The study shows that extensive replication of an RNA virus in a constant biological environment does not limit exploration of sequence space and adaptive options. There was no convergence toward a restricted set of adapted genomes. Mutational waves and mutant spectrum broadening affected infectious genomes. Therefore, profound modifications of mutant spectrum composition and consensus sequence diversification are not exclusively dependent on environmental alterations or the intervention of population bottlenecks.
KEYWORDS: RNA virus evolution, viral quasispecies, sequence space, adaptation, mutant spectrum, mutational waves
ABSTRACT
Previous studies documented that long-term hepatitis C virus (HCV) replication in human hepatoma Huh-7.5 cells resulted in viral fitness gain, expansion of the mutant spectrum, and several phenotypic alterations. In the present work, we show that mutational waves (changes in frequency of individual mutations) occurred continuously and became more prominent as the virus gained fitness. They were accompanied by an increasing proportion of heterogeneous genomic sites that affected 1 position in the initial HCV population and 19 and 69 positions at passages 100 and 200, respectively. Analysis of biological clones of HCV showed that these dynamic events affected infectious genomes, since part of the fluctuating mutations became incorporated into viable genomes. While 17 mutations were scored in 3 biological clones isolated from the initial population, the number reached 72 in 3 biological clones from the population at passage 200. Biological clones differed in their responses to antiviral inhibitors, indicating a phenotypic impact of viral dynamics. Thus, HCV adaptation to a specific constant environment (cell culture without external influences) broadens the mutant repertoire and does not focus the population toward a limited number of dominant genomes. A retrospective examination of mutant spectra of foot-and-mouth disease virus passaged in cell cultures suggests a parallel behavior here described for HCV. We propose that virus diversification in a constant environment has its basis in the availability of multiple alternative mutational pathways for fitness gain. This mechanism of broad diversification should also apply to other replicative systems characterized by high mutation rates and large population sizes.
IMPORTANCE The study shows that extensive replication of an RNA virus in a constant biological environment does not limit exploration of sequence space and adaptive options. There was no convergence toward a restricted set of adapted genomes. Mutational waves and mutant spectrum broadening affected infectious genomes. Therefore, profound modifications of mutant spectrum composition and consensus sequence diversification are not exclusively dependent on environmental alterations or the intervention of population bottlenecks.
INTRODUCTION
Quasispecies dynamics is a key determinant of virus adaptability to changing environments. High mutation rates and large population sizes convert viral populations in ample reservoirs of continuously changing mutant spectra, also termed mutant swarms or mutant clouds, that include phenotypic variants among their minority components (a recent overview is given in reference 1). The involvement of quasispecies dynamics in virus adaptation has often been approached by determining fitness variation of the viral population in response to gradual or abrupt environmental changes, in some cases with conflicting results (for examples, see references 2 and 3 and references quoted therein). When the underlying variations in mutant spectrum composition have been studied, the results point to virus adaptation being associated with the replacement of mutant clouds by others, with a critical contribution of high mutation rates and intramutant spectrum interactions among genomes or their expression products (4–14).
We are interested in understanding the molecular basis of the quasispecies dynamics of hepatitis C virus (HCV) due to its ranking among the most variable pathogens and the several implications of its genetic variation for clinical practice. Indeed, quasispecies features of HCV in vivo have been related to disease progression, response to antiviral agents, and treatment failures through selection of inhibitor-escape mutants (15–21). The availability of cell culture systems for the sustained multiplication of HCV (22–25), rendered genome variation, and adaptive mechanisms of this important human pathogen are amenable to experimental analysis under controlled (constant or changing) environments.
In a previous study on genotypic and phenotypic HCV evolution, we subjected HCV rescued from a molecular clone encoded in plasmid Jc1FLAG2(p7-nsGluc2A) (26) to 100 serial infections of human hepatoma Huh-7.5 cells in the absence or presence of different amounts of interferon alpha (IFN-α) (27). We obtained IFN-α-resistant populations associated with alternative mutations at multiple genomic sites (27). Unexpectedly, IFN-α resistance was also observed in parallel lineages of HCV passaged in the absence of IFN-α. This result was extended to show resistance of the same HCV populations to additional antiviral agents and led to the conclusion that a multidrug resistance phenotype was acquired by HCV while gaining fitness in Huh-7.5 cells (27–30). The observation alerted us to a remarkable change in virus population composition and behavior only as a consequence of monotonous replication in cell culture without perturbations derived from external selective constraints or bottleneck events. When infection in the absence of any drug was extended to 200 passages (equivalent to about 700 days of continuous virus replication), the viral population did not approach a steady mutant spectrum, since comparison of the populations at passages 45, 100, and 200 revealed variations in mutant frequency that we termed mutational waves (defined as individual mutations whose frequency increased or decreased between the populations under comparison) (30). This observation raised two questions: (i) how soon in the course of HCV passaging can mutational waves be detected, and (ii) what is their biological role in the diversification of viable HCV genomes. The latter is a relevant issue given the generation of defective HCV genomes during virus multiplication (31, 32) and their effective propagation in Huh-7.5 cells (33).
Here, we provide evidence that the process of HCV adaptation to Huh-7.5 cells results in a remarkable increase of mutant spectrum amplitude (average number of mutations per genome), with mutational waves discernible even between successive passages. Adaptation was also associated with a diversification of the consensus sequence that includes viable genomes with altered phenotypes. The results imply that replication conditions that could favor a focused selection of limited sets of genomes adapted to a specific cellular environment may have the opposite effect of generating a large repertoire of divergent sequences. We consider two main possibilities to explain this class of viral genome diversification: host cell heterogeneity as a fine-grained environment that provides a mosaic of nonidentical selective constraints in cell culture experiments, and availability of multiple adaptive pathways even in a constant cellular environment, with critical dependence on HCV high mutation rates and large population sizes.
RESULTS
Time frame of mutational waves.
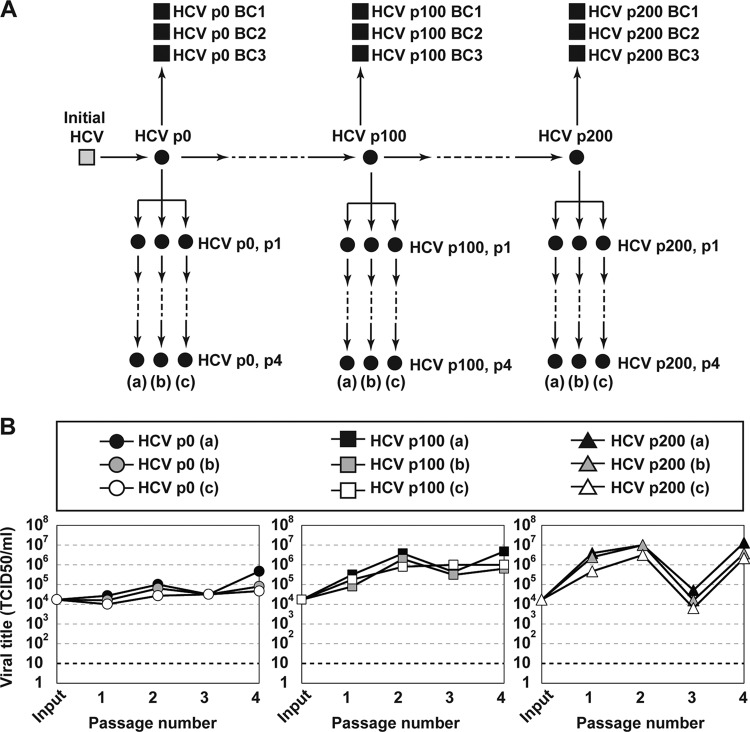
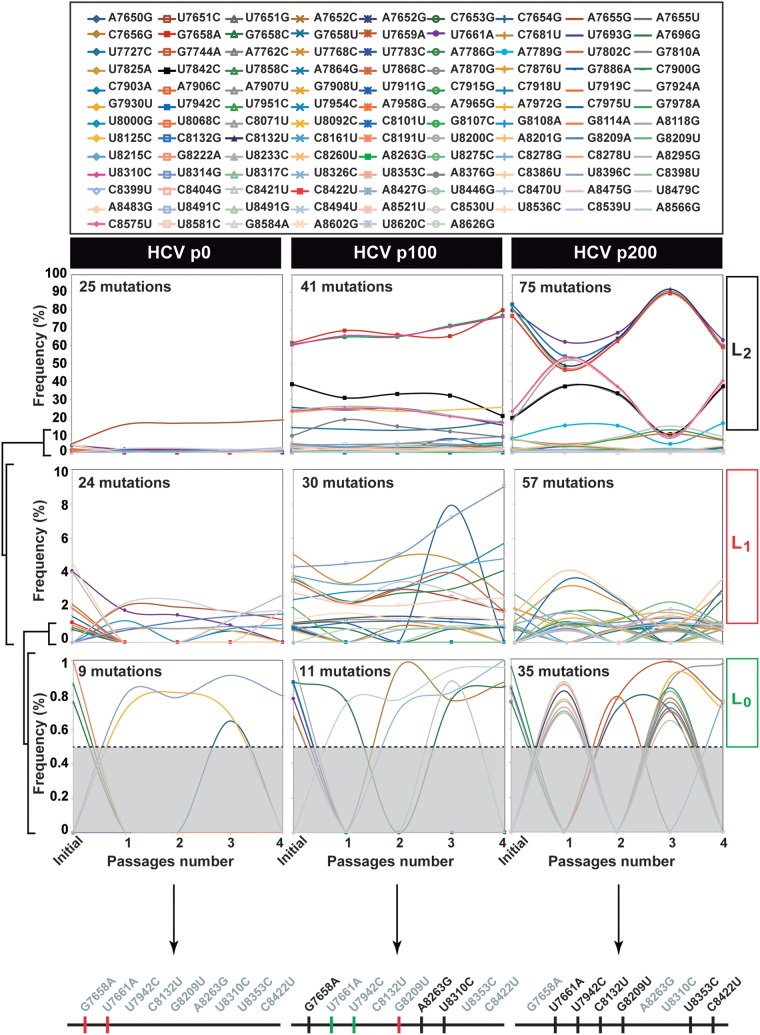
To investigate if changes of mutant frequencies (mutational waves) were observed in successive infections in human hepatoma cells, triplicate populations of HCV p0, HCV p100, and HCV p200 (p indicates passage number, and p0 refers to the initial population obtained as described in reference 27) were subjected to four passages in Huh-7.5 cells (Fig. 1). The mutant spectra of the triplicate series [termed (a), (b), (c)] were analyzed by deep sequencing of three NS5B amplicons, and the mutations were ranked according to their change in frequency (Fig. 2 and Fig. S1 and S2 at http://babia.cbm.uam.es/~lab121/SupplMatGallego3). Mutational waves were observed even in successive passages of each population, and the three replicas exhibited similar dynamic behavior, albeit with differences in some of the participating mutations. Taking replica (a) for a description of mutant spectrum dynamics, the total number of mutations (counted relative to Jc1FLAG2[p7-nsGluc2A]) identified in the course of the four passages was 25, 41, and 70 for HCV p0, HCV p100, and HCV p200, respectively (Table S1 [http://babia.cbm.uam.es/~lab121/SupplMatGallego3]). The majority of them (100% for HCV p0, all replicas; 93% to 95% for HCV p100, all replicas; 100% for HCV p200, all replicas) exhibited a read frequency that varied by more than 0.5% (the cutoff-validated mutant frequency) in any of the four passages. Mutations with such read frequency variations are defined as participating in mutational waves (Fig. 2 and Table S1 at http://babia.cbm.uam.es/~lab121/SupplMatGallego3). For each population, mutations that participated in waves were divided into three levels: level L0 (mutations whose mutant frequency did not exceed 1% but that, at some passage, attained a frequency above the 0.5% cutoff value), level L1 (mutations whose mutant frequency did not exceed 10%), and level L2 (mutations whose mutant frequency reached, at some passage, a frequency higher than 10%) (Fig. 2). Replicas (b) and (c) exhibited a parallel behavior to replica (a) regarding the number of mutations that participated in mutational waves and their level distribution (Fig. 2; Fig. S1 and S2 and Table S1, S2, and S3 at http://babia.cbm.uam.es/~lab121/SupplMatGallego3).
FIG 1.
Experimental scheme and infectious progeny production of HCV populations. (A) Passage history and origin of HCV populations (filled circles) and biological clones (filled squares) used in the present study. The initial HCV (gray square) was derived by transcription of plasmid Jc1FLAG2(p7-nsGluc2A). Virus passages are indicated by arrows; p denotes passage number; and (a), (b), and (c) indicate three replicate passages. (B) Extracellular infectious viral titer produced by the triplicate populations shown in the box above the panels. Procedures and infection conditions are detailed in Materials and Methods.
FIG 2.
Mutant spectrum dynamics of HCV p0, HCV p100, and HCV p200 during four passages in Huh-7.5 reporter cells. Frequency variation of mutations (color and symbol code in the upper box) identified by deep sequencing of amplicons from the NS5B-coding region (residues 7649 to 8653). The results correspond to replica (a) of Fig. 1. Genomic residues are numbered according to HCV reference isolate JFH-1 (GenBank accession number AB047639). Color lines join the mutation frequency values determined for the initial virus (described in reference 80) and subsequent passages 1 to 4. Mutational waves have been divided into levels L0 (with the cutoff frequency limit of 0.5% delimited by the discontinuous horizontal lines), L1, and L2, according to the mutant frequency range indicated in ordinates. For completeness, at each level, mutations belonging to lower levels are maintained. The three thick horizontal lines below the frequency panels represent the consensus sequence of the region covered by the NS5B amplicons of each virus; mutations written in black or gray on top of the lines correspond to the mutations present or absent, respectively, in the consensus sequence of each population; and the color of the vertical lines represents the level at which the mutations belong: green, L0; red, L1; black, L2. Deep sequencing procedures and data processing are described in Materials and Methods.
The level distinction revealed an increase of L1 and L2 mutations in HCV p100 and HCV p200 relative to HCV p0. The average wave amplitude between HCV passages 200 and 204 was significantly larger than between passages 100 to 104 for the 15 mutations shared by HCV p100 and HCV p200 (a 5.2-fold difference, P = 0.0039; t test); similar results were obtained for replicas (b) and (c) (Fig. S3 [http://babia.cbm.uam.es/~lab121/SupplMatGallego3]). The total number of mutations found in HCV p0, HCV p100, and HCV p200 was 141, 131, and 143 for replicas (a), (b), and (c), respectively (compare Fig. 2 and Fig. S1 and S2 at http://babia.cbm.uam.es/~lab121/SupplMatGallego3); the number of different mutations (counting each mutation only once despite being present in different haplotypes) was 114, 108, and 110, respectively. Of these, 87 were found in the 3 replicas and 13 in 2 replicas, while 23, 9, and 13 mutations were present only in replicas (a), (b), and (c), respectively. When mutations common to HCV p100 and HCV p200 were compared, 47%, 50%, and 42% of them in replicas (a), (b), and (c), respectively, changed significantly in frequency between the two populations (Fig. S3 [http://babia.cbm.uam.es/~lab121/SupplMatGallego3]). Of the different mutations that participated in waves along the virus passages (average of the three replicas), 35% were nonsynonymous, and 65% were synonymous; for mutations in level L2 waves, the corresponding values were 59% and 41%, respectively. The possible significance of these differences is under study. Thus, dynamics of mutant frequency change occurs in successive HCV passages in human hepatoma cells, and wave amplitudes were larger for high-fitness HCV.
Heterogeneity points in the consensus genomic sequences.
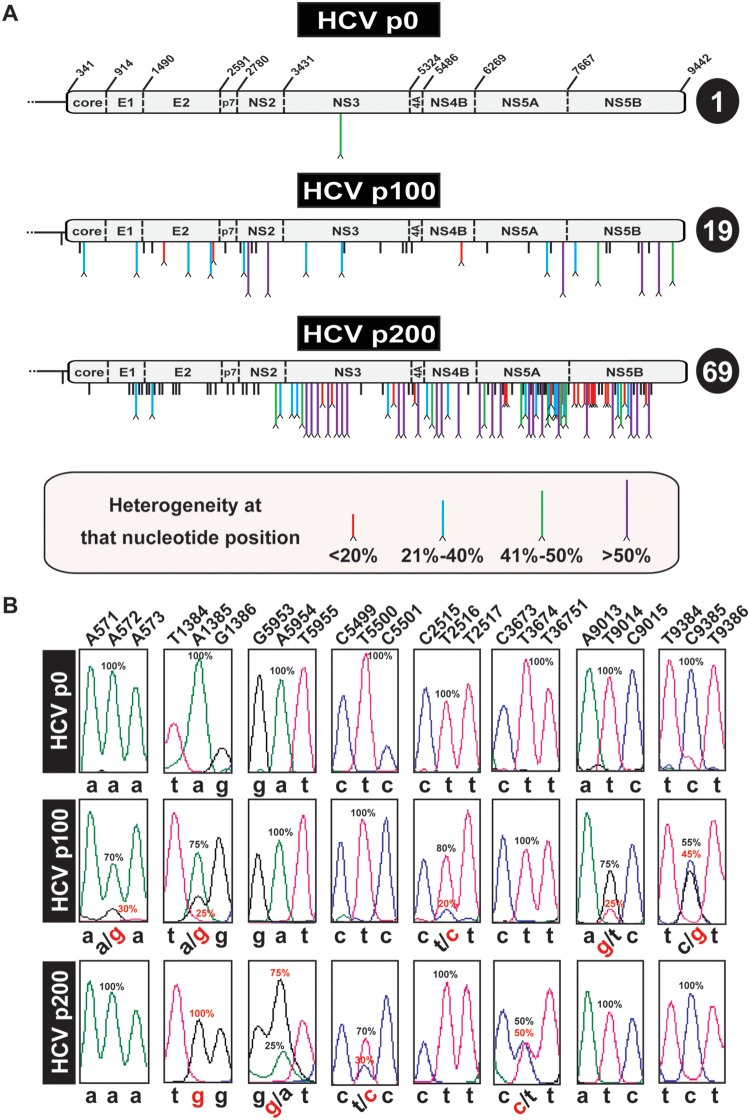
To investigate if the amplitude of molecular waves found in the NS5B gene was associated with multiple dominant or nearly dominant genomes in the high-fitness HCV populations, the number of heterogeneity sites in the complete consensus sequences previously determined for HCV p0, HCV p100, and HCV p200 using real-time PCR (RT-PCR) amplification and Sanger sequencing (30) was reevaluated. Each genomic site where two peaks were present in the sequence pattern was ranked according to the frequency level of the mutant nucleotide in the mixture, expressed as a percentage. The results show a remarkable increase in the number of heterogeneous genomic positions in HCV p100 and HCV p200 with respect to HCV p0 at all frequency levels (Fig. 3). For HCV p200, a significantly larger number of heterogeneity points was noted in the nonstructural protein-coding than in the structural protein-coding region (P = 9.2 × 10−7; proportion test); no such difference was seen for HCV p100 (P = 0.356; proportion test), and the increase in the number of heterogeneous sites at the nonstructural protein-coding region upon evolution of HCV p100 to HCV p200 was also significant (P = 0.017; proportion test). Thus, multiple dominant or nearly dominant individual mutations were present in HCV p100 but not in HCV p0, and the number increased 3.6-fold from HCV p100 to HCV p200.
FIG 3.
Heterogeneities in the consensus sequence of HCV p0, HCV p100, and HCV p200. (A) Schematic of the HCV p0, HCV p100, and HCV p200 genomes. In HCV p0, the RNA residue numbers (reference isolate, JFH-1) that delimit the encoded proteins are indicated. Mutations (relative to the HCV sequence in plasmid Jc1FLAG2[p7-nsGluc2A]) are identified by vertical lines below the genome. Mutations at sites with no evidence of heterogeneity are indicated by short black lines. A caret at the tip of a line represents heterogeneity at that nucleotide position (two peaks in the same sequence position). The percentage range (length and color code given in the box at the bottom) indicates the proportion of the mutant in the chromatogram sequence, estimated by triangulation, as explained in Materials and Methods. The total number of heterogeneity points per genome is indicated in the circle at the right of each genome. (B) Examples of nucleotide positions (shown at the top) where heterogeneities have been found. For simplicity, below each panel, the nucleotides are written in lowercase letters; heterogeneities are depicted by two nucleotides separated by a slash. Frequencies (percentage) are written next to the relevant peaks.
To examine the stability of genome sites displaying heterogeneity, the complete consensus nucleotide sequence of HCV p0, p4; HCV p100, p4; and HCV p200, p4 was determined by Sanger sequencing. The results show that 0%, 61%, and 72% of the heterogeneous genomic sites present in the parental HCV p0, HCV p100, and HCV p200 populations, respectively, were maintained after the four passages. In addition, there were sites at which the heterogeneity was lost (100%, 18.4%, and 17.8%, respectively) or gained (0%, 20.4%, and 10.1%, respectively) (Fig. S4 [http://babia.cbm.uam.es/~lab121/SupplMatGallego3]). Therefore, despite dynamics of altered mutation dominances, the prevalence of genome heterogeneity sites in high-fitness HCV was maintained upon further viral replication.
Sequence divergence among molecular clones.
The increased genetic heterogeneity of high-fitness HCV anticipated a higher divergence among molecular clones sampled from the corresponding populations. This prediction was confirmed by Sanger sequencing of the NS5A-coding region of 27, 29, and 30 molecular clones from HCV p0, HCV p100, and HCV p200, respectively. The percentage of NS5A sites without mutations (counted relative to the consensus of HCV p0) was 98%, 94.3%, and 90.3% for HCV p0, HCV p100, and HCV p200, respectively (Fig. S4 and Table S6 [http://babia.cbm.uam.es/~lab121/SupplMatGallego3]); these differences are all highly significant (P = 4.1 × 10−7 for HCV p0 versus HCV p100; P < 2.2 × 10−16 for HCV p0 versus HCV p200; and P = 3.5 × 10−5 for HCV p100 versus HCV p200; proportion test). The number of mutations that are repeated in more than 50% of the clones, and that consequently are responsible for a modification of the consensus sequence, amount to 0%, 7.6%, and 13.9% of the sites with mutations in HCV p0, HCV p100, and HCV p200, respectively. The divergence among clones from HCV p0 represented a minimal mutation frequency (Mfmin) of 7.4 × 10−4 substitutions per nucleotide (s/nt) and a maximum mutation frequency (Mfmax) of 7.9 × 10−4 s/nt; the corresponding values for HCV p100 were Mfmin = 1.8 × 10−3 s/nt, and Mfmax = 5.0 × 10−3 s/nt, and the values for HCV p200 were Mfmin = 2.8 × 10−3 s/nt, and Mfmax = 5.2 × 10−3 s/nt. The increase reached statistical significance (P = 5.2 × 10−12 and P < 2.2 × 10−16 for the increase of Mfmin and Mfmax between HCV p0 and HCV p200, respectively; proportion test).
Sequence divergence among biological clones.
Genome heterogeneities quantified in consensus sequences or in molecular clones are not necessarily present in infectious genomes. Defective genomes—defined as those that in isolation cannot produce infectious progeny—are increasingly recognized as present in RNA virus populations, including HCV, with multiple biological implications (34–36). To ascertain that the mutant spectrum broadening affected viable HCV genomes, three biological clones (BC1, BC2, and BC3) were isolated from each HCV p0 and HCV p200 population, minimally amplified in Huh-7.5 cells (from the initial focus on the monolayer to about 1.4 ± 0.1 × 104 50% tissue culture infective dose [TCID50] for the HCV p0 clones and 1.5 ± 1.5 × 107 TCID50 for the HCV p200 clones), and their complete genomic nucleotide sequence was determined by RT-PCR amplification and Sanger sequencing (Fig. 4 and 5 and Table S7 and S8 at http://babia.cbm.uam.es/~lab121/SupplMatGallego3).
FIG 4.
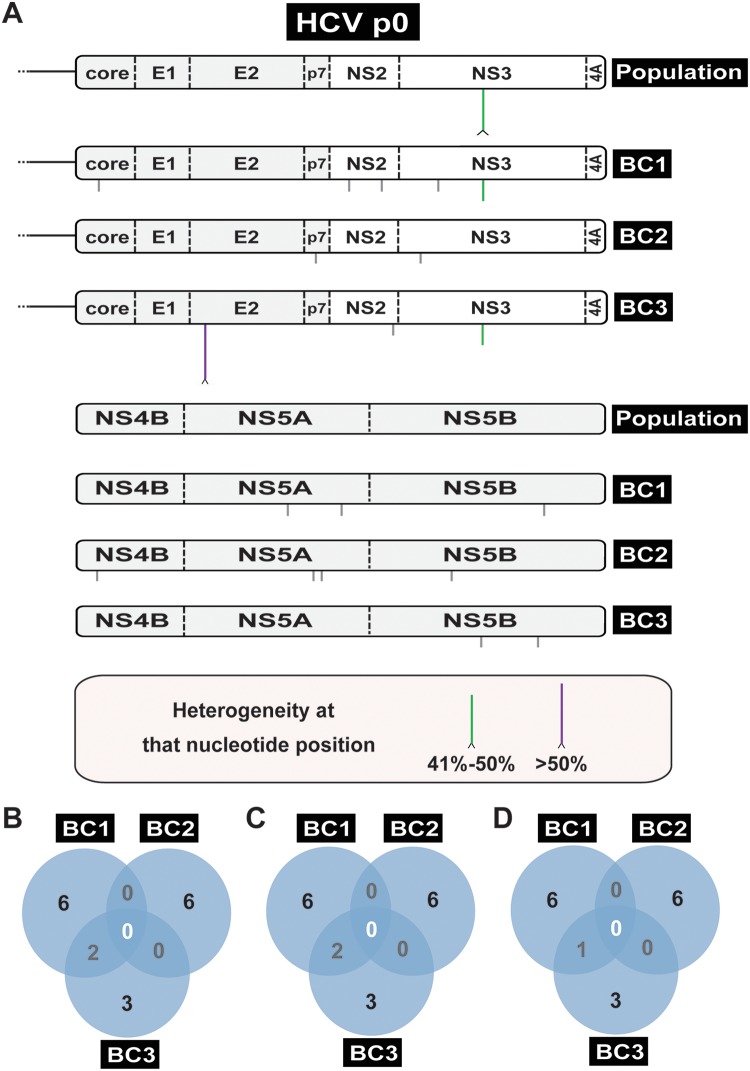
Consensus genomic sequence of the HCV p0 population and three biological clones isolated from the population. (A) Schematic of the HCV genome divided into two parts, with the genome under study indicated on the right (population is the uncloned HCV p0; BC1, BC2, and BC3 are three biological clones from HCV p0). A caret at the tip of a line represents heterogeneity at that nucleotide position (two peaks in the same sequence position, with the frequency range given in the bottom box). The NS3 mutation indicated by a vertical green line present in BC1 and BC3 is the result of segregation of a nucleotide mixture in the uncloned population. Mutations in the biological clones that were not detected in the corresponding parental population are indicated by vertical gray lines below the genome. (B) Venn diagram of mutations (counted relative to the plasmid Jc1FLAG2[p7-nsGluc2A] sequence) that are shared among the biological clones. (C) Diagram of mutations (counted relative to the consensus sequence of HCV p0 population) that are shared among the biological clones. (D) Diagram indicating new mutations (those that were not found in the parental HCV p0 population) that are shared among the biological clones.
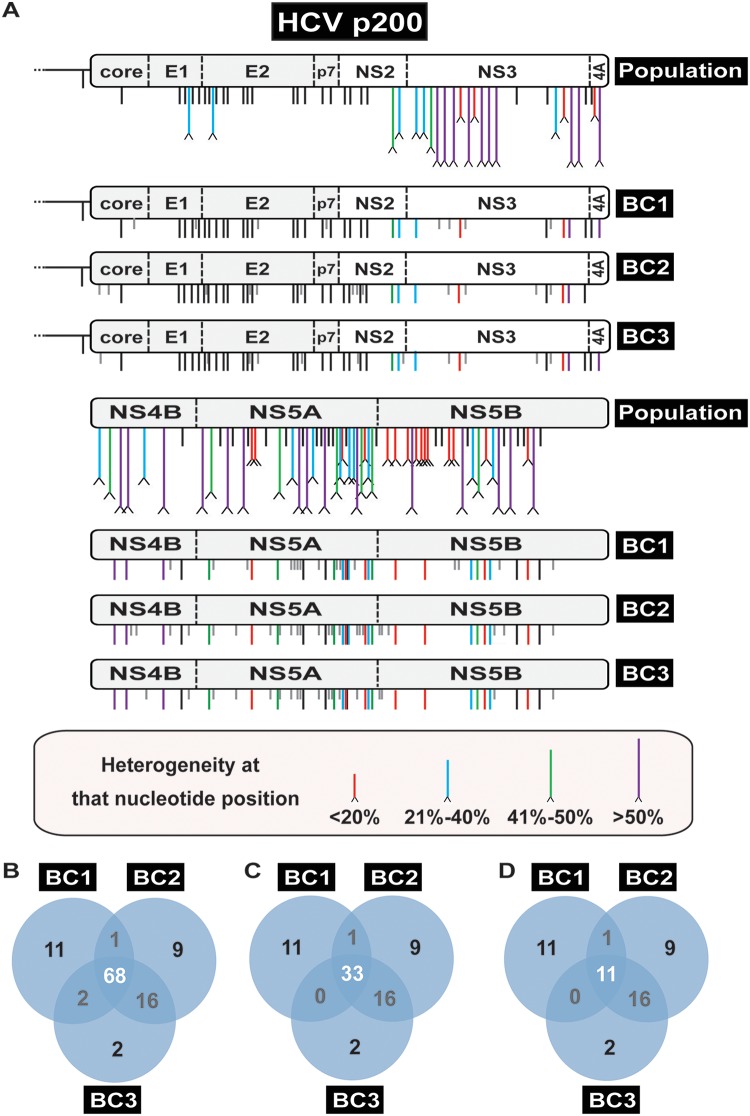
FIG 5.
Comparison of the consensus sequence of the HCV p200 population and three biological clones isolated from the population. (A) Schematic of the HCV genome divided into two parts, with the genome under study indicated on the right (population is the uncloned HCV p200; BC1, BC2, BC3 are three biological clones from HCV p200). Mutations relative to the sequence of Jc1FLAG2(p7-nsGluc2A) are indicated by vertical black lines below the genome; mutations in the biological clones that were not detected in the corresponding parental population are indicated by shorter vertical gray lines. A caret at the tip of a line represents heterogeneity at that nucleotide position (two peaks in the same sequence position, with the frequency range given in the bottom box). Some points of heterogeneity yield mutations in individual biological clones. (B) Venn diagram of mutations (counted relative to the plasmid Jc1FLAG2[p7-nsGluc2A] sequence) that are shared among the biological clones. (C) Diagram of mutations (counted relative to the consensus sequence of HCV p200 population) that are shared among the biological clones. (D) Diagram indicating new mutations (those that were not found in the parental HCV p200 population) that are shared among the biological clones.
Comparison of the number of mutations and their distribution among the three biological clones derived from HCV p0 and the three clones derived from HCV p200 revealed significant differences. The total number of mutations (counted relative to the genomic HCV sequence in plasmid Jc1FLAG2[p7-nsGluc2A] [26]) found in the three biological clones from HCV p0 was 17; of those, 16 were not present in the parental HCV p0 population, some were unique to one clone, and others were shared by two clones (Fig. 4). In contrast, the total number of mutations found in the HCV p200 biological clones reached 72, and of those, 50 were not present in the parental HCV p200 population, some were unique to one clone, and others were present in two or three clones (Fig. 5). The eight mutations shared by the three biological clones from HCV p200 participated in level L2 mutational waves, with identical results in replicas (a), (b), and (c). In addition, in replica (c), a mutation that fluctuated al level L0 was present in one of the biological clones from HCV p200 (see Fig. S2 and Table S3 and S8 the URL mentioned above). Five and 28 points of heterogeneity present in HCV p100 and HCV p200, respectively, contributed mutations that were fixed in the three biological clones from HCV p200. As expected, the only heterogeneity found in the population of HCV p0 was segregated in the biological clones from HCV p0.
Mfmin and Mfmax, calculated for the entire genome, were 6.1 × 10−4 s/nt and 6.7 × 10−4 s/nt, respectively, for clones derived from HCV p0, and 3.9 × 10−3 s/nt and 9.4 × 10−3 s/nt, respectively, for clones derived from HCV p200. These comparisons evidence a broadening of the mutant spectrum among viable genomes. The Mfmin and Mfmax values for the NS5A-coding region were slightly larger for biological clones than for molecular clones (Table S9 [http://babia.cbm.uam.es/~lab121/SupplMatGallego3]), indicating that the mutant spectrum broadening in the evolution from HCV p0 to HCV p200 involved viable genomes.
Phenotypic differences among populations and clones.
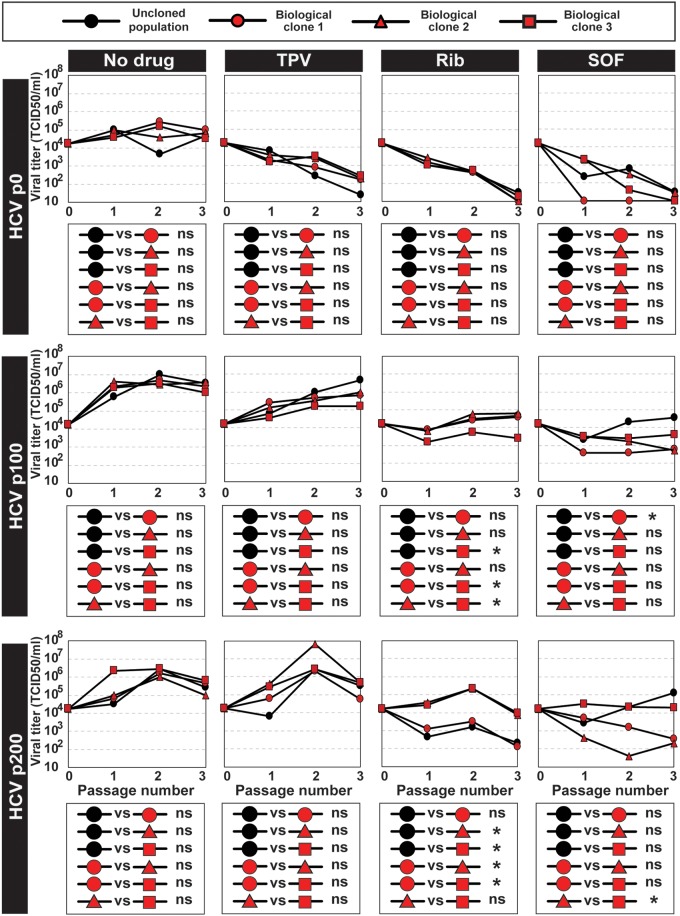
HCV fitness is a determinant of drug resistance (27, 28, 33). To explore if a phenotypic difference could be identified among biological clones, the responses of molecular clones derived from HCV p0, from HCV p100, and from HCV p200 to telaprevir, ribavirin, and sofosbuvir were determined. Significant differences were quantified between some clones within a population and between some populations and biological clones (Fig. 6). Examination of amino acid substitutions in NS5B of HCV populations and biological clones (Table S4, S7, and S8 [http://babia.cbm.uam.es/~lab121/SupplMatGallego3]) does not provide an interpretation of differences in drug sensitivity; such differences might relate to changes elsewhere in the genome or to fitness differences, as previously documented for uncloned HCV populations (28, 29). The mutational dynamics of HCV and the diversification of the mutant spectrum affected infectious viral genomes and entailed at least a phenotypic variation.
FIG 6.
Response of biological clones to antiviral agents. The origin of the populations and biological clones is depicted in Fig. 1. Infectious progeny production upon infection of Huh-7.5 reporter cells with HCV p0, HCV p100, HCV p200, or the three biological clones derived from each population (indicated on the left of the panel groups, with symbols given at the top box) at an MOI of 0.03 TCID50/cell, in the absence of any drug (No drug) or the presence of 600 nM telaprevir (TPV), 100 μM ribavirin (Rib), or 800 nM sofosbuvir (SOF). The statistical significance of pairwise differences is given in the boxes below the corresponding virus progeny production plots; they are based on the three progeny titer values using the ANOVA test (*, P < 0.05; ns, not significant). Procedures for infection in the absence and presence of drugs are given in Materials and Methods.
DISCUSSION
Environmental modifications generally alter the composition of mutant spectra of viral populations toward genomes that can improve survival in the new environment (see references 6 to 8, 12, and 37, to ,39, among other studies). Prolonged viral replication in the same noncoevolving cellular environment, as is the case with our experimental design with HCV, might have focused the mutant spectrum toward a subset of viral genomes that best multiply in such a cellular environment as a result of stabilizing selection (40, 41). However, the genomic analyses reported here, based on standard molecular cloning and Sanger sequencing, as well as on deep sequencing, denote an expanding mutant spectrum as the virus gained fitness, achieved through continuous mutational waves. Mutant spectrum expansion was such that each of the NS5A molecular clones analyzed from HCV p100 and HCV p200 exhibited a unique constellation of mutations. A total of 508 different mutations have been scored in all the analyses reported here, and we are currently analyzing their presence in HCV genomic sequences compiled in data banks.
Neither mutational waves nor coexistence of alternative dominant mutations can be attributed to defective genomes known to be present in HCV populations (35) because genomic divergence affected biological clones similarly to molecular clones sampled from the same populations. Our approach to analyzing molecular and biological clones solved one of the deficiencies of deep sequencing when applied to bulk RNA isolated from viral populations: the incapacity to distinguish mutations belonging to defective genomes from those present in viable genomes. In addition, the analysis of biological clones permitted the assignment of a constellation of mutations to the same HCV genomic molecule. This partially solved the problem of mutation linkage that could not be established for other mutations that participated in mutational waves whose quantification is based on short genomic reads. Noticeably, mutant spectrum expansion is not the only possible outcome of RNA virus replication; a mutant spectrum compression was previously observed in foot-and-mouth disease virus (FMDV) under ribavirin treatment (42).
The diversifying selection of HCV in human hepatoma cells described here is different from other types of positive selection such as stabilizing, directional, or disruptive selection, previously described in general genetics (40, 41, 43). Two main nonexclusive possibilities could explain the observed HCV diversification: (i) the Huh-7.5 cells may provide a nonevolving but heterogeneous environment, inherent to transformed cells in culture (44–46), in such a way that the individual nonidentical cellular environments may favor replication of different HCV variant sets; environmental heterogeneity in the cell culture could also arise from nutrient depletion, pH decrease, waste product accumulation, or differences in the cell cycle stage of individuals cells; and (ii) even if cellular heterogeneity were not relevant to HCV replication parameters, the virus may be able to explore multiple mutational pathways to reach comparable fitness levels in the Huh-7.5 environment. As the exploration of sequence space increases with passage number, additional mutational pathways are open to scrutiny for fitness gain. An increased choice of mutational pathways can have an effect akin to relaxation of purifying selection. The possibility of a virus to follow multiple pathways for fitness gain has been previously documented with other virus-host systems (47–49). We favor a more important contribution of alternative mutational pathways [model (ii)] than of cellular heterogeneity [model (i)] for the observed HCV diversification. The main reason is that both mutational waves and points of genome heterogeneity were far more prominent during the infections with HCV p100 and HCV p200 than with HCV p0. As yet another possibility, population bottlenecks may be also considered as promoters of virus diversification. However, bottlenecks alone cannot explain mutational waves in our experiments because bottleneck intensity did not increase from early to late passages (see Materials and Methods). We cannot exclude participation of HCV recombination in the observed wave dynamics, genome diversification, and fitness gain; examination of the possible involvement of recombination would require comparative analyses of longer sequence reads derived from the HCV populations at different passages.
Retrospective examination of consensus genomic sequences of FMDV subjected to 240 serial passages in BHK-21 cells, which eventually resulted in FMDV genome segmentation (50–52), revealed 0, 1, 11, and 9 genomic sites of heterogeneity (equivalent to the double nucleotide peaks described here for HCV) at passages 0, 100, 200, and 260, respectively (derived from the sequences published in reference 53). Thus, there is evidence of broad diversification in a constant cellular environment at least for another virus-host system.
Additional observations previously made with HCV are also consistent with mutant spectrum broadening as a source of viral adaptability: (i) IFN-α resistance was associated with alternative mutations in different genomic regions (27); (ii) for IFN-free, direct-acting antiviral (DAA) treatments, alternative combinations of mutations have been attributed to resistance to the same inhibitor combination (19, 20, 54); (iii) there is no evidence that long-term adaptation of HCV to a specific human liver decreases the probability of reinfection of the grafted liver following liver transplantation (55); and (iv) the potential phenotypic flexibility associated with sequence space exploration was illustrated by the selection of HCV mutations that permitted viral replication in the absence of the microRNA miR-122 (56). The different responses of some biological clones to anti-HCV inhibitors described here adds to previously reported phenotypic alterations (increased cytopathology, virion density, and capacity to shut off host cell protein synthesis) that accompanied HCV adaptation to the Huh-7.5 cells (30).
In contrast to multiple pathways for fitness gain, other studies involving different viruses have documented the acquisition of identical mutations during adaptive processes in parallel viral lineages (57–65). Parallel genome modifications as a repeatable and predictable trait in viral evolution (66) appear contrary to the proposals of broadly diversifying selection in a constant cellular environment reported here with HCV. To reconcile the two groups of evidence, it should be considered that the nature of the selective constraint and the population size of the virus that participates in the response to the constraint may influence the number of alternative evolutionary outcomes. Also, the nucleotide sequencing methodology at the time of some of the early reported studies may have restrained the identification of possible nearly dominant sequences in the relevant populations in the absence of clonal analyses.
According to our evidence and models, broad diversification should operate in biological systems characterized by high effective population sizes and elevated mutation rates, and it implies that environmental constancy does not impair prospects for adaptation. Such requirements are not exclusive to viruses; they are, at least, also fulfilled by cancer cells in large tumors (67), as well as in large microbial populations, in particular by their mutator versions (68). For multicellular organisms with their usual population sizes, low mutation rates, and characteristic functional constraints, broad diversification is not attainable. The standard diversifying selection depicted as a single fitness peak evolving toward two or a few peaks (42, 43) would be the counterpart for differentiated organisms of the broadly diversifying selection characterized in our study.
MATERIALS AND METHODS
Cells, viruses, and infections.
The origin of Huh-7.5, Huh-7-Lunet, and Huh-7.5 reporter cell lines and procedures for cell growth in Dulbecco’s modified Eagle’s medium (DMEM) have been previously described (27, 69, 70); cells were cultured at 37°C and 5% CO2. Huh-7.5 cells were used for titration of virus infectivity, while Huh-7.5 reporter cells were used for standard infections and serial passages of HCV. Cells were periodically thawed from a large frozen stock and passaged a maximum of 30 times at a split ratio of 1:3 before use in the experiments.
The initial HCV was rescued from plasmid Jc1FLAG2(p7-nsGluc2A) (a chimera of J6 and JFH-1 from genotype 2a) and amplified to yield HCV p0 as described (26), and from plasmid GNNFLAG2(p7-nsGluc2A), termed GNN (which carries a mutation in the NS5B RNA-dependent RNA polymerase that renders the virus replication defective) (26). GNN was used as a negative infection control. HCV p100 and HCV p200 resulted from population HCV p0 passaged 100 and 200 times, respectively, in Huh-7.5 reporter cells, as described (28, 30). To control for the absence of contamination, the supernatants of mock-infected cells, which were maintained in parallel with the infected cultures, were titrated; no infectivity in the mock-infected or GNN-infected cultures was detected in any of the experiments.
Biological clones were isolated from HCV populations by limiting dilution followed by growth in Huh 7.5 cells. Briefly, HCV p0, HCV p100, and HCV p200 populations were treated with sodium deoxycholate (0.01%) for 10 min at room temperature to minimize particle aggregation (71). Dilutions (10−3 to 10−7) were used to infect Huh-7.5 cells in 96-well plates; at 72 h postinfection, supernatants were collected, and the infected cells were visualized by immunostaining with an anti-NS5A antibody (9E10) (22). Viruses collected from wells containing a single infected cell focus were passaged twice on fresh Huh-7.5 cells, yielding clonal populations; each clonal preparation was then subjected to another round of mild detergent treatment and limiting dilution as above to maximize clonality, yielding infectious virus stocks with titers of about 105 to 106 TCID50/ml. These clonal preparations were amplified to a total of about 1 × 104 to 1 × 107 TCID50 for genotypic and phenotypic (drug resistance) analyses.
For titration of infectious HCV, cell culture supernatants were serially diluted and applied to Huh-7.5 cells that had been seeded in 96-well plates at 6,400 cells/well 16 h earlier. Three days postinfection, cells were washed with phosphate-buffered saline (PBS), fixed with ice-cold methanol, and stained to detect NS5A using an anti-NS5A monoclonal antibody 9E10, as described previously (22, 27). Virus titers are expressed as TCID50/ml (72).
Serial passage of HCV.
Serial passages of HCV p0 were carried out as previously described (27). Briefly, 4 × 105 Huh-7.5 reporter cells were infected with HCV p0 at a multiplicity of infection (MOI) of 0.03 TCID50/cell; after a virus adsorption period of 5 h at 37°C, the inoculum was removed, and 2 ml of medium was added to the cell monolayer. The infected cells were further incubated at 37°C for 72 h or 96 h; for each subsequent passage, 4 × 105 Huh-7.5 reporter cells were infected as indicated above using 0.5 ml of cell culture supernatant from the previous passage.
RNA extraction, cDNA synthesis, and PCR amplification.
Intracellular viral RNA was extracted from infected cells using the Qiagen RNeasy kit according to the manufacturer’s instructions (Qiagen, Valencia, CA, USA). RT-PCR amplification was carried out using AccuScript (Agilent), as specified by the manufacturer. Amplification products were analyzed by agarose gel electrophoresis, using Gene Ruler 1 KB Plus DNA Ladder (Thermo Scientific) as a molar mass standard. Negative controls without template RNA were included to ascertain the absence of contaminating templates. Oligonucleotide primers used in the present study are listed in Table S10 (http://babia.cbm.uam.es/~lab121/SupplMatGallego3).
Sanger sequencing and determination of genomic sites with mixed nucleotides.
Amplified DNA was sequenced using the 23 ABI 3730 XLS sequencer (Macrogen, Inc.). The oligonucleotide primers used for nucleotide sequencing are listed in Table S10 (http://babia.cbm.uam.es/~lab121/SupplMatGallego3). The proportion of a mutant nucleotide at a given genomic position was calculated by triangulation and approximation to the triangle height (73).
Deep sequencing and mutational waves.
For Illumina deep sequencing, PCR products were purified (QIAquick gel extraction kit; Qiagen), quantified (Qubit dsDNA assay kit; Thermo Fisher Scientific), and tested for quality (Bioanalyzer DNA 1000 LabChip; Agilent). Three amplicons of NS5B spanning genomic residues 7626 to 7962, 7941 to 8257, and 8229 to 8653 were analyzed from intracellular viral RNA using the Illumina MiSeq platform, with the 2 × 300-bp mode with v3 chemistry; the fastq files were processed as previously described (74–76) to obtain the forward and reverse consensus haplotypes with abundances at or above 0.1% and median coverage of 99,694 reads per amplicon. Procedures for read cleaning, as well as controls for reliable mutant detection, and to ensure that two different amplifications of the same sample yield comparable read compositions, have been described (74, 76–78). The cutoff value for mutant detection was set at 0.5%, and only mutations found in two DNA strands were considered. The resulting median coverage was of 89,398 reads per amplicon.
Quantification of HCV RNA by real-time RT-PCR.
Prior to any amplification, viral RNA was quantified by real-time quantitative RT-PCR (qRT-PCR) using the Light Cycler RNA Master SYBR green I kit (Roche) (27). The 5′ untranslated region (UTR) of the HCV genome was amplified using as primers oligonucleotides HCV-5UTR-F2 and HCV-5UTR-R2 (Table S10 [http://babia.cbm.uam.es/~lab121/SupplMatGallego3]). Quantification was relative to a standard curve obtained with known amounts of HCV RNA synthesized by in vitro transcription of plasmid GNNFLAG2(p7-nsGluc2A) (26). The specificity of the reaction was monitored by the denaturation curve of the amplified DNAs. Negative controls (without template RNA, and using RNA from mock-infected cells) were run in parallel with each amplification reaction to ascertain the absence of contamination with undesired templates.
Phenotypic assays.
Drug resistance assays with populations and biological clones were carried out as previously described (27, 28).
Statistical analyses.
The statistical methods to quantify the significances of differences in RNA or infectivity values have been described (30, 79). In brief, to determine the statistical significance of differences in infectivity and RNA, two-way analysis of variance (ANOVA) was carried out using Prism 7 software (GraphPad). The statistical significance of differences between mutation frequencies or mutation types was evaluated by the proportion test. The statistical significance of differences in wave amplitude was evaluated by the t test.
Data availability.
The consensus genomic nucleotide sequences of HCV p0, HCV p100, and HCV p200 have been deposited in GenBank with accession numbers KC595606, KC595609, and KY123743. The Illumina data have been deposited in the NCBI BioSample database under accession numbers SAMN13531332 to SAMN13531367 (BioProject accession number PRJNA593382).
ACKNOWLEDGMENTS
We thank Charles M. Rice for the supply of plasmid Jc1FLAG2(p7-nsGluc2A), Huh-7.5, and Huh-7.5 reporter cell lines, anti-NS5A monoclonal antibody 9E10, and helpful advice for the implementation of HCV cell culture system. We thank Mercedes Dávila for expert technical assistance.
The work at CBMSO was supported by grants SAF2014-52400-R from Ministerio de Economía y Competitividad (MINECO); SAF2017-87846-R and BFU2017-91384-EXP from Ministerio de Ciencia, Innovación y Universidades (MICIU); and S2013/ABI-2906 (PLATESA from Comunidad de Madrid [CAM]/FEDER), S2018/BAA-4370 (PLATESA2 from CAM/FEDER), and PI18/00210 from Instituto de Salud Carlos III. C.P. is supported by the Miguel Servet program of the Instituto de Salud Carlos III (CP14/00121), cofinanced by the European Regional Development Fund (ERDF). CIBERehd is funded by Instituto de Salud Carlos III. Institutional grants from the Fundación Ramón Areces and Banco Santander to the CBMSO are also acknowledged. The team at CBMSO belongs to the Global Virus Network (GVN). The work in Barcelona was supported by Instituto de Salud Carlos III, cofinanced by the European Regional Development Fund (ERDF) (grant numbers PI16/00337) and by the Centro para el Desarrollo Tecnológico Industrial (CDTI) from the Spanish Ministry of Economy and Business (grant number IDI-20151125). Work at CAB was supported by MINECO grant BIO2016-79618-R (funded by the European Union under the FEDER program) and by the Spanish State Research Agency (AEI) through project number MDM-2017-0737 Unidad de Excelencia “María de Maeztu”-Centro de Astrobiologia (CSIC-INTA).
REFERENCES
- 1.Domingo E, Perales C. 2019. Viral quasispecies. PLoS Genet 15:e1008271. doi: 10.1371/journal.pgen.1008271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morley VJ, Turner PE. 2017. Dynamics of molecular evolution in RNA virus populations depend on sudden versus gradual environmental change. Evolution 71:872–883. doi: 10.1111/evo.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Somovilla P, Manrubia S, Lazaro E. 2019. Evolutionary dynamics in the RNA bacteriophage Qbeta Depends on the pattern of change in selective pressures. Pathogens 8:80. doi: 10.3390/pathogens8020080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfeiffer JK, Kirkegaard K. 2005. Increased fidelity reduces poliovirus fitness and virulence under selective pressure in mice. PLoS Pathog 1:e11. doi: 10.1371/journal.ppat.0010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. 2006. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 439:344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsibris AM, Korber B, Arnaout R, Russ C, Lo CC, Leitner T, Gaschen B, Theiler J, Paredes R, Su Z, Hughes MD, Gulick RM, Greaves W, Coakley E, Flexner C, Nusbaum C, Kuritzkes DR. 2009. Quantitative deep sequencing reveals dynamic HIV-1 escape and large population shifts during CCR5 antagonist therapy in vivo. PLoS One 4:e5683. doi: 10.1371/journal.pone.0005683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischer W, Ganusov VV, Giorgi EE, Hraber PT, Keele BF, Leitner T, Han CS, Gleasner CD, Green L, Lo CC, Nag A, Wallstrom TC, Wang S, McMichael AJ, Haynes BF, Hahn BH, Perelson AS, Borrow P, Shaw GM, Bhattacharya T, Korber BT. 2010. Transmission of single HIV-1 genomes and dynamics of early immune escape revealed by ultra-deep sequencing. PLoS One 5:e12303. doi: 10.1371/journal.pone.0012303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cale EM, Hraber P, Giorgi EE, Fischer W, Bhattacharya T, Leitner T, Yeh WW, Gleasner C, Green LD, Han CS, Korber B, Letvin NL. 2011. Epitope-specific CD8+ T lymphocytes cross-recognize mutant simian immunodeficiency virus (SIV) sequences but fail to contain very early evolution and eventual fixation of epitope escape mutations during SIV infection. J Virol 85:3746–3757. doi: 10.1128/JVI.02420-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domingo E, Sheldon J, Perales C. 2012. Viral quasispecies evolution. Microbiol Mol Biol Rev 76:159–216. doi: 10.1128/MMBR.05023-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andino R, Domingo E. 2015. Viral quasispecies. Virology 479–480:46–51. doi: 10.1016/j.virol.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farci P. 2011. New insights into the HCV quasispecies and compartmentalization. Semin Liver Dis 31:356–374. doi: 10.1055/s-0031-1297925. [DOI] [PubMed] [Google Scholar]
- 12.Kortenhoeven C, Joubert F, Bastos AD, Abolnik C. 2015. Virus genome dynamics under different propagation pressures: reconstruction of whole genome haplotypes of West Nile viruses from NGS data. BMC Genomics 16:118. doi: 10.1186/s12864-015-1340-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borderia AV, Rozen-Gagnon K, Vignuzzi M. 2016. Fidelity variants and RNA quasispecies. Curr Top Microbiol Immunol 392:303–322. doi: 10.1007/82_2015_483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donohue RC, Pfaller CK, Cattaneo R. 2019. Cyclical adaptation of measles virus quasispecies to epithelial and lymphocytic cells: to V, or not to V. PLoS Pathog 15:e1007605. doi: 10.1371/journal.ppat.1007605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bukh J. 2016. The history of hepatitis C virus (HCV): basic research reveals unique features in phylogeny, evolution and the viral life cycle with new perspectives for epidemic control. J Hepatol 65:S2–S21. doi: 10.1016/j.jhep.2016.07.035. [DOI] [PubMed] [Google Scholar]
- 16.Pawlotsky JM. 2006. Hepatitis C virus population dynamics during infection. Curr Top Microbiol Immunol 299:261–284. doi: 10.1007/3-540-26397-7_9. [DOI] [PubMed] [Google Scholar]
- 17.Farci P, Shimoda A, Coiana A, Diaz G, Peddis G, Melpolder JC, Strazzera A, Chien DY, Munoz SJ, Balestrieri A, Purcell RH, Alter HJ. 2000. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science 288:339–344. doi: 10.1126/science.288.5464.339. [DOI] [PubMed] [Google Scholar]
- 18.Farci P, Strazzera R, Alter HJ, Farci S, Degioannis D, Coiana A, Peddis G, Usai F, Serra G, Chessa L, Diaz G, Balestrieri A, Purcell RH. 2002. Early changes in hepatitis C viral quasispecies during interferon therapy predict the therapeutic outcome. Proc Natl Acad Sci U S A 99:3081–3086. doi: 10.1073/pnas.052712599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dietz J, Susser S, Vermehren J, Peiffer KH, Grammatikos G, Berger A, Ferenci P, Buti M, Mullhaupt B, Hunyady B, Hinrichsen H, Mauss S, Petersen J, Buggisch P, Felten G, Huppe D, Knecht G, Lutz T, Schott E, Berg C, Spengler U, von Hahn T, Berg T, Zeuzem S, Sarrazin C, European HCV Resistance Study Group. 2018. Patterns of resistance-associated substitutions in patients with chronic HCV infection following treatment with direct-acting antivirals. Gastroenterology 154:976–988 e974. doi: 10.1053/j.gastro.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Di Maio VC, Cento V, Lenci I, Aragri M, Rossi P, Barbaliscia S, Melis M, Verucchi G, Magni CF, Teti E, Bertoli A, Antonucci F, Bellocchi MC, Micheli V, Masetti C, Landonio S, Francioso S, Santopaolo F, Pellicelli AM, Calvaruso V, Gianserra L, Siciliano M, Romagnoli D, Cozzolongo R, Grieco A, Vecchiet J, Morisco F, Merli M, Brancaccio G, Di Biagio A, Loggi E, Mastroianni CM, Pace Palitti V, Tarquini P, Puoti M, Taliani G, Sarmati L, Picciotto A, Vullo V, Caporaso N, Paoloni M, Pasquazzi C, Rizzardini G, Parruti G, Craxi A, Babudieri S, Andreoni M, Angelico M, Perno CF, Ceccherini-Silberstein F, HCV Italian Resistance Network Study Group. 2017. Multiclass HCV resistance to direct-acting antiviral failure in real-life patients advocates for tailored second-line therapies. Liver Int 37:514–528. doi: 10.1111/liv.13327. [DOI] [PubMed] [Google Scholar]
- 21.Perales C. 2018. Quasispecies dynamics and clinical significance of hepatitis C virus (HCV) antiviral resistance. Int J Antimicrob Agents 10:105562. doi: 10.1016/j.ijantimicag.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 22.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 23.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. 2005. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A 102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gottwein JM, Pham LV, Mikkelsen LS, Ghanem L, Ramirez S, Scheel TKH, Carlsen THR, Bukh J. 2018. Efficacy of NS5A inhibitors against hepatitis C virus genotypes 1–7 and escape variants. Gastroenterology 154:1435–1448. doi: 10.1053/j.gastro.2017.12.015. [DOI] [PubMed] [Google Scholar]
- 26.Marukian S, Jones CT, Andrus L, Evans MJ, Ritola KD, Charles ED, Rice CM, Dustin LB. 2008. Cell culture-produced hepatitis C virus does not infect peripheral blood mononuclear cells. Hepatology 48:1843–1850. doi: 10.1002/hep.22550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perales C, Beach NM, Gallego I, Soria ME, Quer J, Esteban JI, Rice C, Domingo E, Sheldon J. 2013. Response of hepatitis C virus to long-term passage in the presence of alpha interferon: multiple mutations and a common phenotype. J Virol 87:7593–7607. doi: 10.1128/JVI.02824-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheldon J, Beach NM, Moreno E, Gallego I, Pineiro D, Martinez-Salas E, Gregori J, Quer J, Esteban JI, Rice CM, Domingo E, Perales C. 2014. Increased replicative fitness can lead to decreased drug sensitivity of hepatitis C virus. J Virol 88:12098–12111. doi: 10.1128/JVI.01860-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gallego I, Sheldon J, Moreno E, Gregori J, Quer J, Esteban JI, Rice CM, Domingo E, Perales C. 2016. Barrier-independent, fitness-associated differences in sofosbuvir efficacy against hepatitis C virus. Antimicrob Agents Chemother 60:3786–3793. doi: 10.1128/AAC.00581-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moreno E, Gallego I, Gregori J, Lucia-Sanz A, Soria ME, Castro V, Beach NM, Manrubia S, Quer J, Esteban JI, Rice CM, Gomez J, Gastaminza P, Domingo E, Perales C. 2017. Internal disequilibria and phenotypic diversification during replication of hepatitis C Virus in a noncoevolving cellular environment. J Virol 91:e02505-16. doi: 10.1128/JVI.02505-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noppornpanth S, Smits SL, Lien TX, Poovorawan Y, Osterhaus AD, Haagmans BL. 2007. Characterization of hepatitis C virus deletion mutants circulating in chronically infected patients. J Virol 81:12496–12503. doi: 10.1128/JVI.01059-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Q, Tong Y, Xu Y, Niu J, Zhong J. 2018. Genetic analysis of serum-derived defective hepatitis C virus genomes revealed novel viral cis elements for virus replication and assembly. J Virol 92:e02182-17. doi: 10.1128/JVI.02182-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pacini L, Graziani R, Bartholomew L, De Francesco R, Paonessa G. 2009. Naturally occurring hepatitis C virus subgenomic deletion mutants replicate efficiently in Huh-7 cells and are trans-packaged in vitro to generate infectious defective particles. J Virol 83:9079–9093. doi: 10.1128/JVI.00308-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rezelj VV, Levi LI, Vignuzzi M. 2018. The defective component of viral populations. Curr Opin Virol 33:74–80. doi: 10.1016/j.coviro.2018.07.014. [DOI] [PubMed] [Google Scholar]
- 35.Karamichali E, Chihab H, Kakkanas A, Marchio A, Karamitros T, Pogka V, Varaklioti A, Kalliaropoulos A, Martinez-Gonzales B, Foka P, Koskinas I, Mentis A, Benjelloun S, Pineau P, Georgopoulou U. 2018. HCV defective genomes promote persistent infection by modulating the viral life cycle. Front Microbiol 9:2942. doi: 10.3389/fmicb.2018.02942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vignuzzi M, Lopez CB. 2019. Defective viral genomes are key drivers of the virus-host interaction. Nat Microbiol 4:1075–1087. doi: 10.1038/s41564-019-0465-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mercuri L, Thomson EC, Hughes J, Karayiannis P. 2018. Quasispecies changes with distinctive point mutations in the hepatitis C virus internal ribosome entry site (IRES) derived from PBMCs and plasma. Adv Virol 2018:4835252. doi: 10.1155/2018/4835252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gisder S, Mockel N, Eisenhardt D, Genersch E. 2018. In vivo evolution of viral virulence: switching of deformed wing virus between hosts results in virulence changes and sequence shifts. Environ Microbiol 20:4612–4628. doi: 10.1111/1462-2920.14481. [DOI] [PubMed] [Google Scholar]
- 39.Olszewska-Tomczyk M, Dolka I, Świętoń E, Śmietanka K. 2018. Genetic changes in pigeon paramyxovirus type-1 induced by serial passages in chickens and microscopic lesions caused by the virus in various avian hosts. J Vet Res 62:447–455. doi: 10.2478/jvetres-2018-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams GC. 1992. Natural selection. domains, levels, and challenges. Oxford University Press, New York, NY. [Google Scholar]
- 41.Kirkpatrick M. 1996. Genes and adaptation: a pocket guide to the theory, p 123–146. In Rose MR, Lauder GV (ed), Adaptation. Academic Press, San Diego, CA. [Google Scholar]
- 42.Ojosnegros S, Agudo R, Sierra M, Briones C, Sierra S, Gonzalez-Lopez C, Domingo E, Cristina J. 2008. Topology of evolving, mutagenized viral populations: quasispecies expansion, compression, and operation of negative selection. BMC Evol Biol 8:207. doi: 10.1186/1471-2148-8-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thoday JM. 1972. Disruptive selection. Proc R Soc Lond B Biol Sci 182:109–143. doi: 10.1098/rspb.1972.0070. [DOI] [PubMed] [Google Scholar]
- 44.Wild AT, Gandhi N, Chettiar ST, Aziz K, Gajula RP, Williams RD, Kumar R, Taparra K, Zeng J, Cades JA, Velarde E, Menon S, Geschwind JF, Cosgrove D, Pawlik TM, Maitra A, Wong J, Hales RK, Torbenson MS, Herman JM, Tran PT. 2013. Concurrent versus sequential sorafenib therapy in combination with radiation for hepatocellular carcinoma. PLoS One 8:e65726. doi: 10.1371/journal.pone.0065726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ren Q, Li C, Yuan P, Cai C, Zhang L, Luo GG, Wei W. 2015. A dual-reporter system for real-time monitoring and high-throughput CRISPR/Cas9 library screening of the hepatitis C virus. Sci Rep 5:8865. doi: 10.1038/srep08865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peng J, Zhou Y, Zhu S, Wei W. 2015. High-throughput screens in mammalian cells using the CRISPR-Cas9 system. FEBS J 282:2089–2096. doi: 10.1111/febs.13251. [DOI] [PubMed] [Google Scholar]
- 47.Escarmís C, Dávila M, Domingo E. 1999. Multiple molecular pathways for fitness recovery of an RNA virus debilitated by operation of Muller’s ratchet. J Mol Biol 285:495–505. doi: 10.1006/jmbi.1998.2366. [DOI] [PubMed] [Google Scholar]
- 48.Nguyen AH, Molineux IJ, Springman R, Bull JJ. 2012. Multiple genetic pathways to similar fitness limits during viral adaptation to a new host. Evolution 66:363–374. doi: 10.1111/j.1558-5646.2011.01433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arenas M, Lorenzo-Redondo R, Lopez-Galindez C. 2016. Influence of mutation and recombination on HIV-1 in vitro fitness recovery. Mol Phylogenet Evol 94:264–270. doi: 10.1016/j.ympev.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 50.García-Arriaza J, Manrubia SC, Toja M, Domingo E, Escarmís C. 2004. Evolutionary transition toward defective RNAs that are infectious by complementation. J Virol 78:11678–11685. doi: 10.1128/JVI.78.21.11678-11685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ojosnegros S, Garcia-Arriaza J, Escarmis C, Manrubia SC, Perales C, Arias A, Mateu MG, Domingo E. 2011. Viral genome segmentation can result from a trade-off between genetic content and particle stability. PLoS Genet 7:e1001344. doi: 10.1371/journal.pgen.1001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moreno E, Ojosnegros S, Garcia-Arriaza J, Escarmis C, Domingo E, Perales C. 2014. Exploration of sequence space as the basis of viral RNA genome segmentation. Proc Natl Acad Sci U S A 111:6678–6683. doi: 10.1073/pnas.1323136111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.García-Arriaza J, Ojosnegros S, Dávila M, Domingo E, Escarmís C. 2006. Dynamics of mutation and recombination in a replicating population of complementing, defective viral genomes. J Mol Biol 360:558–572. doi: 10.1016/j.jmb.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 54.Pawlotsky JM. 2016. Hepatitis C virus resistance to direct-acting antiviral drugs in interferon-free regimens. Gastroenterology 151:70–86. doi: 10.1053/j.gastro.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 55.Chopra KB, Demetris AJ, Blakolmer K, Dvorchik I, Laskus T, Wang LF, Araya VR, Dodson F, Fung JJ, Rakela J, Vargas HE. 2003. Progression of liver fibrosis in patients with chronic hepatitis C after orthotopic liver transplantation. Transplantation 76:1487–1491. doi: 10.1097/01.TP.0000088668.28950.7C. [DOI] [PubMed] [Google Scholar]
- 56.Hopcraft SE, Azarm KD, Israelow B, Leveque N, Schwarz MC, Hsu TH, Chambers MT, Sourisseau M, Semler BL, Evans MJ. 2016. Viral determinants of miR-122-independent hepatitis C virus replication. mSphere 1:e00009-15. doi: 10.1128/mSphere.00009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de la Torre JC, Giachetti C, Semler BL, Holland JJ. 1992. High frequency of single-base transitions and extreme frequency of precise multiple-base reversion mutations in poliovirus. Proc Natl Acad Sci U S A 89:2531–2535. doi: 10.1073/pnas.89.7.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Borrego B, Novella IS, Giralt E, Andreu D, Domingo E. 1993. Distinct repertoire of antigenic variants of foot-and-mouth disease virus in the presence or absence of immune selection. J Virol 67:6071–6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martín Hernández AM, Carrillo EC, Sevilla N, Domingo E. 1994. Rapid cell variation can determine the establishment of a persistent viral infection. Proc Natl Acad Sci U S A 91:3705–3709. doi: 10.1073/pnas.91.9.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mateu MG, Hernández J, Martínez MA, Feigelstock D, Lea S, Pérez JJ, Giralt E, Stuart D, Palma EL, Domingo E. 1994. Antigenic heterogeneity of a foot-and-mouth disease virus serotype in the field is mediated by very limited sequence variation at several antigenic sites. J Virol 68:1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ruiz-Jarabo CM, Miller E, Gómez-Mariano G, Domingo E. 2003. Synchronous loss of quasispecies memory in parallel viral lineages: a deterministic feature of viral quasispecies. J Mol Biol 333:553–563. doi: 10.1016/j.jmb.2003.08.054. [DOI] [PubMed] [Google Scholar]
- 62.Herrera M, Grande-Perez A, Perales C, Domingo E. 2008. Persistence of foot-and-mouth disease virus in cell culture revisited: implications for contingency in evolution. J Gen Virol 89:232–244. doi: 10.1099/vir.0.83312-0. [DOI] [PubMed] [Google Scholar]
- 63.Couderc T, Guedo N, Calvez V, Pelletier I, Hogle J, Colbere-Garapin F, Blondel B. 1994. Substitutions in the capsids of poliovirus mutants selected in human neuroblastoma cells confer on the Mahoney type 1 strain a phenotype neurovirulent in mice. J Virol 68:8386–8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu Z, Rezapkin GV, Douthitt MP, Ran Y, Asher DM, Levenbook IS, Chumakov KM. 1996. Limited genetic changes in the Sabin 1 strain of poliovirus occurring in the central nervous system of monkeys. J Gen Virol 77:273–280. doi: 10.1099/0022-1317-77-2-273. [DOI] [PubMed] [Google Scholar]
- 65.Brown EG, Liu H, Kit LC, Baird S, Nesrallah M. 2001. Pattern of mutation in the genome of influenza A virus on adaptation to increased virulence in the mouse lung: identification of functional themes. Proc Natl Acad Sci U S A 98:6883–6888. doi: 10.1073/pnas.111165798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gutierrez B, Escalera-Zamudio M, Pybus OG. 2019. Parallel molecular evolution and adaptation in viruses. Curr Opin Virol 34:90–96. doi: 10.1016/j.coviro.2018.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martincorena I, Raine KM, Gerstung M, Dawson KJ, Haase K, Van Loo P, Davies H, Stratton MR, Campbell PJ. 2017. Universal patterns of selection in cancer and somatic tissues. Cell 171:1029–1041 e1021. doi: 10.1016/j.cell.2017.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mehta HH, Prater AG, Beabout K, Elworth RAL, Karavis M, Gibbons HS, Shamoo Y, Mehta HH, Prater AG, Beabout K, Elworth RAL, Karavis M, Gibbons HS, Shamoo Y. 2019. The essential role of hypermutation in rapid adaptation to antibiotic stress. Antimicrob Agents Chemother 63:e00744-19. doi: 10.1128/AAC.00744-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Blight KJ, McKeating JA, Rice CM. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol 76:13001–13014. doi: 10.1128/jvi.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jones CT, Catanese MT, Law LM, Khetani SR, Syder AJ, Ploss A, Oh TS, Schoggins JW, MacDonald MR, Bhatia SN, Rice CM. 2010. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat Biotechnol 28:167–171. doi: 10.1038/nbt.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sobrino F, Dávila M, Ortín J, Domingo E. 1983. Multiple genetic variants arise in the course of replication of foot-and-mouth disease virus in cell culture. Virology 128:310–318. doi: 10.1016/0042-6822(83)90258-1. [DOI] [PubMed] [Google Scholar]
- 72.Reed LJ, Muench H. 1938. A simple method for estimating fifty per cent endpoints. Am J Hyg 27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 73.Dyson N. 1998. Chromatographic integration methods, 2nd ed The Royal Society of Chemistry, Cambridge, UK. [Google Scholar]
- 74.Gregori J, Perales C, Rodriguez-Frias F, Esteban JI, Quer J, Domingo E. 2016. Viral quasispecies complexity measures. Virology 493:227–237. doi: 10.1016/j.virol.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 75.Gallego I, Soria ME, Gregori J, de Ávila AI, García-Crespo C, Moreno E, Gadea I, Esteban J, Fernández-Roblas R, Esteban JI, Gómez J, Quer J, Domingo E, Perales C, Gallego I, Soria ME, Gregori J, de Ávila AI, García-Crespo C, Moreno E, Gadea I, Esteban J, Fernández-Roblas R, Esteban JI, Gómez J, Quer J, Domingo E, Perales C. 2019. Synergistic lethal mutagenesis of hepatitis C virus. Antimicrob Agents Chemother 63:e01653-19. doi: 10.1128/AAC.01653-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gregori J, Salicrú M, Domingo E, Sanchez A, Esteban JI, Rodríguez-Frías F, Quer J. 2014. Inference with viral quasispecies diversity indices: clonal and NGS approaches. Bioinformatics 30:1104–1111. doi: 10.1093/bioinformatics/btt768. [DOI] [PubMed] [Google Scholar]
- 77.Quer J, Rodríguez-Frias F, Gregori J, Tabernero D, Soria ME, García-Cehic D, Homs M, Bosch A, Pintó RM, Esteban JI, Domingo E, Perales C. 2017. Deep sequencing in the management of hepatitis virus infections. Virus Res 239:115–125. doi: 10.1016/j.virusres.2016.12.020. [DOI] [PubMed] [Google Scholar]
- 78.Soria ME, Gregori J, Chen Q, Garcia-Cehic D, Llorens M, de Avila AI, Beach NM, Domingo E, Rodriguez-Frias F, Buti M, Esteban R, Esteban JI, Quer J, Perales C. 2018. Pipeline for specific subtype amplification and drug resistance detection in hepatitis C virus. BMC Infect Dis 18:446. doi: 10.1186/s12879-018-3356-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.de Avila AI, Moreno E, Perales C, Domingo E. 2017. Favipiravir can evoke lethal mutagenesis and extinction of foot-and-mouth disease virus. Virus Res 233:105–112. doi: 10.1016/j.virusres.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 80.Gallego I, Gregori J, Soria ME, García-Crespo C, García-Álvarez M, Gómez-González A, Valiergue R, Gómez J, Esteban JI, Quer J, Domingo E, Perales C. 2018. Resistance of high fitness hepatitis C virus to lethal mutagenesis. Virology 523:100–109. doi: 10.1016/j.virol.2018.07.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The consensus genomic nucleotide sequences of HCV p0, HCV p100, and HCV p200 have been deposited in GenBank with accession numbers KC595606, KC595609, and KY123743. The Illumina data have been deposited in the NCBI BioSample database under accession numbers SAMN13531332 to SAMN13531367 (BioProject accession number PRJNA593382).