HIV-1 Env conformation is one of the key parameters determining viral infectivity. The flow virometry-based assay developed in this study allows for the characterization of proteins incorporated in HIV-1 particles. We studied the conformation of HIV-1 Env and the impact that the viral protein Nef and the cellular proteins CD4 and SERINC5 have on Env accessibility to antibodies. Our assay permitted us to highlight some noticeable differences in the conformation of Env between producer cells and viral particles. It contributes to a better understanding of the actual composition of HIV-1 particles.
KEYWORDS: Env, flow virometry, HIV-1, broadly neutralizing antibodies, nonneutralizing antibodies
ABSTRACT
The HIV-1 Env protein is exposed at the surface of virions and infected cells. Env fluctuates between different closed and open structural states and these conformations influence both viral infectivity and sensitivity to antibody binding and neutralization. We established a flow virometry assay to visualize Env proteins at the surface of human immunodeficiency virus type 1 (HIV-1) virions. The assay is performed on ultracentrifuged fluorescent viral particles that are stained with a panel of broadly neutralizing antibodies (bNAbs) and nonneutralizing antibodies (nnAbs) that probe different epitopes of Env. We used this assay to compare Env at the surface of producer cells and viral particles and to analyze the effect of Nef, CD4, and SERINC5 on Env accessibility to antibodies. We studied the laboratory-adapted strain NL4-3 and two transmitted/founder viruses, THRO and CH058. We confirm that antibody accessibility varies between viral strains and show that Nef, CD4, and SERINC5 additively impact Env conformations. We further demonstrate that the Env accessibility profile on virions is globally similar to that observed on HIV-1-infected cells, with some noticeable differences. For instance, nnAbs bind to virions more efficiently than to producer cells, likely reflecting changes in Env conformational states on mature viral particles. This test complements other techniques and provides a convenient and simple tool for quantifying and probing the structure of Env at the virion surface and to analyze the impact of viral and cellular proteins on these parameters.
IMPORTANCE HIV-1 Env conformation is one of the key parameters determining viral infectivity. The flow virometry-based assay developed in this study allows for the characterization of proteins incorporated in HIV-1 particles. We studied the conformation of HIV-1 Env and the impact that the viral protein Nef and the cellular proteins CD4 and SERINC5 have on Env accessibility to antibodies. Our assay permitted us to highlight some noticeable differences in the conformation of Env between producer cells and viral particles. It contributes to a better understanding of the actual composition of HIV-1 particles.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) particles are surrounded by an envelope, which consists of the host cell-derived membrane, cellular proteins, and the viral envelope glycoproteins (Env) (1). Env mediates binding to the viral receptor CD4 and the coreceptors CCR5 and CXCR4, which are expressed on target cells. The interaction between Env and CD4/coreceptors is required for fusion between viral and cellular membranes and the intracellular delivery of the viral genome (2). Env is the only viral protein exposed at the surface of both infected cells and viral particles, and it is one of the main targets of host antiviral antibody responses. Antibodies neutralize both cell-free and cell-associated viral transmission (3, 4). Antibodies also exert Fc-mediated antiviral functions such as antibody-dependent cellular cytotoxicity, complement-dependent cytotoxicity, and antibody-dependent cellular phagocytosis (5–11).
Gp160 Env glycoprotein precursors are synthesized in the endoplasmic reticulum (ER), transit through the Golgi compartments, and are cleaved by furin to generate mature gp120/gp41 complexes that trimerize and then directed to the plasma membrane. Between 0 and 20 Gp120 trimers are incorporated into single virions (12). A fraction of Gp160 escapes correct glycosylation and/or processing and is expressed at the cell surface but not incorporated in viral particles (13). Steady-state surface Env levels depend on various factors, such as synthesis and recycling rates, and interaction with Gag (14). The number and conformation of Env trimers influence virion infectivity (15). Moreover, during viral capsid maturation, the trimers which are dispersed across the surface of the virion relocalize and become more concentrated (16).
Various approaches have been developed to analyze Env structure. Recombinant stable Env trimers reconstituted in vitro (such as SOSIP), coupled with X-ray crystallography or cryo-electron microscopy analysis, are widely used tools (17–19). The use of anti-Env antibodies and viral capture assays (VCAs), neutralization assays, single-molecule fluorescence resonance energy transfer, or double electron-electron resonance spectroscopy and compared results to soluble and virion-bound Env structures also provided information about the conformations of virion-associated or recombinant soluble Env (6, 20–24). Recently, flow virometry has been used to analyze particles from different viral species, including vaccinia, Junin, and Nipah viruses and HIV-1 (25–30). Flow virometry can also be used to assess the binding of some anti-Env antibodies (30, 31). However, with HIV, the binding of virions to synthetic nanoparticles was required for efficient detection (31).
Env proteins fluctuate between closed and open conformations. Env proteins in closed conformation are in a native form and are unable to fuse. This state is recognized by broadly neutralizing antibodies (bNAbs) and generally not by nonneutralizing antibodies (nnAbs). The interaction of Env with CD4 induces a conformational change that opens the Env trimer to expose the regions required for fusion (19, 20, 23, 32). HIV-1 isolates are divided into distinct tiers based on their neutralization sensitivity (33). Laboratory-adapted strains, such as NL4-3, are highly sensitive to neutralizing antibodies and belong to tiers 1A and 1B. Primary strains and transmitted/founder (T/F) viruses are more resistant to neutralization and belong to tiers 2 and 3.
Various viral and cellular factors influence Env synthesis, processing, conformation or incorporation into viral particles. For instance, the HIV-1 Nef protein enhances infectivity at an early stage of the viral life cycle (34–37). Interestingly, the effect of Nef on viral infectivity depends on the variable region of Env and is inversely correlates with Env sensitivity to neutralization (37), suggesting that Nef indirectly affects Env conformation. The cellular restriction factor SERINC5 decreases viral fusion/entry and is targeted by Nef (38–40). SERINC5-mediated viral restriction also depends on Env, and tier 1 Env appears to be more sensitive than tier 2/3 Env (41, 42). It has been suggested that SERINC5 selectively targets open Env trimers (43).
In this study, we established a simple flow virometry assay to visualize HIV and detect Env at the surface of virions. We then used our assay to compare the binding of a panel of anti-Env broadly neutralizing and nonneutralizing antibodies to producer cells and virions. We studied the laboratory adapted strain NL4-3 and two transmitted/founder, tier 2/3, THRO and CH058 (44, 45). Because Nef, SERINC5 and CD4 proteins impact viral infectivity, we evaluated their effects on Env both in producer cells and in virions.
RESULTS
Detection of HIV-1 particles by flow virometry.
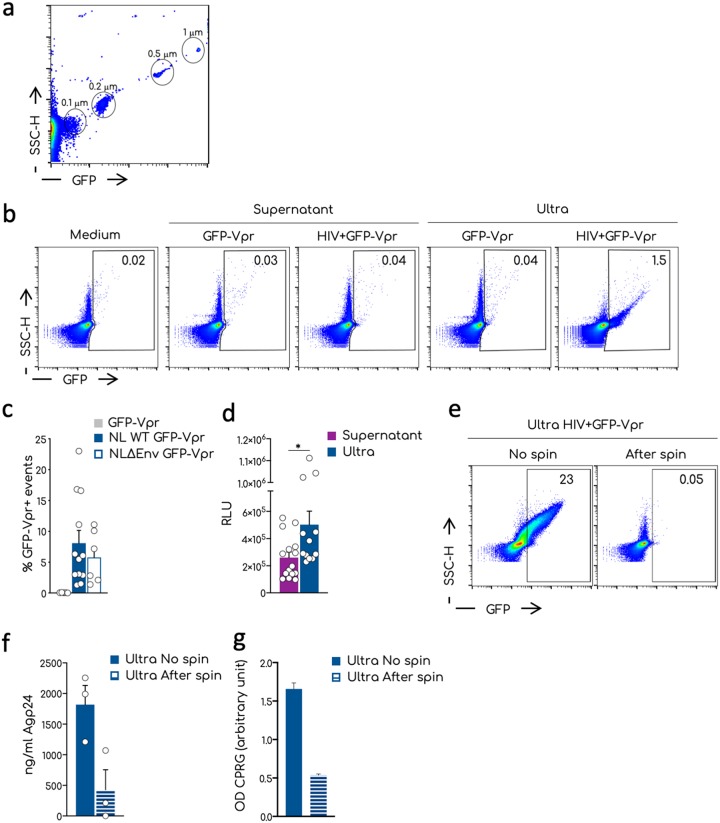
HIV-1 particles are around 120 nm in size. To evaluate whether our flow cytometer, the Attune NTx, could detect and analyze these virions, we first tested commercially available green fluorescent protein (GFP)-positive nanobeads of different sizes, ranging from 100 to 1,000 nm (Fig. 1a). The detection of nanoparticles by standard cytometer requires coupling of light scattering (SSC-H) and fluorescence (46). Beads ≥200 nm were readily detected by both SSC-H and GFP. The 100-nm beads were within the SSC-H background but were visible in the GFP channel.
FIG 1.
Visualization by flow virometry of nanoparticles and HIV-1 aggregates. (a) Representative dot plot analysis of the commercial nanoparticles GFP+ acquired on the standard cytometer Attune NTx. Different sizes of the beads are indicated in the gates. (b) Representative dot plots of the analysis of viral productions by flow virometry. Culture medium, supernatants, or ultracentrifuged supernatants (ultra) were acquired at the Attune NTx using the same setting established for the nanobeads in panel a. Numbers in the gates indicate the percentages of the GFP-Vpr+ events. (c) Mean and standard errors of the mean (SEM) of the percentages of the GFP-Vpr+ events in the ultra of multiple independent viral stocks of the NL4-3 and NL4-3ΔEnv viruses. Negative control corresponds to the ultra of the GFP-Vpr alone supernatants. Each dot corresponds to one independent viral preparation. (d) Infectivity of the viral supernatants versus the corresponding ultra. TZM-bl cells were exposed to the same amount of Gag-p24 and luciferase activity measured 36 h after infection. The means and SEM of at least three independent experiments conducted in triplicate are shown as relative luminescence units (RLU). (e) Representative dot plots of the procedure used to remove aggregates from ultra. Ultra were left untreated (No spin) or spun for 15 min at 20,000 × g. The supernatant devoid of aggregated particles was recovered (After spin) and analyzed by flow virometry. Numbers indicate the percentages of GFP+ events in the gate. (f) Quantification by ELISA of the HIV-1 Gag-p24 associated with the ultra treated as shown in panel e of three independent experiments. (g) Infectivity of the viral particles of the samples measured in panel f. Means and SEM are shown. Statistical analysis was performed using the Prism software and a multiple unpaired t test. *, P < 0.05.
We then tested the detection of HIV-1 particles. The HIV-1 Vpr protein is incorporated into the virion during viral assembly through its interaction with Gag p6 (47). A GFP-Vpr fusion protein is also incorporated, rendering the virions fluorescent and allowing for instance the visualization of viral entry by immunofluorescence (48). GFP-Vpr is incorporated in 40 to 80% of virions, without changing infectivity (12, 48–50). Moreover, the GFP-Vpr signal allows us to distinguish between viral particles and microvesicles released into the supernatant (12).
We produced fluorescent HIV-1 by cotransfecting 293T cells with proviral NL4-3 and GFP-Vpr plasmids. As controls, we used supernatants from cells transfected only with GFP-Vpr. Supernatants filtered through a 0.45-μm filter were analyzed by flow cytometry (Fig. 1b). None of the samples showed a significant number of GFP+ events. Hence, the GFP-Vpr signal is not sufficient to allow the visualization of single virions. We reasoned that we could increase the sensitivity of the technique by concentrating and aggregating the viral particles by ultracentrifugation and thus increasing their fluorescence (25). Filtered supernatants were ultracentrifuged for 1 to 2 h at 100,000 × g, and the pellets were gently resuspended in 1:100 of the original volume. This procedure allowed the recovery of about 50 to 80% of Gag p24 levels present in the supernatants. Figure 1b shows representative dot plots of ultracentrifuged supernatants (termed Ultra). The culture medium and concentrated supernatants of cells expressing only GFP-Vpr remained negative. With HIV, we observed a GFP+ population which likely corresponds to the aggregated GFP-Vpr+ viral particles. GFP+ events were distributed with a proportional increase of both SSC-H and fluorescence intensities. This suggests that viral particles assemble in aggregates of different sizes.
The frequency of the GFP+ events were variable across viral preparations (Fig. 1c). A similar signal was observed with NL4-3ΔEnv GFP-Vpr+ (Fig. 1c), indicating that Env is not required to generate detectable viral aggregates.
The infectivity of ultracentrifuged virions, normalized for Gag p24 levels and measured in the TZM-bl reporter assay (51), was slightly increased compared to nonultracentrifuged supernatants (Fig. 1d). We repelleted ultracentrifuged virions with a moderate spin (20,000 × g for 15 min) that should not bring down single viral particles. After the spin, we collected the supernatants and analyzed their content by flow virometry, Gag p24 levels and infectivity. The fluorescent signal, as well as 80% of Gag p24, disappeared after the moderate spinning (Fig. 1e). Moreover, the aggregated fraction contained most of viral infectivity (Fig. 1f and g).
Therefore, our protocol allows the detection of ultracentrifuged and aggregated HIV-1 virions by flow virometry.
Sorting and characterization of viral aggregates.
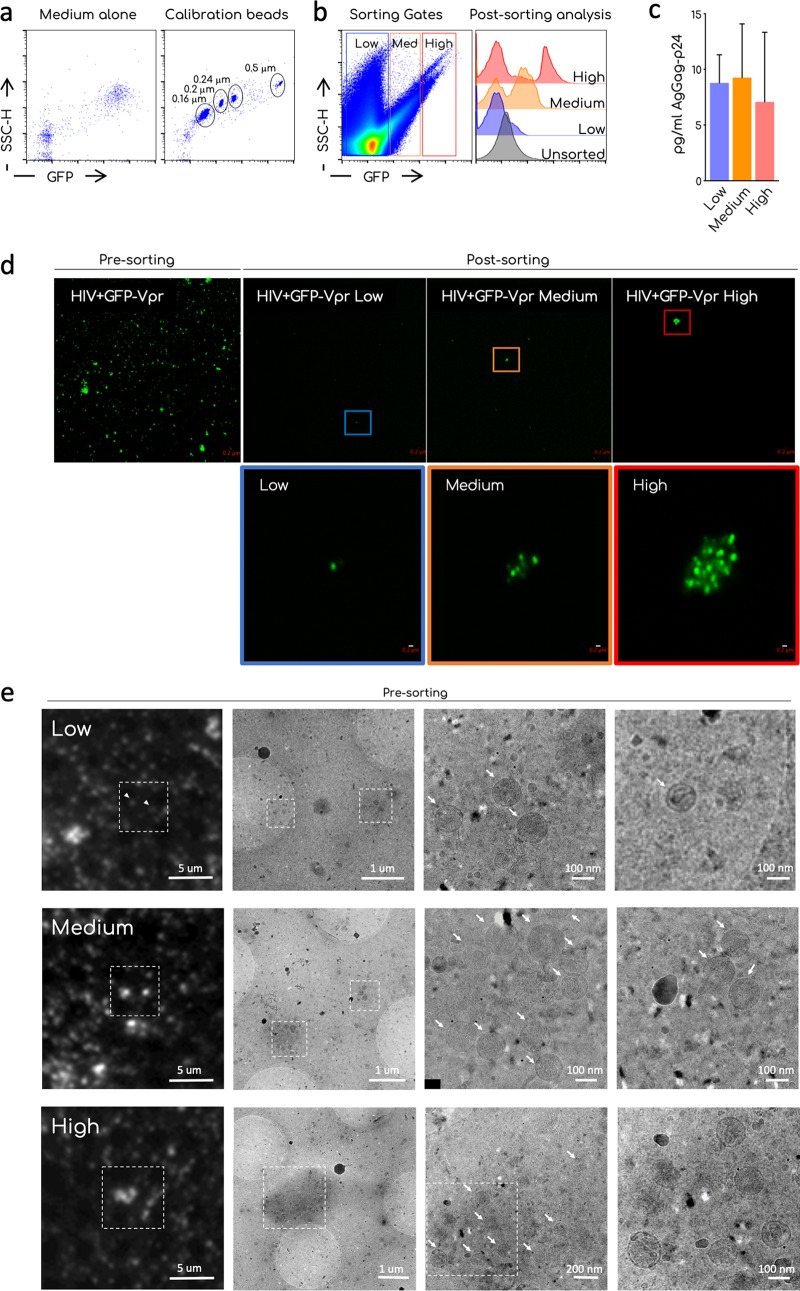
We further characterized ultracentrifuged viral particles. We verified that the GFP-Vpr+ population was composed of aggregates of various sizes. For this purpose, we sorted the GFP-Vpr+ population according to their GFP fluorescence and analyzed them by confocal microscopy. To sort GFP-Vpr+ aggregates, we used a MoFlo Astrios cytometer. We verified that this cytometer detects nanoparticles. GFP+ commercial beads from 160 to 500 nm were readily detected on both SSC and GFP channels (Fig. 2a). Figure 2b shows the gating strategy and results from one representative sorting experiment. We divided ultracentrifuged particles in negative/low, medium, and high GFP+ populations. A postsorting analysis demonstrated the enrichment of medium and high GFP+ particles in the respective populations. Of note, in medium and high sorted population, a low GFP population was present after sorting and corresponded to the background noise of the instrument. Alternatively, this may also correspond to GFP+ viral aggregates that disassembled during or after sorting. We detected similar levels of Gag p24 by ultrasensitive enzyme-linked immunosorbent assay (ELISA; SIMOA) (52) in each fraction (Fig. 2c).
FIG 2.
Sorting and microscopy analysis of ultracentrifuged viral preparations. (a) Representative dot plot analysis of the commercial nanoparticles GFP+ acquired on the standard cytometer MoFLO Astrios. The background noise of the instrument (medium alone) and the calibration beads are shown. Different sizes of the beads are indicated in the gates. (b) Sorting strategy. Ultracentrifuged viral preparations (Ultra) were acquired on a MoFLO Astrios cytometers and arbitrary gates on the GFP– and GFP+ samples were designed to sort aggregates accordingly with their relative fluorescence (low, medium, and high). Histogram overlay shows the efficiency of sorting. Unsorted ultra and postsorting low, medium, and high GFP samples are compared. A representative sorting out of at least four is depicted. (b) Quantification by ultrasensitive SIMOA digital ELISA of the HIV-1 Gag-p24 associated with the sorted fractions. Means and SEM of two independent quantifications are shown. (c) Confocal analysis of unsorted ultra (presorting) and postsorting fraction (low, medium, and high). Images were taken using an LSM-700 confocal microscope and a 63× objective. The magnification shows the 10× digital zoom on the indicated aggregates. Scale bar, 0.2 μm. (d) CLEM analysis of unsorted ultra. Ultra were left to settle on a TEM grid. GFP low, medium, or high aggregates were identified in the sample and imaged using a fluorescence microscope. The grid was then processed on a TEM and sample imaged to identify viral particles in the aggregates. Arrows indicates single virions. Scale bars, 1 μm to 100 nm.
Confocal microscopy analysis showed that, before sorting, ultracentrifuged GFP-Vpr-tagged virions were observed as clusters of different sizes, ranging from the limit of the resolution of the confocal microscope (around 200 nm), to up to 1 μm (Fig. 2d). After sorting, the size of GFP+ events measured by confocal microscopy increased in the “medium” fraction and to a higher extent in the “high” fractions, compared to the “low” fraction.
Transfected cells also secrete extracellular vesicles incorporating cellular and viral proteins. To test whether viral aggregates contained extracellular vesicles that could impact the subsequent analysis of Env, we performed a correlative light electron microscopy (CLEM) analysis of viral preparations (53, 54) (Fig. 2e). We analyzed by fluorescence microscopy the unsorted ultracentrifuged virions, and selected spots with increasing brightness and size. Low GFP+ spots corresponded to single virions and aggregates composed of up to three virions. Medium GFP+ spots contained up to 12 viral particles and high GFP spots contained up to 20 viral particles. No or very few extracellular vesicles were detected in the aggregates. Of note, viral particles appeared to be held together by a relatively dense structure. We did not investigate its composition, but we speculate that it could be derived from the culture medium or the producer cells.
Analysis of Env proteins at the cell surface and on viral particles.
We sought to determine whether our assay can be used to study Env proteins at the surface of viral particles. Since HIV-1 Nef protein is known to enhance viral infectivity by acting at an early postbinding step of the viral cycle (38, 39, 42, 55), we compared the conformation of Env on WT and Nef deletion viruses.
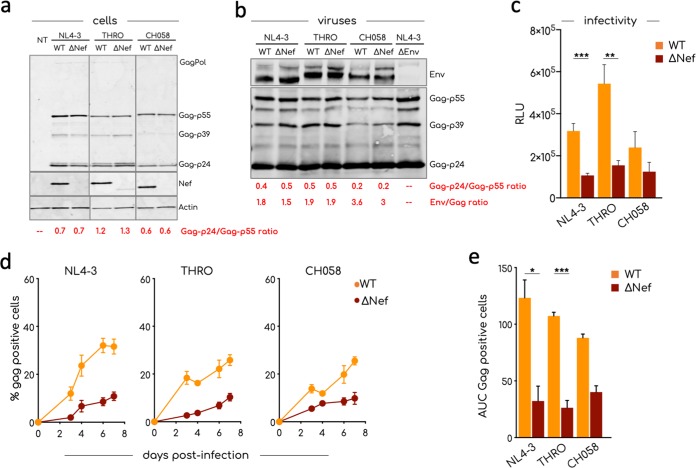
For this pilot study, we selected three different HIV-1 strains, the laboratory-adapted NL4-3 virus, two transmitted founder (T/F) viruses, THRO and CH058 (45). We first characterized the three viruses by analyzing their protein content, their infectivity in TZM-bl cells, and their replication kinetics in primary CD4+ T cells. Wild-type (WT) and Nef-deleted (ΔNef) NL4-3, THRO, and CH058 were generated by transfection of 293T cells. We did not observe a defect in viral release in the absence of Nef (not shown). We analyzed lysates of transfected 293T cells and released virions by Western blotting (Fig. 3a and b). The immature (Gag-Pol, p55, and p39) and mature (p24) forms of Gag were similarly detected in cell lysates, irrespective of the presence of Nef, suggesting that Gag processing is not affected (Fig. 3a). The Nef protein was only present in cells infected with WT viruses. Moreover, there was no significant difference in Gag-p24/Gag-p55 ratios and in Env/Gag ratios between WT and ΔNef virions (Fig. 3b). Their infectivity in TZM-bl cells was reduced in the absence of Nef (Fig. 3c). In activated CD4+ T cells, efficient viral replication required the presence of Nef (Fig. 3d), with differences between strains: Nef significantly enhanced replication of NL4-3 and THRO, whereas the effect was less marked for CH058. The three pairs of viruses were then used for further analysis.
FIG 3.
Characterization of viral isolates. (a) Western blot analysis of cell lysates of 293T cells transfected with the indicated viral strains. 293T cells were transfected, and cell lysates were prepared 48 h after transfection. The expression of Gag, Nef, and actin in cells was determined with specific antibodies. A lysate from nontransfected (NT) cells was used as negative control. A representative Western blot of two is shown. (b) Western blot analysis of purified aggregated viral particles. The incorporation of HIV-1 Env and Gag was determined with specific antibodies. The ratio Env/Gag indicated at the bottom of each lane was calculated after the analysis of the fluorescent signal associated with Gag and Env band using the Image Studio Lite software. One of two blots is shown. (c) Infectivity of viral particles. A TZM-bl cell-based infectivity assay was used to calculate the infectivity of the indicated viral strains. The means and SEM of the RLU in at least three independent assays, each conducted in triplicate, are shown. (d) The means and SEM of the replication of the indicated viral strains in activated primary CD4+ T cells derived from four healthy donors are shown. Primary CD4+ T cells were infected, and viral replication was monitored by flow cytometry over a week as the percentage of Gag+ cells. (e) Area under the curve (AUC) of graph plotted in panel d. Statistical analysis was performed using the Prism software and a multiple unpaired t test (c) and an unpaired t test (e). *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
When we compared different viral strains (both laboratory-adapted and T/F viruses), we found that variations in antibody binding are due to multiple parameters, including Env sequence variability, expression levels, or conformations. To analyze Env proteins on producer cells and on viral particles, we selected four broadly neutralizing antibodies (bNAbs; PG16, 3BNC117, NIH45-46, and 10-1074) and four nonneutralizing antibodies (nnAbs; 5-25, 4-8, 1-262, and 2-59) that cover some of the major domains of Env (Table 1). Selected antibodies showed different neutralization capacities depending on the viral strain (9, 56). We produced ultracentrifuged virions from transfected 293T cells as described above. As controls, we used an IgG1 isotype for the staining, as well as an Env deletion HIV-1 (NL4-3ΔEnv). The producer cells were collected and stained for Env and Gag.
TABLE 1.
Epitope specificity of the antibodies used in this study
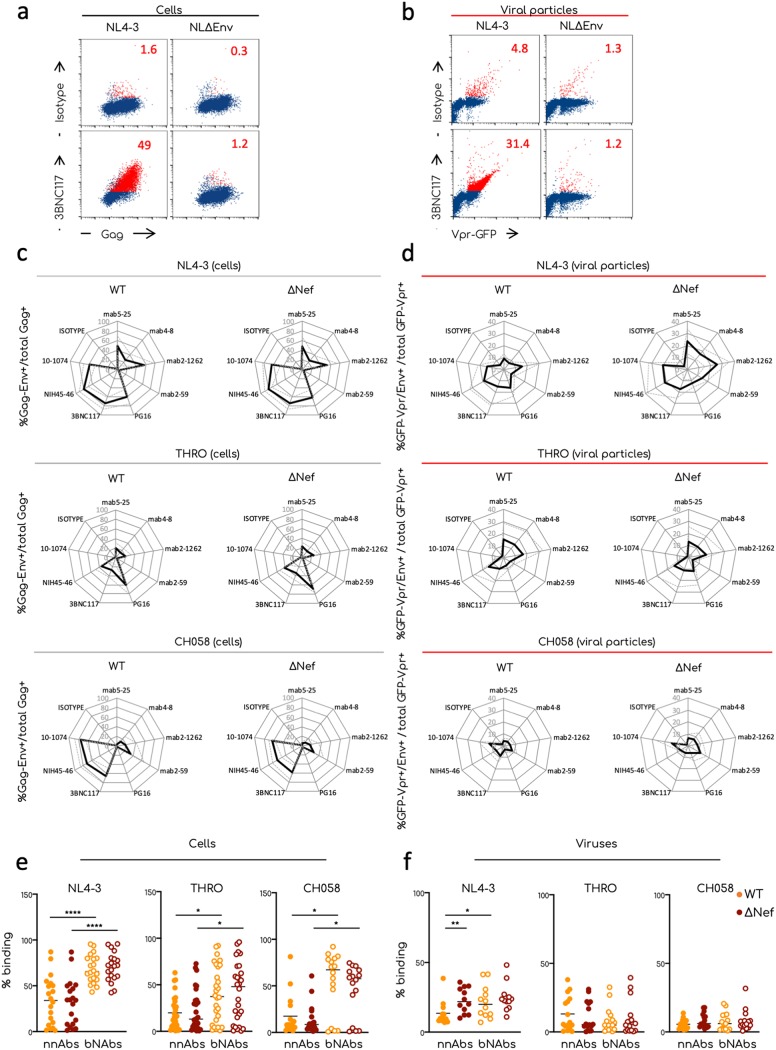
Figure 4a and b shows an example of Env staining of NL4-3 and NL4-3ΔEnv-expressing cells and virions with 3BNC117. The isotype, as expected, did not stain the cells and generated a slight background on NL4-3 and NL4-3ΔEnv virions. 3BNC117 bound to NL4-3-transfected cells, with 50% of double Gag+ and Env+ cells and did not bind to NL4-3ΔEnv-transfected cells. The antibody bound to 30% of NL4-3 virions and to <2% of NL4-3ΔEnv particles without promoting further viral aggregation. Similar results were obtained with the other antibodies tested (not shown).
FIG 4.
Comparative analysis of Env conformation at the surface of producer cells and viral particles. Transfected 293T cells and aggregated viral particles were stained for the expression of Env using the indicated antibodies. (a) Representative dot plot analyses of 293T cells transfected with the NL4-3-WT or -ΔEnv proviruses and stained for Gag and Env expression. 293T cells were plotted as Gag+/Env+ events. Numbers indicate the percentages of Gag/Env double-positive events (in red in the plots). (b) Representative dot plot analysis of viral aggregates stained to visualize Env at the surface of the virions. Numbers indicate the percentages of GFP-Vpr/Env double-positive events (in red in the plots). (c and d) Radar plot analysis showing the means (plain lines) and standard deviations (SD; dotted lines) of the Env profiles of the indicated transfected cells (c) and viral aggregates (d). The percentages were calculated by flow cytometry or flow virometry, respectively. The means and SD of at least three independent viral preparation for each viral isolate were determined. (e and f) Comparison of the binding efficiency of nnAbs and bNAbs on viral particles and producer cells. Raw data of the percentages of anti-Env antibodies binding on transfected cells (e) and on viral aggregates (f). Each dot corresponds to an individual independent staining. All bNAbs or nnAbs listed in Fig. 4 have been grouped together. Statistical analysis was performed using Prism software and the Mann-Whitney t test. *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
Therefore, our assay allows the specific detection of Env on viral aggregates.
Impact of Nef on Env profile at the surface of cells and virions.
We then calculated for each antibody the percentage of Env+ cells among Gag+ cells and of Env+ particles among GFP-Vpr+ virions. The bindings were represented with radar plots to easily visualize the “Env profile” of a given viral strain (Fig. 4c and d). To have a global vision of the binding differences between cells and viral particles we compared for each virus the mean percentage of binding of nnAbs and bNAbs (Fig. 4e and f). As we previously reported (8, 9), the CD4 binding site bNAbs 3BNC117 and NIH45-46 recognized cells infected with the three strains, whereas 10-1074 recognized NL4-3- and CH058-transfected cells. The V1/V2-specific PG16 bNAb was unable to bind cells producing CH058, but recognized NL4-3 and THRO. Nef did not alter the Env profile detected by bNAbs on transfected cells. For all viral strains, the binding of nnAbs to infected cells was significantly lower than that of the bNAbs (Fig. 4e). nnAbs 5-25 and 2-1262 (recognizing a gp41-immunodominant epitope and the CD4 binding site, respectively) recognized 50 and 20% of NL4-3- and THRO-expressing cells, respectively, whereas CH058-expressing cells were poorly or not recognized by these antibodies. The V3-specific nnAb 2-59 was unable to bind to NL4-3- or THRO-transfected cells and poorly recognized CH058 transfected cells. The CD4-induced antibody 4-8, inefficiently bound Gag+ cells. Nef did not change the Env profile recognized by nnAbs.
Env staining of viral particles was globally similar to that on transfected cells, with noticeable differences. First, we observed that NL4-3 and THRO virions were easily recognized by both nonneutralizing and neutralizing antibodies, whereas CH058 particles were barely detected (Fig. 4d and f). This may suggest that CH058 incorporate less Env trimers than the other strains. Moreover, nnAbs bound to viral particles almost as efficiently as the bNAbs, whereas this was not the case on Env-expressing cells (Fig. 4c to f). This difference was particularly evident with T/F viruses. Second, some differences in the intensity of the binding at the cell surface were lost on the viral particles. For instance, the nnAb 2-59, which inefficiently bound NL4-3-infected cells, displayed a moderate but consistent binding to viral particles. NL4-3ΔNef virions bound nnAbs significantly more efficiently than WT virions, a phenomenon that was not visible on producer cells (Fig. 4c to f). However, this difference was not observed with the two T/F viruses. bNAb binding was also slightly different on virions and infected cells. PG16 recognized less than 20% of WT and ΔNef NL4-3 virions and was almost unable to bind to WT THRO virions, whereas cells infected by those viruses are efficiently detected. PG16 binding to ΔNef THRO was consistently stronger than to WT particles. 3BNC117, 10-1074, and NIH45-46 efficiently bound WT and ΔNef NL4-3 and THRO virions. 10-1074 and NIH45-46 both barely bound CH058 virions, while displaying a strong binding to transfected cells.
Thus, antibody binding to infected cells is generally similar to that observed on virions, with some variations depending on the viral strain or the antibody used. Some tested nnAbs efficiently bind virions and not infected cells.
Impact of SERINC5 and CD4 expression in producer cells on Env detection.
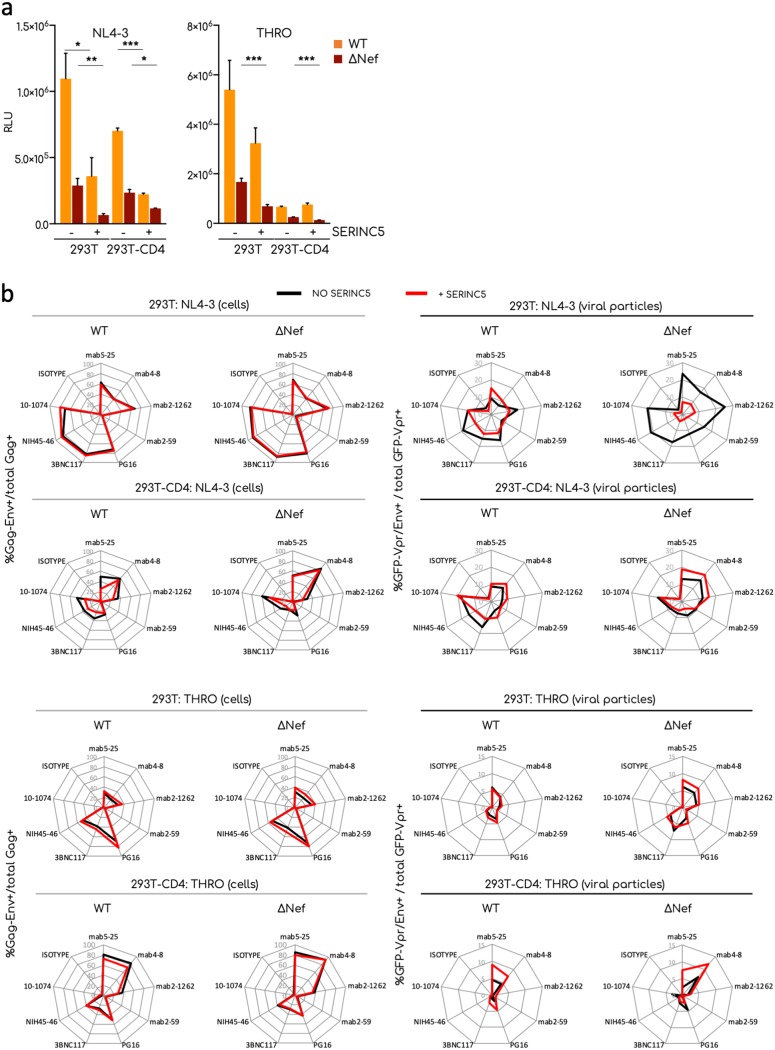
We then investigated the impact of CD4 and SERINC5 on the detection of Env proteins. SERINC5 strongly affected ΔNef and, to a lower extent, WT infectivity (39). CD4 expression has been recently suggested as modulating the effect of SERINC5 on Env fusion (43). We produced fluorescent NL4-3 and THRO virions in 293T or 293T cells stably expressing CD4 (293T-CD4), with or without transient transfection of SERINC5. To limit possible artifacts due to excessive expression of SERINC5 in producer cells, we used 20-fold less SERINC5 plasmid than proviral DNA (57). We tested the infectivity of viral particles and stained producer cells and ultracentrifuged viral particles as described above.
NL4-3 is known to be more restricted by SERINC5 than T/F viruses (41). As expected, SERINC5 reduced the infectivity of WT viruses and inhibited ΔNef virions (Fig. 5a). NL4-3 was more inhibited by SERINC5 than by THRO (27 and 63% of the residual relative infectivity, respectively) (Fig. 5a). CD4 expression in producer cells strongly reduced viral infectivity, irrespective of Nef or SERINC5. However, in the presence of CD4, SERINC5 further affected NL4-3, but not THRO, infectivity (Fig. 5a).
FIG 5.
Effect of CD4 and SERINC5 on Env conformation. 293T cells expressing or not CD4 were transfected with the indicated provirus alone or cotransfected with a plasmid expressing SERINC5-HA. (a) Infectivity of viral particles tested using the TZM-bl infectivity assay. The means and SEM of two independent experiments are shown. (b) Radar plot analyses of the Env profile in transfected cells (left) or viral aggregates (right) in the presence (red) or absence (black) of SERINC5. The percentages were calculated by flow cytometry or flow virometry, respectively. The means of two independent experiments are shown. Statistical analysis was performed using Prism software and a multiple unpaired t test. *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
Figure 5b shows the Env profiles of infected 293T cells expressing or not SERINC5 or CD4. The presence of SERINC5 did not affect these profiles, with neither WT nor ΔNef viruses. As expected, the presence of CD4 impacted the Env profile. The binding of the nnAb 4-8 was strongly enhanced for NL4-3- and THRO-transfected cells, as predicted by the fact that it recognizes a CD4-induced epitope. Binding of PG16, 3BNC117, NIH45-46, and 10-1074 bNAbs was reduced by CD4 in NL4-3-transfected cells but less in THRO-transfected cells. This reduction was more evident with NL4-3ΔNef. Interestingly, in 293T-CD4+ cells, SERINC5 induced an Env profile of NL4-3 WT similar to that observed with the ΔNef virus. THRO-expressing 293T-CD4+ cells bound anti-Env nnAbs highly efficiently, and SERINC5 expression did not affect the binding. The absence of Nef did not change this profile.
A different pattern was observed on viral particles. NL4-3ΔNef virions produced by 293T cells were significantly less recognized by both nnAbs and bNAbs in the presence of SERINC5. This suggests that Env proteins incorporated in virions have been impacted by SERINC5 in the absence of Nef. However, this effect of SERINC5 was less visible in the presence of CD4. With THRO virions produced in the absence of CD4, the binding of anti-Env antibodies was not affected by SERINC5. THRO WT or ΔNef virions produced in 293T-CD4+ cells were slightly more sensitive to anti-Env antibodies in the presence of SERINC5.
Altogether, our results indicate that SERINC5 and CD4 independently or additively change the Env profile of NL4-3 and THRO isolates in a Nef-dependent manner, reflecting the impact of the two cellular proteins on viral infectivity. However, the effect of SERINC5 and CD4 on Env accessibility varies with the viral strain and is more marked in viral particles than in infected cells.
DISCUSSION
In past years, technology improvements opened the possibility to visualize 100- to 200-nm nanoparticles using flow cytometers by coupling light scatter and fluorescence detection (26, 27, 46). Mature HIV-1 virions have sizes of between 120 and 200 nm (58) and should therefore be above the limit of detection.
About a dozen Env trimers and a few cellular proteins are present on the viral envelope (1, 12). Variation in the number of Env trimers or in Env conformation can positively or negatively impact viral infectivity (15). Cellular proteins incorporated into virions also affect viral infectivity. For instance, virion-associated ICAM-1 improves infectivity (59–61), whereas restriction factors such as SERINC5 or IFITMs proteins reduce it (38, 39, 62–64). The exact mechanism used by SERINC5 to restrict HIV-1 entry is not completely understood. A correlation between Env sensitivity to antibody neutralization and to SERINC5 has been reported, suggesting that SERINC5 activity may depend on Env conformations (41–43).
We developed a simple method to stain Env proteins at the virion surface and to analyze their levels by flow virometry. Previous flow virometry-based studies characterized the incorporation of some cellular proteins (28) or the conformation of Env proteins (30, 31) at the surfaces of individual viral particles. Fluorescent magnetic nanobeads coated with anti-Env antibodies were used to capture HIV-1 particles. One caveat of this previous approach was that only virions carrying proteins recognized by anti-Env antibody were analyzed. Here, we established a protocol based on ultracentrifugation that can be applied to any viral isolate and could be used to characterize any protein or other component incorporated in the viral membrane. The aggregation of virions by ultracentrifugation enhanced the fluorescent signal and the size of the objects to analyze, making them detectable by cytometry. We then used this technique to compare the conformation of Env at the surface of producer cells and on viral particles and to analyze the impact of CD4, Nef, and SERINC5. We used a panel of eight antibodies that covers different domains of Env to generate a so-called Env profile that was established both on infected cells and on virions.
We initially conducted several experiments to validate our protocol. We confirmed that ultracentrifugation did not decrease infectivity of the viruses and that aggregated viruses corresponded to approximately 80% of the viral population. We could not exclude that particles that are not aggregated (GFP-Vpr low) are somehow different from those that aggregate. However, aggregation did not require the presence of Env at the virion surface. Viral aggregates contained most of the infectivity of the viral preparation, further suggesting that there is not a preselection of a particular subset of viruses through the ultracentrifugation. Confocal and electron microscopy demonstrated that the objects detected by flow virometry are mainly composed of aggregated mature viral particles held together through a matrix-like structure with occasional extracellular vesicles included. This is reminiscent of the biofilm-like structure of virions reported for HTLV-1 (65). Moreover, electron microscopy indicated that aggregates did not contain detectable extracellular vesicles incorporating Env, or other cellular proteins, that could bias subsequent analyses. The method to stain viral particles is similar to that used for cells. However, instead of washing antibodies, they are progressively diluted. We confirmed the specificity of the binding by using ΔEnv HIV-1, which triggered only a background signal. Of note, incubation with anti-Env antibodies did not induce further aggregation, strongly suggesting that viral particles are not cross-linked by antibodies. Moreover, the test is robust and the Env profile of a given viral strain remains consistent from one preparation to another. We noticed that not all GFP-Vpr+ aggregates were positive for Env staining. This could be due to the variability in the number of Env proteins incorporated into virions (12) or to the presence of particles displaying defectives form of Env (i.e., gp41 stumps). These viruses are expected to be randomly present in aggregates, explaining why we observed Env– events scattered along the GFP-Vpr+ region.
As for all methods, there are some limitations associated with this flow virometry technique. The requirement of Vpr-GFP incorporation into viral particles limits its use to easy-to-transfect cells. We are currently developing other methods of staining with fluorescent dyes that may solve this problem. The current relatively low sensitivity of the technique was overcome by ultracentrifugation of virions, which is labor-intensive and may modify their properties. Ultracentrifugation, however, did not impair but rather increased viral infectivity, suggesting that this procedure did not induce Gp120 shedding from viral particles (66). Moreover, because of the acquisition settings, we were able to analyze only a fraction of the total viral particles present inside each preparation.
Despite its limits, our technique allowed an extensive and comparative analysis of the native conformation of Env in producer cells and viral particles and its accessibility to a large panel of antibodies and revealed unexpected differences.
We then sought to determine whether Nef, SERINC5, and CD4 impact Env conformations in virions and in cells. Modifying the cellular proteins present in 293T cells changed the Env profile in virions, in a manner that was, at least partially, expected. For instance, binding of nnAb 4-8, recognizing a CD4-induced epitope, was higher in viruses produced in the presence of CD4. Furthermore, ΔNef viruses were more recognized by nnAb 4-8 than WT virions. This is in agreement with a recent publication showing that viral particles produced in the presence of CD4 do bind more efficiently to anti-CD4 induced antibodies in a VCA (24). As expected, in producer cells the bNAbs were better binders than nnAbs, irrespective of the presence or absence of CD4 and SERINC5. Binding of the nnAbs was higher for NL4-3 than for the T/F viruses (9). The expression of CD4 induced the transition of Env proteins from a closed to an open state: CD4-induced epitopes were more accessible, in particular in THRO-transfected cells. Some bNAbs, in particular those targeting the CD4bs, were less efficient in the presence of CD4. ΔNef viruses showed a marked shift in their Env profile in the presence of CD4. This was also expected since CD4 is not downmodulated in the absence of Nef. The presence of SERINC5 in producer cells affected the Env profile of NL4-3 only when cells were CD4+. THRO was less sensitive to SERINC5. This is partially in agreement with recent results showing a direct interplay between SERINC5 and Env, in the presence of CD4, by a bimolecular fluorescence complementation assay (43).
Our analysis of the Env profile of virions revealed interesting features. For example, nnAbs bound as efficiently as bNAbs to viral aggregates, whereas this was not the case at the cell surface. This suggests that open Env forms are more represented on virions than in producer cells. This may be in part explained by the presence of uncleaved Env monomers at the cell surface that are not incorporated into virions (13). In addition, the reorganization of Env trimers at the virion surface required for optimal infectivity may modify the accessibility of antibodies (16). We also noticed a link between viral infectivity and the breadth of the Env profile: (i) NL4-3 and THRO virions bound to anti-Env antibodies more efficiently than CH058 virions and were two to three times more infectious; (ii) the Env profile of virions produced in the presence of CD4 was generally narrower than in the absence of CD4; (iii) NL4-3ΔNef virions produced in the presence of SERINC5 were poorly infectious and almost inaccessible to antibodies; and (iv) the amplitude of the Env profile appeared to follow the tiers categorization of the viruses analyzed, with the NL4-3 profile being wider than that of the two T/F viruses. Of note, the high levels of antibody binding to NL4-3-infected cells and viral particles might reflect the fact that this strain has been selected for efficient replication in culture in the absence of an immune pressure that may have reduced Env exposure.
Future work with a larger number of viral strains and an extended panel of anti-Env antibodies will be necessary to confirm these observations. In particular, it will be interesting to investigate directly how SERINC5 affects the exposure of other Env domains, such as the Gp41/Gp120 interface or MPER. Previous studies suggested that SERINC5 significantly increases the neutralization sensitivity to these anti-Env antibodies (41, 57, 67). Also, including Nef mutants unable to counteract SERINC5 (68) will help dissecting the complex interplay between Nef, Env, and SERINC5 proteins.
Interestingly, von Bredow et al. recently showed that some bNAbs displayed potent antibody-dependent cellular cytotoxicity activity without neutralizing viral particles and vice versa (69, 70). These results are in line with our study and suggest that differences in Env conformation at the cell surface and on viral particles may exist, with functional consequences.
To gain further insight on the differences between cells and virions, it will be interesting to investigate whether the conformation of Env at the sites of viral budding are different from other membrane domains. Gag proteins present at the plasma membrane, and in immature viral particles, are mostly unprocessed. The interactions between Gag and Env change following Gag maturation, and this could modify the conformation of Env proteins. It will be worth comparing Env profiles of mature and immature viral particles to determine whether the latter recapitulate Env conformations observed in producer cells.
MATERIALS AND METHODS
Cells.
293T cells, 293T-CD4+ cells, and TZM-bl cells were grown at 37°C and 5% CO2 in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum and antibiotics (100 U/ml penicillin, 100 μg/ml streptomycin, complete DMEM). Primary CD4+ T cells were isolated from human peripheral blood (obtained anonymously from the Etablissement Français du Sang) by Ficoll density gradient followed by positive selection with anti-CD4 magnetic beads (Miltenyi Biotec), activated with phytohemagglutinin (1 μg/ml; Oxoid) for 24 h and then cultivated in complete RPMI medium containing interleukin-2 (IL-2; 50 IU/ml; R&D Systems).
Molecular constructs.
The HIV-1 T/F infectious molecular clones pCH058 and pTHRO were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH, from John Kappes and Christina Ochsenbauer (catalog number 11919), and the pNL4-3 plasmid was described previously (71). The ΔNef plasmids were obtained with a QuikChange II XL site-directed mutagenesis kit (Agilent Technologies) according to the manufacturer’s instructions. Mutations were inserted in order to replace the first methionine and the fourth lysine of Nef by two stop codons. The clones were verified by digestion with XhoI/HindIII and BglII and sequencing. The NL4-3-GagGFP plasmid was kindly provided by H. G. Krausslich (72). The GFP-Vpr plasmid was provided by the NIH AIDS Reagent program (catalog number 11386). The pcDNA-SERINC5-HA plasmid was kindly provided by M. Pizzato (39).
Virus production.
Viruses were produced by transfection of 293T cells, either expressing or deficient in CD4, using the Turbofect reagent (Thermo Fisher Scientific). Proviral DNA and, when indicated, the GFP-Vpr plasmid (at a 1/0.3 ratio) were used. To produce viral particles in the presence of SERINC5, cells were cotransfected with the proviral DNA, pGFP-Vpr plasmid, and pSERINC5-HA plasmid at a 1/0.3/0.05 ratio, respectively. The pcDNA3.1-SERINC5-HA plasmid has been described (39). The day after the transfection, the medium was replaced with fresh medium, and viral supernatants were collected the following day. The culture medium was previously filtered (0.1 μm) to remove microparticles that could interfere with subsequent analyses. When indicated, viral supernatants were ultracentrifuged at 100,000 × g for 1 h in a SW41 rotor (Beckman Coulter). The viral pellet was resuspended in phosphate-buffered saline (PBS), filtered (0.1-μm pore size), and concentrated 100-fold. To homogenize viral resuspension, the pellets were gently shaken at 4°C for 1 h at 750 rpm with a Thermomixer (Eppendorf). Ultracentrifuged particles were then aliquoted and stored at −80°C until use. The amount of virus produced was quantified by ELISA according to the manufacturer’s instructions.
Infections.
Primary CD4+ T cells, at day 3 postactivation, were infected with the indicated virus (from 20 to 150 ng/ml of p24, depending on the virus) for 4 h at 37°C under gentle agitation in a Thermomixer. After infection, the cells were washed to remove unbound viral particles and left in culture in medium supplemented with IL-2.
Western blotting.
Transfected 293T cells or viral particles were lysed on ice in 0.1% Triton X-based buffer containing protease inhibitors (73). Lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a nitrocellulose membrane. The following primary antibodies were used: mouse monoclonal anti-HIV-1 Gag (clone 183-H12-5C [NIH AIDS Reagents Program], catalog number 1513; diluted 1:1,000), rabbit polyclonal anti-HIV-1 Nef (NIH AIDS Reagents Program; catalog number 2949; diluted 1:1,000), rabbit anti-actin (N-21 [Santa Cruz]; diluted 1:10,000), and sheep anti-HIV-1 Env (clone gp120b [NIH AIDS Reagents Program]; catalog number 288; diluted 1:1,000). Species-specific fluorescent secondary antibodies were used. Fluorescent signals were detected with a LI-COR Odyssey scanner and analyzed using ImageStudioLite software.
Analysis of viral infectivity.
The TZM-bl cell line (originally called JC.53-BL) was generated from the clone JC.53 (51) expressing a high level of CD4 and CCR5 and endogenously expressing CXCR4. Separate integrated copies of the luciferase and β-galactosidase genes under the control of the HIV-1 promoter were introduced into the JC.53 cell clone (74), and this cell lines is highly sensitive to infection by most viral strains. TZM-bl based assay was performed as previously described, with minor modifications (75). Briefly, TZM-bl cells were infected in triplicate with equivalent amount of Gag p24 (10 ng/ml) of WT or ΔNef viruses. At 48 h after infection, the relative viral infectivity was measured by a luciferase assay using Bright-Glo (Promega) reagent according to the manufacturer’s instructions. When indicated, the infectivity was measured by monitoring β-galactosidase expression using a CPRG assay (Roche).
Flow cytometry.
The anti-Env antibodies used in this study do not bind Env proteins after they have been fixed in paraformaldehyde (PFA). For this reason, we stained Env protein on unfixed cells. Transfected cells (104 to 2 × 104 cells per well) were incubated with the indicated anti-Env antibodies or isotype control (15 μg/ml) in culture medium for 45 min at 4°C. The cells were then washed and incubated for 30 min at 4°C with anti-human IgG1(H+L) Alexa Fluor-647 (1:400 dilution; Life Technologies). The cells were then washed and fixed with 4% PFA. To quantify the percentages of Gag+ cells, fixed cells were permeabilized and stained with anti-HIV-1 p24 KC57-RD1 (Beckman Coulter; diluted 1:500 in PBS supplemented with 1% bovine serum albumin [BSA], 0.05% saponin [Sigma], and 0.01% sodium azide staining buffer) for 30 min at room temperature. After washing and resuspension in PBS, the samples were acquired on a FACSCanto II (BD Biosciences) and analyzed with FlowJo software v10.
Anti-Env antibodies.
Anti-Env nnAbs and bNAbs, as well as the isotypic control mGO53, were produced as recombinant monoclonal antibodies carrying the same human IgG1 Fc region by the cotransfection of 293T or 293F cells (obtained from the American Type Culture Collection) as previously described (76). Antibodies were purified by batch/gravity-flow affinity chromatography using protein G-Sepharose 4 Fast-Flow beads (GE Healthcare).
Flow virometry.
The amount of Gag-p24 used in a single experiment depended on the final yield of the viral preparation and was the same (30 or 50 ng of Gag-p24) for all virus compared. Of note, the anti-Env antibodies used do not bind to Env proteins after PFA fixation. We thus stained Env protein before fixation of the samples and acquisition at the cytometer. A “virus master mix” was prepared in a final volume of 100 to 150 μl of culture medium supplemented with 20% fetal bovine serum (FBS) previously filtered through a 0.1-μm filter. First, 10 to 15 μl/well of virus was distributed in a 96-well round-bottom plate. Each antibody was diluted at 30 μg/ml (2× concentrated), and 10 to 15 μl was distributed into each well to yield a final concentration of 15 μg/ml. After 30 to 45 min at 4°C, 20 to 30 μl of the anti-human IgG1(H+L) Alexa Fluor 647 (1:200 dilution [Life Technologies] in medium 20% FBS) was added to a final dilution of 1:400 and left 30 min at 4°C. After the staining, we fixed our samples with 6 to 8.5 μl of a stock of PFA 32% to yield a final concentration of 4%. Samples were left for 10 min at room temperature, and then the volume was adjusted to 180 μl/well with complete medium 20% FBS. Viral particles were acquired on an Attune NxT cytometer (Thermofisher), and the results were analyzed with FlowJo 10 software.
Confocal microscopy.
PFA-fixed ultracentrifuged viral particles (pre- or postsorting) were allowed to settle onto poly-l-lysine (Sigma)-coated coverslips for 1 to 2 h. Samples were then mounted between slide and coverslip in gel mounting medium (Fluoromount-G; Invitrogen) and analyzed on an LSM-700 (Zeiss, Germany) confocal microscope equipped with a 63× Plan-Apochromat oil immersion objective and, when indicated, a 10× digital zoom. GFP-Vpr+ events were detected by exciting samples at a wavelength of 488 nm. Images were analyzed using Fiji software and assembled with the MagicMontage plugin.
Cryo-electron microscopy sample preparation.
A solution of BSA-gold tracer containing 10-nm-diameter colloidal gold particles was added to a suspension of briefly vortexed purified HIV-1 particles with a final ratio of 1:1. A small portion (5 μl) of the sample was applied to carbon-coated copper grids (Quantifoil R2/2), previously glow discharged at 2 mA and 150 × 10−1 to 180 × 10−1 Pa for 1 min in an ELMO (Corduan) glow discharge system, operated at 18°C and 95% humidity. The sample was then vitrified in a Leica EMGP system. Briefly, the excess liquid was removed by blotting with filter paper the back side of the grids for 5 s at 18°C and 95% humidity, and then the sample was rapidly frozen by plunging it in liquid ethane. The grids were stored in liquid nitrogen until image acquisition.
Correlative light and cryo-electron microscopy.
Vitrified transmission electron microscopy (TEM) grids containing fluorescently labeled HIV-1 particles were imaged at liquid nitrogen temperature on a commercial cryo-FM system (EM Cryo-CLEM; Leica), equipped with a 50 × 0.9-NA long working distance lens objective (Cryo CLEM Objective HCX PL APO 50×/0.9, Leica) and a metal halide HXP120 light source. The samples were transferred from liquid nitrogen storage only when the temperature of the cryo-correlative stage of the microscope was below −180°C. Fluorescent data were acquired using the standard GFP filter cube of the microscope system (excitation, 470/40; dichroic, 495 low pass; emission, 525/50) and an ORCA Flash 4.0LT CCD camera (Hamamatsu).
After cryo-fluorescence imaging, the vitrified samples on grids were stored in liquid nitrogen until they were used for cryo-TEM. In all images, the brightness and contrast were adjusted in order to highlight HIV-1 virions.
Cryo-electron microscopy.
Cryo-electron microscopy was performed on a Tecnai 20 equipped with a field emission gun and operated at 200 kV (Thermo Fisher company). Images were recorded using SerialEM software on a 4k × 4k camera (Ultrascan from Gatan) and a Falcon II (FEI, Thermo Fisher) direct electron detector, with a 14 μm pixel size. Digital images were acquired at magnifications of 6,700 or 50,000, binning 2, corresponding to pixel sizes of 15.52 and 1.99 nm, respectively. For high-magnification images, the defocus was −3 μm.
Sorting of viral particles.
The sorting was carried out on a MoFLO Astrios “Beckman Coulter” sorter equipped with an EQ module specifically developed to detect nanoparticles and with 488- and 561-nm lasers at 200 mW. The sorting was on an SSC parameter of laser 561, with the threshold set to 0.012% in order to have maximum 300 eps with buffer filtered at 0.02 μm. An M2 mask was added in front of the FSC. All SSC and FSC parameters are viewed in logarithmic mode. The sorting was carried out with a 70-μm nozzle at a pressure of 60 lb/in2 and a differential pressure with the sample of 0.3 to 0.4 lb/in2. The sheath liquid NaCl 0.9% (Revol Company) was filtered on a 0.04-μm filter. Gag levels in sorted viral fractions were measured by an ultrasensitive home-made duplex digital ELISA Simoa assay (Quanterix).
ACKNOWLEDGMENTS
We thank M. M. Rajah for critical readings and editing of the manuscript. We thank members of the Virus and Immunity Unit for helpful discussions and assistance. We thank the Imagopole (Institut Pasteur) for help in image acquisition. We especially thank J. Elis for his support.
Work in the Virus and Immunity Unit is funded by the Institut Pasteur, ANRS, Sidaction, the Vaccine Research Institute (ANR-10-LABX-77), Labex IBEID (ANR-10-LABX-62-IBEID), “TIMTAMDEN” ANR-14-CE14-0029, “CHIKV-Viro-Immuno” ANR-14-CE14-0015-01, and the Gilead HIV Cure Program.
REFERENCES
- 1.Burnie J, Guzzo C, Burnie J, Guzzo C. 2019. The incorporation of host proteins into the external HIV-1 envelope. Viruses 11:85. doi: 10.3390/v11010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen B. 2019. Molecular mechanism of HIV-1 entry. Trends Microbiol 27:878–891. doi: 10.1016/j.tim.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malbec M, Porrot F, Rua R, Horwitz J, Klein F, Halper-Stromberg A, Scheid JF, Eden C, Mouquet H, Nussenzweig MC, Schwartz O. 2013. Broadly neutralizing antibodies that inhibit HIV-1 cell to cell transmission. J Exp Med 210:2813–2821. doi: 10.1084/jem.20131244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dufloo J, Bruel T, Schwartz O. 2018. HIV-1 cell-to-cell transmission and broadly neutralizing antibodies. Retrovirology 15:51. doi: 10.1186/s12977-018-0434-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burton DR, Mascola JR. 2015. Antibody responses to envelope glycoproteins in HIV-1 infection. Nat Immunol 16:571–576. doi: 10.1038/ni.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anand SP, Prevost J, Baril S, Richard J, Medjahed H, Chapleau JP, Tolbert WD, Kirk S, Smith AB III, Wines BD, Kent SJ, Hogarth PM, Parsons MS, Pazgier M, Finzi A. 2019. Two families of Env antibodies efficiently engage Fc-gamma receptors and eliminate HIV-1-infected cells. J Virol 93:e01823-18. doi: 10.1128/JVI.01823-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richard J, Prevost J, Alsahafi N, Ding S, Finzi A. 2018. Impact of HIV-1 envelope conformation on ADCC responses. Trends Microbiol 26:253–265. doi: 10.1016/j.tim.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 8.Bruel T, Guivel-Benhassine F, Amraoui S, Malbec M, Richard L, Bourdic K, Donahue DA, Lorin V, Casartelli N, Noel N, Lambotte O, Mouquet H, Schwartz O. 2016. Elimination of HIV-1-infected cells by broadly neutralizing antibodies. Nat Commun 7:10844. doi: 10.1038/ncomms10844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruel T, Guivel-Benhassine F, Lorin V, Lortat-Jacob H, Baleux F, Bourdic K, Noel N, Lambotte O, Mouquet H, Schwartz O. 2017. Lack of ADCC breadth of human nonneutralizing anti-HIV-1 antibodies. J Virol 91:e02440-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis GK, Pazgier M, Evans DT, Ferrari G, Bournazos S, Parsons MS, Bernard NF, Finzi A. 2017. Beyond viral neutralization. AIDS Res Hum Retroviruses 33:760–764. doi: 10.1089/AID.2016.0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richard J, Ding S, Finzi A. 2017. Unlocking HIV-1 Env: implications for antibody attack. AIDS Res Ther 14:42. doi: 10.1186/s12981-017-0168-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pang Y, Song H, Kim JH, Hou X, Cheng W. 2014. Optical trapping of individual human immunodeficiency viruses in culture fluid reveals heterogeneity with single-molecule resolution. Nat Nanotechnol 9:624–630. doi: 10.1038/nnano.2014.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moulard M, Hallenberger S, Garten W, Klenk HD. 1999. Processing and routage of HIV glycoproteins by furin to the cell surface. Virus Res 60:55–65. doi: 10.1016/s0168-1702(99)00002-7. [DOI] [PubMed] [Google Scholar]
- 14.Checkley MA, Luttge BG, Freed EO. 2011. HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. J Mol Biol 410:582–608. doi: 10.1016/j.jmb.2011.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeSantis MC, Kim JH, Song H, Klasse PJ, Cheng W. 2016. Quantitative correlation between infectivity and Gp120 density on HIV-1 virions revealed by optical trapping virometry. J Biol Chem 291:13088–13097. doi: 10.1074/jbc.M116.729210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chojnacki J, Staudt T, Glass B, Bingen P, Engelhardt J, Anders M, Schneider J, Muller B, Hell SW, Krausslich HG. 2012. Maturation-dependent HIV-1 surface protein redistribution revealed by fluorescence nanoscopy. Science 338:524–528. doi: 10.1126/science.1226359. [DOI] [PubMed] [Google Scholar]
- 17.Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. 2013. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. 2013. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward AB, Wilson IA. 2017. The HIV-1 envelope glycoprotein structure: nailing down a moving target. Immunol Rev 275:21–32. doi: 10.1111/imr.12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stadtmueller BM, Bridges MD, Dam KM, Lerch MT, Huey-Tubman KE, Hubbell WL, Bjorkman PJ. 2018. DEER spectroscopy measurements reveal multiple conformations of HIV-1 SOSIP envelopes that show similarities with envelopes on native virions. Immunity 49:235–246. doi: 10.1016/j.immuni.2018.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stieh DJ, King DF, Klein K, Aldon Y, McKay PF, Shattock RJ. 2015. Discrete partitioning of HIV-1 Env forms revealed by viral capture. Retrovirology 12:81. doi: 10.1186/s12977-015-0207-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alsahafi N, Bakouche N, Kazemi M, Richard J, Ding S, Bhattacharyya S, Das D, Anand SP, Prevost J, Tolbert WD, Lu H, Medjahed H, Gendron-Lepage G, Ortega Delgado GG, Kirk S, Melillo B, Mothes W, Sodroski J, Smith AB III, Kaufmann DE, Wu X, Pazgier M, Rouiller I, Finzi A, Munro JB. 2019. An asymmetric opening of HIV-1 envelope mediates antibody-dependent cellular cytotoxicity. Cell Host Microbe 25:578–587. doi: 10.1016/j.chom.2019.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bruel T, Schwartz O. 2019. HIV-1 envelope FRETted over by antibodies. Cell Host Microbe 25:767–768. doi: 10.1016/j.chom.2019.05.009. [DOI] [PubMed] [Google Scholar]
- 24.Ding S, Gasser R, Gendron-Lepage G, Medjahed H, Tolbert WD, Sodroski J, Pazgier M, Finzi A. 2019. CD4 incorporation into HIV-1 viral particles exposes envelope epitopes recognized by CD4-induced antibodies. J Virol doi: 10.1128/JVI.01403-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang VA, Renner TM, Varette O, Le Boeuf F, Wang J, Diallo JS, Bell JC, Langlois MA. 2016. Single-particle characterization of oncolytic vaccinia virus by flow virometry. Vaccine 34:5082–5089. doi: 10.1016/j.vaccine.2016.08.074. [DOI] [PubMed] [Google Scholar]
- 26.Zamora JLR, Aguilar HC. 2018. Flow virometry as a tool to study viruses. Methods 134-135:87–97. doi: 10.1016/j.ymeth.2017.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lippe R. 2018. Flow virometry: a powerful tool to functionally characterize viruses. J Virol 92:e01765-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arakelyan A, Fitzgerald W, Margolis L, Grivel JC. 2013. Nanoparticle-based flow virometry for the analysis of individual virions. J Clin Invest 123:3716–3727. doi: 10.1172/JCI67042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaudin R, Barteneva NS. 2015. Sorting of small infectious virus particles by flow virometry reveals distinct infectivity profiles. Nat Commun 6:6022. doi: 10.1038/ncomms7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landowski M, Dabundo J, Liu Q, Nicola AV, Aguilar HC. 2014. Nipah virion entry kinetics, composition, and conformational changes determined by enzymatic virus-like particles and new flow virometry tools. J Virol 88:14197–14206. doi: 10.1128/JVI.01632-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arakelyan A, Fitzgerald W, King DF, Rogers P, Cheeseman HM, Grivel JC, Shattock RJ, Margolis L. 2017. Flow virometry analysis of envelope glycoprotein conformations on individual HIV virions. Sci Rep 7:948. doi: 10.1038/s41598-017-00935-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu M, Ma X, Castillo-Menendez LR, Gorman J, Alsahafi N, Ermel U, Terry DS, Chambers M, Peng D, Zhang B, Zhou T, Reichard N, Wang K, Grover JR, Carman BP, Gardner MR, Nikic-Spiegel I, Sugawara A, Arthos J, Lemke EA, Smith AB III, Farzan M, Abrams C, Munro JB, McDermott AB, Finzi A, Kwong PD, Blanchard SC, Sodroski JG, Mothes W. 2019. Associating HIV-1 envelope glycoprotein structures with states on the virus observed by smFRET. Nature 568:415–419. doi: 10.1038/s41586-019-1101-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montefiori DC, Roederer M, Morris L, Seaman MS. 2018. Neutralization tiers of HIV-1. Curr Opin HIV AIDS 13:128–136. doi: 10.1097/COH.0000000000000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laguette N, Bregnard C, Benichou S, Basmaciogullari S. 2010. Human immunodeficiency virus (HIV) type-1, HIV-2 and simian immunodeficiency virus Nef proteins. Mol Aspects Med 31:418–433. doi: 10.1016/j.mam.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 35.Fackler OT, Baur AS. 2002. Live and let die: Nef functions beyond HIV replication. Immunity 16:493–497. doi: 10.1016/s1074-7613(02)00307-2. [DOI] [PubMed] [Google Scholar]
- 36.Schiavoni I, Trapp S, Santarcangelo AC, Piacentini V, Pugliese K, Baur A, Federico M. 2004. HIV-1 Nef enhances both membrane expression and virion incorporation of Env products: a model for the Nef-dependent increase of HIV-1 infectivity. J Biol Chem 279:22996–23006. doi: 10.1074/jbc.M312453200. [DOI] [PubMed] [Google Scholar]
- 37.Usami Y, Gottlinger H. 2013. HIV-1 Nef responsiveness is determined by Env variable regions involved in trimer association and correlates with neutralization sensitivity. Cell Rep 5:802–812. doi: 10.1016/j.celrep.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Usami Y, Wu Y, Gottlinger HG. 2015. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature 526:218–223. doi: 10.1038/nature15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosa A, Chande A, Ziglio S, De Sanctis V, Bertorelli R, Goh SL, McCauley SM, Nowosielska A, Antonarakis SE, Luban J, Santoni FA, Pizzato M. 2015. HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature 526:212–217. doi: 10.1038/nature15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fackler OT. 2015. Spotlight on HIV-1 Nef: SERINC3 and SERINC5 identified as restriction factors antagonized by the pathogenesis factor. Viruses 7:6730–6738. doi: 10.3390/v7122970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beitari S, Ding S, Pan Q, Finzi A, Liang C. 2017. Effect of HIV-1 Env on SERINC5 Antagonism. J Virol 91:e02214-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sood C, Marin M, Chande A, Pizzato M, Melikyan GB. 2017. SERINC5 protein inhibits HIV-1 fusion pore formation by promoting functional inactivation of envelope glycoproteins. J Biol Chem 292:6014–6026. doi: 10.1074/jbc.M117.777714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X, Shi J, Qiu X, Chai Q, Frabutt DA, Schwartz RC, Zheng Y-H, Zhang X, Shi J, Qiu X, Chai Q, Frabutt DA, Schwartz RC, Zheng Y-H. 2019. CD4 expression and Env conformation are critical for HIV-1 restriction by SERINC5. J Virol 93:e00544-19. doi: 10.1128/JVI.00544-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol 84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ochsenbauer C, Edmonds TG, Ding H, Keele BF, Decker J, Salazar MG, Salazar-Gonzalez JF, Shattock R, Haynes BF, Shaw GM, Hahn BH, Kappes JC. 2012. Generation of transmitted/founder HIV-1 infectious molecular clones and characterization of their replication capacity in CD4 T lymphocytes and monocyte-derived macrophages. J Virol 86:2715–2728. doi: 10.1128/JVI.06157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bonar MM, Tilton JC. 2017. High sensitivity detection and sorting of infectious human immunodeficiency virus (HIV-1) particles by flow virometry. Virology 505:80–90. doi: 10.1016/j.virol.2017.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paxton W, Connor RI, Landau NR. 1993. Incorporation of Vpr into human immunodeficiency virus type 1 virions: requirement for the p6 region of gag and mutational analysis. J Virol 67:7229–7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McDonald D, Vodicka MA, Lucero G, Svitkina TM, Borisy GG, Emerman M, Hope TJ. 2002. Visualization of the intracellular behavior of HIV in living cells. J Cell Biol 159:441–452. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Campbell EM, Perez O, Melar M, Hope TJ. 2007. Labeling HIV-1 virions with two fluorescent proteins allows identification of virions that have productively entered the target cell. Virology 360:286–293. doi: 10.1016/j.virol.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muthumani K, Montaner LJ, Ayyavoo V, Weiner DB. 2000. Vpr-GFP virion particle identifies HIV-infected targets and preserves HIV-1Vpr function in macrophages and T-cells. DNA Cell Biol 19:179–188. doi: 10.1089/104454900314564. [DOI] [PubMed] [Google Scholar]
- 51.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol 72:2855–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Passaes CPB, Bruel T, Decalf J, David A, Angin M, Monceaux V, Muller-Trutwin M, Noel N, Bourdic K, Lambotte O, Albert ML, Duffy D, Schwartz O, Saez-Cirion A. 2017. Ultrasensitive HIV-1 p24 assay detects single infected cells and differences in reservoir induction by latency reversal agents. J Virol 91:e02296-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sartori A, Gatz R, Beck F, Rigort A, Baumeister W, Plitzko JM. 2007. Correlative microscopy: bridging the gap between fluorescence light microscopy and cryo-electron tomography. J Struct Biol 160:135–145. doi: 10.1016/j.jsb.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 54.Hampton CM, Strauss JD, Ke Z, Dillard RS, Hammonds JE, Alonas E, Desai TM, Marin M, Storms RE, Leon F, Melikyan GB, Santangelo PJ, Spearman PW, Wright ER. 2017. Correlated fluorescence microscopy and cryo-electron tomography of virus-infected or transfected mammalian cells. Nat Protoc 12:150–167. doi: 10.1038/nprot.2016.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Campbell EM, Nunez R, Hope TJ. 2004. Disruption of the actin cytoskeleton can complement the ability of Nef to enhance human immunodeficiency virus type 1 infectivity. J Virol 78:5745–5755. doi: 10.1128/JVI.78.11.5745-5755.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, Phogat A, Chakrabarti B, Li Y, Connors M, Pereyra F, Walker BD, Wardemann H, Ho D, Wyatt RT, Mascola JR, Ravetch JV, Nussenzweig MC. 2009. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 57.Schulte B, Selyutina A, Opp S, Herschhorn A, Sodroski JG, Pizzato M, Diaz-Griffero F. 2018. Localization to detergent-resistant membranes and HIV-1 core entry inhibition correlate with HIV-1 restriction by SERINC5. Virology 515:52–65. doi: 10.1016/j.virol.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Briggs JA, Wilk T, Welker R, Krausslich HG, Fuller SD. 2003. Structural organization of authentic, mature HIV-1 virions and cores. EMBO J 22:1707–1715. doi: 10.1093/emboj/cdg143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tardif MR, Tremblay MJ. 2003. Presence of host ICAM-1 in human immunodeficiency virus type 1 virions increases productive infection of CD4+ T lymphocytes by favoring cytosolic delivery of viral material. J Virol 77:12299–12309. doi: 10.1128/jvi.77.22.12299-12309.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bounou S, Giguere JF, Cantin R, Gilbert C, Imbeault M, Martin G, Tremblay MJ. 2004. The importance of virus-associated host ICAM-1 in human immunodeficiency virus type 1 dissemination depends on the cellular context. FASEB J 18:1294–1296. doi: 10.1096/fj.04-1755fje. [DOI] [PubMed] [Google Scholar]
- 61.Bounou S, Leclerc JE, Tremblay MJ. 2002. Presence of host ICAM-1 in laboratory and clinical strains of human immunodeficiency virus type 1 increases virus infectivity and CD4+-T-cell depletion in human lymphoid tissue, a major site of replication in vivo. J Virol 76:1004–1014. doi: 10.1128/jvi.76.3.1004-1014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Firrito C, Bertelli C, Vanzo T, Chande A, Pizzato M. 2018. SERINC5 as a new restriction factor for human immunodeficiency virus and murine leukemia virus. Annu Rev Virol 5:323–340. doi: 10.1146/annurev-virology-092917-043308. [DOI] [PubMed] [Google Scholar]
- 63.Compton AA, Bruel T, Porrot F, Mallet A, Sachse M, Euvrard M, Liang C, Casartelli N, Schwartz O. 2014. IFITM proteins incorporated into HIV-1 virions impair viral fusion and spread. Cell Host Microbe 16:736–747. doi: 10.1016/j.chom.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tartour K, Appourchaux R, Gaillard J, Nguyen XN, Durand S, Turpin J, Beaumont E, Roch E, Berger G, Mahieux R, Brand D, Roingeard P, Cimarelli A. 2014. IFITM proteins are incorporated onto HIV-1 virion particles and negatively imprint their infectivity. Retrovirology 11:103. doi: 10.1186/s12977-014-0103-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pais-Correia AM, Sachse M, Guadagnini S, Robbiati V, Lasserre R, Gessain A, Gout O, Alcover A, Thoulouze MI. 2010. Biofilm-like extracellular viral assemblies mediate HTLV-1 cell-to-cell transmission at virological synapses. Nat Med 16:83–89. doi: 10.1038/nm.2065. [DOI] [PubMed] [Google Scholar]
- 66.Chertova E, Bess JW Jr, Crise BJ, Sowder IR, Schaden TM, Hilburn JM, Hoxie JA, Benveniste RE, Lifson JD, Henderson LE, Arthur LO. 2002. Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), Is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. J Virol 76:5315–5325. doi: 10.1128/jvi.76.11.5315-5325.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lai RP, Yan J, Heeney J, McClure MO, Gottlinger H, Luban J, Pizzato M. 2011. Nef decreases HIV-1 sensitivity to neutralizing antibodies that target the membrane-proximal external region of TMgp41. PLoS Pathog 7:e1002442. doi: 10.1371/journal.ppat.1002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trautz B, Wiedemann H, Luchtenborg C, Pierini V, Kranich J, Glass B, Krausslich HG, Brocker T, Pizzato M, Ruggieri A, Brugger B, Fackler OT. 2017. The host-cell restriction factor SERINC5 restricts HIV-1 infectivity without altering the lipid composition and organization of viral particles. J Biol Chem 292:13702–13713. doi: 10.1074/jbc.M117.797332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.von Bredow B, Andrabi R, Grunst M, Grandea AG, Le K, Song G, Berndsen ZT, Porter K, Pallesen J, Ward AB, Burton DR, Evans DT. 2019. Differences in the binding affinity of an HIV-1 V2 apex-specific antibody for the SIVsmm/mac envelope glycoprotein uncouple antibody-dependent cellular cytotoxicity from neutralization. mBio 10:e01255-19. doi: 10.1128/mBio.01255-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.von Bredow B, Arias JF, Heyer LN, Moldt B, Le K, Robinson JE, Zolla-Pazner S, Burton DR, Evans DT. 2016. Comparison of antibody-dependent cell-mediated cytotoxicity and virus neutralization by HIV-1 Env-specific monoclonal antibodies. J Virol 90:6127–6139. doi: 10.1128/JVI.00347-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol 59:284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Muller B, Daecke J, Fackler OT, Dittmar MT, Zentgraf H, Krausslich HG. 2004. Construction and characterization of a fluorescently labeled infectious human immunodeficiency virus type 1 derivative. J Virol 78:10803–10813. doi: 10.1128/JVI.78.19.10803-10813.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Qian SB, Bennink JR, Yewdell JW. 2005. Quantitating defective ribosome products. Methods Mol Biol 301:271–281. doi: 10.1385/1-59259-895-1:271. [DOI] [PubMed] [Google Scholar]
- 74.Derdeyn CA, Decker JM, Sfakianos JN, Wu X, O’Brien WA, Ratner L, Kappes JC, Shaw GM, Hunter E. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J Virol 74:8358–8367. doi: 10.1128/jvi.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Montefiori DC. 2009. Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol Biol 485:395–405. doi: 10.1007/978-1-59745-170-3_26. [DOI] [PubMed] [Google Scholar]
- 76.Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, Scheid JF, Halper-Stromberg A, Gnanapragasam PN, Spencer DI, Seaman MS, Schuitemaker H, Feizi T, Nussenzweig MC, Bjorkman PJ. 2012. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A 109:E3268–E3277. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Walker LM, Protocol Group Principal Investigators, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Protocol GPI, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, Hurley A, Myung S, Boulad F, Poignard P, Burton DR, Pereyra F, Ho DD, Walker BD, Seaman MS, Bjorkman PJ, Chait BT, Nussenzweig MC. 2011. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]