binding proteins. Nucleic Acids Res 45:4189–4201. doi: 10.1093/nar/gkw1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anantharaman A, Gholamalamdari O, Khan A, Yoon J-H, Jantsch MF, Hartner JC, Gorospe M, Prasanth SG, Prasanth KV. 2017. RNA-editing enzymes ADAR1 and ADAR2 coordinately regulate the editing and expression of Ctn RNA. FEBS Lett 591:2890–2904. doi: 10.1002/1873-3468.12795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Z, Carmichael GG. 2001. The fate of dsRNA in the nucleus: a p54nrb-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell 106:465–475. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]
- 20.Rueter SM, Dawson TR, Emeson RB. 1999. Regulation of alternative splicing by RNA editing. Nature 399:75–80. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
- 21.Flomen R, Knight J, Sham P, Kerwin R, Makoff A. 2004. Evidence that RNA editing modulates splice site selection in the 5-HT2C receptor gene. Nucleic Acids Res 32:2113–2122. doi: 10.1093/nar/gkh536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paz-Yaacov N, Levanon EY, Nevo E, Kinar Y, Harmelin A, Jacob-Hirsch J, Amariglio N, Eisenberg E, Rechavi G. 2010. Adenosine-to-inosine RNA editing shapes transcriptome diversity in primates. Proc Natl Acad Sci U S A 107:12174–12179. doi: 10.1073/pnas.1006183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paul MS, Bass BL. 1998. Inosine exists in mRNA at tissue-specific levels and is most abundant in brain mRNA. EMBO J 17:1120–1127. doi: 10.1093/emboj/17.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sommer B, Köhler M, Sprengel R, Seeburg PH. 1991. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 67:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- 25.Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB. 1997. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 26.Pfaller CK, Radeke MJ, Cattaneo R, Samuel CE. 2014. Measles virus C protein impairs production of defective copyback double-stranded viral RNA and activation of protein kinase R. J Virol 88:456–468. doi: 10.1128/JVI.02572-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfaller CK, Mastorakos GM, Matchett WE, Ma X, Samuel CE, Cattaneo R. 2015. Measles virus defective interfering RNAs are generated frequently and early in the absence of C protein and can be destabilized by adenosine deaminase acting on RNA-1-like hypermutations. J Virol 89:7735–7747. doi: 10.1128/JVI.01017-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfaller CK, Donohue RC, Nersisyan S, Brodsky L, Cattaneo R. 2018. Extensive editing of cellular and viral double-stranded RNA structures accounts for innate immunity suppression and the proviral activity of ADAR1p150. PLoS Biol 16:e2006577. doi: 10.1371/journal.pbio.2006577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang K-S, Choo Q-L, Weiner AJ, Ou J-H, Najarian RC, Thayer RM, Mullenbach GT, Denniston KJ, Gerin JL, Houghton M. 1986. Structure, sequence and expression of the hepatitis delta (δ) viral genome. Nature 323:508–514. doi: 10.1038/323508a0. [DOI] [PubMed] [Google Scholar]
- 30.Jayan GC, Casey JL. 2002. Inhibition of hepatitis delta virus RNA editing by short inhibitory RNA-mediated knockdown of ADAR1 but not ADAR2 expression. J Virol 76:12399–12404. doi: 10.1128/jvi.76.23.12399-12404.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar M, Carmichael GG. 1997. Nuclear antisense RNA induces extensive adenosine modifications and nuclear retention of target transcripts. Proc Natl Acad Sci U S A 94:3542–3547. doi: 10.1073/pnas.94.8.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garren SB, Kondaveeti Y, Duff MO, Carmichael GG. 2015. Global analysis of mouse polyomavirus infection reveals dynamic regulation of viral and host gene expression and promiscuous viral RNA editing. PLoS Pathog 11:e1005166. doi: 10.1371/journal.ppat.1005166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.George CX, Samuel CE. 2011. Host response to polyomavirus infection is modulated by RNA adenosine deaminase ADAR1 but not by ADAR2. J Virol 85:8338–8347. doi: 10.1128/JVI.02666-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clerzius G, Gelinas J-F, Daher A, Bonnet M, Meurs EF, Gatignol A. 2009. ADAR1 interacts with PKR during human immunodeficiency virus infection of lymphocytes and contributes to viral replication. J Virol 83:10119–10128. doi: 10.1128/JVI.02457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doria M, Neri F, Gallo A, Farace MG, Michienzi A. 2009. Editing of HIV-1 RNA by the double-stranded RNA deaminase ADAR1 stimulates viral infection. Nucleic Acids Res 37:5848–5858. doi: 10.1093/nar/gkp604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weiden MD, Hoshino S, Levy DN, Li Y, Kumar R, Burke SA, Dawson R, Hioe CE, Borkowsky W, Rom WN, Hoshino Y. 2014. Adenosine deaminase acting on RNA-1 (ADAR1) inhibits HIV-1 replication in human alveolar macrophages. PLoS One 9:e108476. doi: 10.1371/journal.pone.0108476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biswas N, Wang T, Ding M, Tumne A, Chen Y, Wang Q, Gupta P. 2012. ADAR1 is a novel multi targeted anti-HIV-1 cellular protein. Virology 422:265–277. doi: 10.1016/j.virol.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doria M, Tomaselli S, Neri F, Ciafrè SA, Farace MG, Michienzi A, Gallo A. 2011. ADAR2 editing enzyme is a novel human immunodeficiency virus-1 proviral factor. J Gen Virol 92:1228–1232. doi: 10.1099/vir.0.028043-0. [DOI] [PubMed] [Google Scholar]
- 39.Tenoever BR, Ng S-L, Chua MA, McWhirter SM, Garcia-Sastre A, Maniatis T. 2007. Multiple functions of the IKK-related kinase IKK in interferon-mediated antiviral immunity. Science 315:1274–1278. doi: 10.1126/science.1136567. [DOI] [PubMed] [Google Scholar]
- 40.Ward SV, George CX, Welch MJ, Liou L-Y, Hahm B, Lewicki H, de la Torre JC, Samuel CE, Oldstone MB. 2011. RNA editing enzyme adenosine deaminase is a restriction factor for controlling measles virus replication that also is required for embryogenesis. Proc Natl Acad Sci U S A 108:331–336. doi: 10.1073/pnas.1017241108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Chassey B, Aublin-Gex A, Ruggieri A, Meyniel-Schicklin L, Pradezynski F, Davoust N, Chantier T, Tafforeau L, Mangeot P-E, Ciancia C, Perrin-Cocon L, Bartenschlager R, André P, Lotteau V. 2013. The interactomes of influenza virus NS1 and NS2 proteins identify new host factors and provide insights for ADAR1 playing a supportive role in virus replication. PLoS Pathog 9:e1003440. doi: 10.1371/journal.ppat.1003440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomaselli S, Galeano F, Locatelli F, Gallo A. 2015. ADARs and the balance game between virus infection and innate immune cell response. Curr Issues Mol Biol 17:37–51. [PubMed] [Google Scholar]
- 43.Briese T, Schneemann A, Lewis AJ, Park YS, Kim S, Ludwig H, Lipkin WI. 1994. Genomic organization of Borna disease virus. Proc Natl Acad Sci U S A 91:4362–4366. doi: 10.1073/pnas.91.10.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ludwig H, Bode L, Gosztonyi G. 1988. Borna disease: a persistent virus infection of the central nervous system. Prog Med Virol 35:107–115. [PubMed] [Google Scholar]
- 45.Bilzer T, Planz O, Lipkin WI, Stitz L. 1995. Presence of CD4+ and CD8+ T cells and expression of MHC class I and MHC class II antigen in horses with Borna disease virus‐induced encephalitis. Brain Pathol 5:223–230. doi: 10.1111/j.1750-3639.1995.tb00598.x. [DOI] [PubMed] [Google Scholar]
- 46.Tomonaga K, Kobayashi T, Ikuta K. 2002. Molecular and cellular biology of Borna disease virus infection. Microbes Infect 4:491–500. doi: 10.1016/s1286-4579(02)01564-2. [DOI] [PubMed] [Google Scholar]
- 47.Ludwig H, Bode L. 2000. Borna disease virus: new aspects on infection, disease, diagnosis and epidemiology. Rev Sci Tech 19:259–288. doi: 10.20506/rst.19.1.1217. [DOI] [PubMed] [Google Scholar]
- 48.Matsumoto Y, Hayashi Y, Omori H, Honda T, Daito T, Horie M, Ikuta K, Fujino K, Nakamura S, Schneider U, Chase G, Yoshimori T, Schwemmle M, Tomonaga K. 2012. Bornavirus closely associates and segregates with host chromosomes to ensure persistent intranuclear infection. Cell Host Microbe 11:492–503. doi: 10.1016/j.chom.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 49.Martin A, Hoefs N, Tadewaldt J, Staeheli P, Schneider U. 2011. Genomic RNAs of Borna disease virus are elongated on internal template motifs after realignment of the 3′ termini. Proc Natl Acad Sci U S A 108:7206–7211. doi: 10.1073/pnas.1016759108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Habjan M, Andersson I, Klingström J, Schümann M, Martin A, Zimmermann P, Wagner V, Pichlmair A, Schneider U, Mühlberger E, Mirazimi A, Weber F. 2008. Processing of genome 5′ termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS One 3:e2032. doi: 10.1371/journal.pone.0002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Son K-N, Liang Z, Lipton HL. 2015. Double-stranded RNA is detected by immunofluorescence analysis in RNA and DNA virus infections, including those by negative-stranded RNA viruses. J Virol 89:9383–9392. doi: 10.1128/JVI.01299-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Higuchi M, Single FN, Köhler M, Sommer B, Sprengel R, Seeburg PH. 1993. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell 75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 53.Lai F, Drakas R, Nishikura K. 1995. Mutagenic analysis of double-stranded RNA adenosine deaminase, a candidate enzyme for RNA editing of glutamate-gated ion channel transcripts. J Biol Chem 270:17098–17105. doi: 10.1074/jbc.270.29.17098. [DOI] [PubMed] [Google Scholar]
- 54.Valente L, Nishikura K. 2007. RNA binding-independent dimerization of adenosine deaminases acting on RNA and dominant negative effects of nonfunctional subunits on dimer functions. J Biol Chem 282:16054–16061. doi: 10.1074/jbc.M611392200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stahl T, Mohr C, Kacza J, Reimers C, Pannicke T, Sauder C, Reichenbach A, Seeger J. 2003. Characterization of the acute immune response in the retina of Borna disease virus infected Lewis rats. J Neuroimmunol 137:67–78. doi: 10.1016/s0165-5728(03)00044-4. [DOI] [PubMed] [Google Scholar]
- 56.Shankar V, Kao M, Hamir AN, Sheng H, Koprowski H, Dietzschold B. 1992. Kinetics of virus spread and changes in levels of several cytokine mRNAs in the brain after intranasal infection of rats with Borna disease virus. J Virol 66:992–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li J, Yu M, Zheng W, Liu W, Li J, Yu M, Zheng W, Liu W. 2015. Nucleocytoplasmic shuttling of influenza A virus proteins. Viruses 7:2668–2682. doi: 10.3390/v7052668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pichlmair A, Schulz O, Tan CP, Näslund TI, Liljeström P, Weber F, Reis e Sousa C. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 59.Liu G, Park H-S, Pyo H-M, Liu Q, Zhou Y. 2015. Influenza A virus panhandle structure is directly involved in RIG-I activation and interferon induction. J Virol 89:6067–6079. doi: 10.1128/JVI.00232-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cattaneo R, Schmid A, Eschle D, Baczko K, ter Meulen V, Billeter MA. 1988. Biased hypermutation and other genetic changes in defective measles viruses in human brain infections. Cell 55:255–265. doi: 10.1016/0092-8674(88)90048-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suspène R, Petit V, Puyraimond-Zemmour D, Aynaud M-M, Henry M, Guetard D, Rusniok C, Wain-Hobson S, Vartanian J-P. 2011. Double-stranded RNA adenosine deaminase ADAR-1-induced hypermutated genomes among inactivated seasonal influenza and live attenuated measles virus vaccines. J Virol 85:2458–2462. doi: 10.1128/JVI.02138-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taylor DR, Puig M, Darnell MER, Mihalik K, Feinstone SM. 2005. New antiviral pathway that mediates hepatitis C virus replicon interferon sensitivity through ADAR1. J Virol 79:6291–6298. doi: 10.1128/JVI.79.10.6291-6298.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nishikura K, Yoo C, Kim U, Murray JM, Estes PA, Cash FE, Liebhaber SA. 1991. Substrate specificity of the dsRNA unwinding/modifying activity. EMBO J 10:3523–3532. doi: 10.1002/j.1460-2075.1991.tb04916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li JB, Levanon EY, Yoon JK, Aach J, Xie B, LeProust E, Zhang K, Gao Y, Church GM. 2009. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science 324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 65.Xiao Y, Dolan PT, Goldstein EF, Li M, Farkov M, Brodsky L, Andino R. 2017. Poliovirus intrahost evolution is required to overcome tissue-specific innate immune responses. Nat Commun 8:375. doi: 10.1038/s41467-017-00354-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schädler R, Diringer H, Ludwig H. 1985. Isolation and characterization of a 14500 molecular weight protein from brains and tissue cultures persistently infected with Borna disease virus. J Gen Virol 66:2479–2484. doi: 10.1099/0022-1317-66-11-2479. [DOI] [PubMed] [Google Scholar]
- 67.Pauli G, Ludwig H. 1985. Increase of virus yields and releases of Borna disease virus from persistently infected cells. Virus Res 2:29–33. doi: 10.1016/0168-1702(85)90057-7. [DOI] [PubMed] [Google Scholar]
- 68.Yanai M, Sakai M, Makino A, Tomonaga K. 2017. Dual function of the nuclear export signal of the Borna disease virus nucleoprotein in nuclear export activity and binding to viral phosphoprotein. Virol J 14:126. doi: 10.1186/s12985-017-0793-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wong SK, Lazinski DW. 2002. Replicating hepatitis delta virus RNA is edited in the nucleus by the small form of ADAR1. Proc Natl Acad Sci U S A 99:15118–15123. doi: 10.1073/pnas.232416799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fujino K, Yamamoto Y, Daito T, Makino A, Honda T, Tomonaga K. 2017. Generation of a non-transmissive Borna disease virus vector lacking both matrix and glycoprotein genes. Microbiol Immunol 61:380–386. doi: 10.1111/1348-0421.12505. [DOI] [PubMed] [Google Scholar]
- 71.Breitling R, Armengaud P, Amtmann A, Herzyk P. 2004. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 72.Huang DW, Sherman BT, Lempicki RA. 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 74.Briese T, de la Torre JC, Lewis A, Ludwig H, Lipkin WI. 1992. Borna disease virus, a negative-strand RNA virus, transcribes in the nucleus of infected cells. Proc Natl Acad Sci U S A 89:11486–11489. doi: 10.1073/pnas.89.23.11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hirai Y, Domai E, Yoshikawa Y, Okamura H, Makino A, Tomonaga K. 2019. Intracellular dynamics of actin affects Borna disease virus replication in the nucleus. Virus Res 264:179–183. doi: 10.1016/j.virusres.2019.02.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We stored the microarray data in the NCBI Gene Expression Omnibus (accession number GSE138927).