Influenza viruses escape immunity through continuous antigenic changes that occur predominantly on the viral hemagglutinin (HA). Induction of broadly neutralizing antibodies (bnAbs) targeting conserved epitopes following vaccination is a goal of universal influenza vaccines and advantageous in protecting hosts against virus evolution and antigenic drift. To date, most of the discovered bnAbs bind either to conserved sites in the stem region or to the sialic acid-binding pocket. Generally, antibodies targeting the stem region offer broader breadth with low potency, while antibodies targeting the sialic acid-binding pocket cover narrower breadth but usually have higher potency. In this study, we identified a novel neutralizing epitope in the head region recognized by a broadly neutralizing human antibody against a broad range of H3N2 with high potency. This epitope may provide insights for future universal vaccine design.
KEYWORDS: H3N2, broadly neutralizing antibody, hemagglutinin, influenza, influenza vaccines
ABSTRACT
The discovery of potent and broadly protective influenza virus epitopes could lead to improved vaccines that are resistant to antigenic drift. Here, we describe human antibody C585, isolated from a vaccinee with remarkable serological breadth as measured by hemagglutinin inhibition (HAI). C585 binds and neutralizes multiple H3N2 strains isolated between 1968 and 2016, including strains that emerged up to 4 years after B cells were isolated from the vaccinated donor. The crystal structure of C585 Fab in complex with the HA from A/Switzerland/9715293/2013 (H3N2) shows that the antibody binds to a novel and well-conserved epitope on the globular head of H3 HA and that it differs from other antibodies not only in its epitope but in its binding geometry and hypermutated framework 3 region, thereby explaining its breadth and ability to mediate hemagglutination inhibition across decades of H3N2 strains. The existence of epitopes such as the one elucidated by C585 has implications for rational vaccine design.
IMPORTANCE Influenza viruses escape immunity through continuous antigenic changes that occur predominantly on the viral hemagglutinin (HA). Induction of broadly neutralizing antibodies (bnAbs) targeting conserved epitopes following vaccination is a goal of universal influenza vaccines and advantageous in protecting hosts against virus evolution and antigenic drift. To date, most of the discovered bnAbs bind either to conserved sites in the stem region or to the sialic acid-binding pocket. Generally, antibodies targeting the stem region offer broader breadth with low potency, while antibodies targeting the sialic acid-binding pocket cover narrower breadth but usually have higher potency. In this study, we identified a novel neutralizing epitope in the head region recognized by a broadly neutralizing human antibody against a broad range of H3N2 with high potency. This epitope may provide insights for future universal vaccine design.
INTRODUCTION
Influenza virus remains a significant, worldwide public health concern, despite the availability of licensed vaccines. Seasonal influenza infects about 1 billion people each year, causing 3 to 5 million severe illnesses and 250,000 to 500,000 deaths globally (World Health Organization [WHO]). Previous pandemics in the 20th century are estimated to have caused between 50 and 100 million deaths and with the continued threat of future pandemics, there is a need for next-generation influenza vaccines that preferentially elicit breadth of protection. While licensed vaccines have the ability to protect against well-matched prevailing seasonal epidemic strains, there is a frequent need to update the vaccine formulation based on epidemiological trends. A “next-generation” influenza vaccine that can focus immunity to key conserved, cross-protective determinants would therefore be a considerable advantage.
Given the pivotal role of hemagglutinin (HA) in the infection process and its antigenic properties (1), HA-based vaccines have been widely used and extensively studied for vaccine design, including the development of novel HA molecules to induce broadly neutralizing antibodies (2, 3). HA is a type 1 transmembrane protein that is assembled as a homotrimer from a precursor polypeptide chain (HA0), which is cleaved by host proteases into HA1 and HA2 chains in order to become fusion competent. After cleavage, HA1 and HA2 remain cross-linked through a single disulfide bond (4, 5). Structurally, HA can be divided into two domains, a membrane-distal globular domain containing the receptor-binding site (RBS) and a membrane-proximal stem structure that undergoes significant conformational change during low-pH-triggered membrane fusion. By far, most of the broadly neutralizing antibodies (bnAbs) described in the literature target two regions of HA, the RBS (6–8) and the stem region (6–12). Antibodies targeting the RBS prevent virus infection by blocking virus attachment to host cells (6, 13–17), while antistem antibodies function by preventing membrane fusion, blocking proteolytic activation of the HA0, or by ADCC (antibody-dependent cellular cytotoxicity) through Fc-mediated interactions (18–24).
Here, we report the isolation and epitope “footprinting” of a human monoclonal antibody (MAb) C585, isolated from a donor after immunization with Fluzone split vaccine, which possesses broadly neutralizing activity against a wide panel of H3N2 viruses dating back to 1968. The crystal structure of C585 Fab in complex with the HA from A/Switzerland/9715293/2013 (A/Sw/13) shows that C585 recognizes a novel epitope on the globular head, distinct from other bnAbs that target the RBS. Sequence analysis together with structural information revealed the likely mechanism of broad neutralizing activity of C585 for most of the H3 strains tested. Overall, these results support the identification of a novel epitope on H3 HA, providing additional insights into the variety of broadly neutralizing antibody responses to influenza virus and structural determinants that may contribute to the development of more broadly protective H3N2-based vaccines.
RESULTS
Serological profiling of Fluzone vaccinees and selection of donor 236 for BCR-SEQ.
MAb C585 was initially isolated from a healthy, 56-year-old male (donor 236) vaccinated in the 2013/2014 season with Fluzone trivalent, inactivated influenza vaccine (TIV) composed of A/California/7/2009 (H1N1), A/Texas/50/2012 (H3N2), and B/Massachusetts/2/2012 (Yamagata lineage). Serological profiling for functional hemagglutinin inhibition (HAI) responses on day 0 (prevaccination) and day 21 (postvaccination) against both H1N1 and H3N2 vaccinal strains deployed that season as well as against a historical panel for influenza A viruses (IAV) supported the identification of several individuals that exhibited breadth of neutralization (25). Donor 236 was confirmed to have elevated and sustained HAI responses against 15/16 H3 vaccinal strains tested and that had been deployed since 1968 (data not shown). As a result of the extensive breadth of neutralizing antibodies, donor 236 was selected as a candidate for high-throughput sequencing of his B-cell receptors (BCR-SEQ) from plasmablasts isolated 7 days after vaccination.
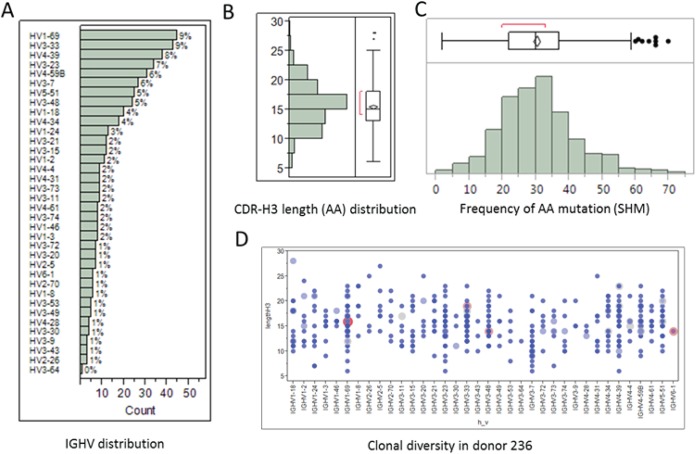
A total of 457 unique VH-VL pairs from donor 236 were identified. We analyzed the germ line gene usage of 36 immunoglobulin heavy chain (IGHV) subfamilies, and 19 immunoglobulin kappa variable (IGKV) and 24 immunoglobulin lambda variable (IGLV) light chains using IMGT/HighV-QUEST (26). Figure 1 shows the frequency distributions of IGHV gene subfamilies, CDR-H3 amino acid (AA) lengths, and number of somatic hypermutations (SHM). We also found that there were significantly diverse IGHV gene families and CDR-H3 length usages in donor 236 (Fig. 1D). Using in silico analysis of the sequences obtained, we set out to select a large diverse group of MAbs for functional characterization. We selected a list of 150 MAbs with diversities in CDR-H3 amino acid length and number of somatic hypermutations and theoretically calculated hydropathy and pI values as depicted in Fig. 2.
FIG 1.
BCR-SEQ analysis of donor 236 vaccinated with Fluzone showing the frequencies of IGHV gene subfamilies (A), total CDR-H3 amino acid (AA) lengths (B), and total number of AA mutations from VH and VL chains (C). (D) A total of 440 unique HCDR3s with frequency in the range of 1 to 4 times were found; there was no significant clonal expansion in donor 0236.
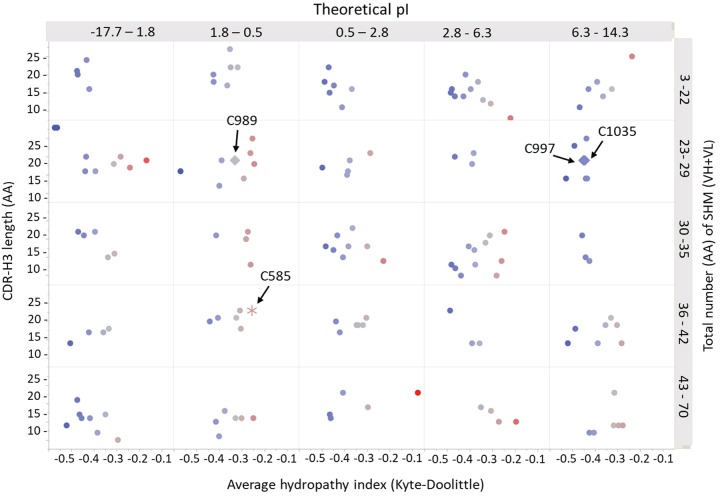
FIG 2.
Scatter plot showing the diverse properties of 150 selected MAbs from donor 236 for binding characterizations. The different properties included CDR-H3 AA lengths, total number of somatic hypermutations (SHM) from both VH and VL, average hydropathy, and calculated pI values. H3-reactive antibodies were indicated in the plot by arrows and had different properties and relatively longer CDR-H3s.
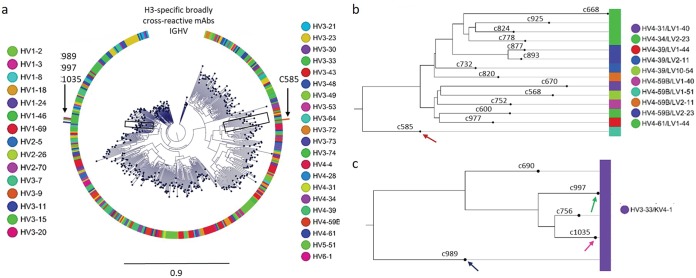
Binding and neutralization assays revealed several H3-reactive antibodies with different sequence properties and relatively longer CDR-H3s: 25 AA for singleton MAb C585 and 21 AA for clonally related MAbs C989, C997, and C1035 (Fig. 2). We concatenated the heavy and light chain sequences of 457 MAbs and aligned them to construct a phylogenetic tree for lineage analysis (Fig. 3). As seen in the circular phylogram, donor 236 had a large clonal diversity in which H3-reactive MAbs, C585, C989, C997, and C1035 were mapped (Fig. 3a). C585 was derived from the HV4-59 heavy chain germ line lineage and cognately paired with light chain germ line IGLV1-51. We also noticed a few other HV4-derived MAbs which had different sequences and paired with diverse lambda chains (Fig. 3b). Additionally, we found three more H3-specific MAbs, C989, C997, and C1035 (data not shown) that had HV3-33/KV4-1 lineages (Fig. 3c). The singleton MAb C585 had a higher SHM of 37 AA changes compared to other H3-reactive clonal families, C989, C997, and C1035 (23, 24, and 28 AA mutations, respectively). This is in line with the previous observation that singleton antibodies possessed a higher frequency of overall mutations than antibodies from clonal families (27). The matured singleton C585 MAb was chosen for studying the structural basis of H3-specific broad cross-reactivity.
FIG 3.
Phylogenetic tree analysis of donor 236 vaccinated with Fluzone. (a) Circular phylogram tree drawn using 457 unique paired heavy and light chain antibody sequences showing a highly diverse plasmablast antibody repertoire at day 7. (b) Singleton MAbs from HV4 family preferably paired with different lambda light chains and were highly mutated, including C585 (shown with an arrow). (c) Other clonally related MAbs C989, C997, and C1035 from HV3-33/KV4-1 lineages that were found to beH3 specific with broad cross-reactivity (indicated by arrows).
C585 is a human broadly neutralizing antibody against influenza virus hemagglutinin H3.
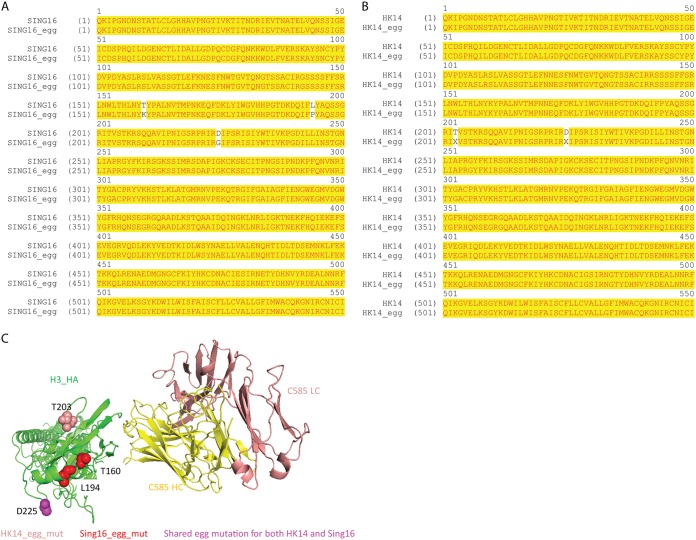
To determine its breadth of binding, C585 IgG was expressed, purified, and analyzed by biolayer interferometry (BLI) against 18 representative H3N2 HAs from 1968 to 2016. The results showed that C585 strongly binds to most of the tested HAs from H3N2 strains with few exceptions (Fig. 4). Thus, C585 can broadly recognize HA molecules representing the diversity of the time period from 1968 to 2016, during which significant antigenic drift occurred in human seasonal H3N2 viruses. In order to test the neutralization activity of MAb C585, hemagglutination inhibition and microneutralization (MN) assays were performed against a panel of representative H3N2 strains in addition to the binding assay. Consistent with the binding profile, C585 showed strong activities in HAI and MN assays against all tested H3N2 strains (Fig. 4), including four newly emerged drift variants (A/Switzerland/9715293/2013, A/HongKong/4108/2014, A/Massachusetts/28/2016, and A/Singapore INFIMH-16-0019/2016), three of which became vaccine strains in the following four seasons after C585 was isolated. Given the fact that egg-derived viruses were used for the HAI assay, we note that there were two and three point mutations identified in the A/HongKong/4108/2014 and A/Singapore INFIMH-16-0019/2016 egg-derived HA sequences, respectively, including the loss of a potential N-linked glycosylation site at N158 due to mutation at position 160 (Fig. 5A and B). Although the mutations are not located within the C585 epitope (Fig. 5C), they may affect the C585 binding through long-distance effects. This could be an explanation of the eightfold difference in the hemagglutination inhibition titers for the 2014 and 2016 viruses compared to the 2012 and 2013 viruses (Fig. 4). Nonetheless, these data suggest that MAb C585 possesses broad-spectrum binding and neutralizing activity for viruses of the H3 subtype, including future drift variants that had not yet emerged. Interestingly, the HAI profile for the clonally unrelated C997 H3-reactive MAb overlapped with many of the strains that were neutralized by C585, and additional binning studies indicate that MAbs C585 and C997 compete for the same binding site (data not shown), although the potency of the response against historical strains was less than that demonstrated by C585.
FIG 4.
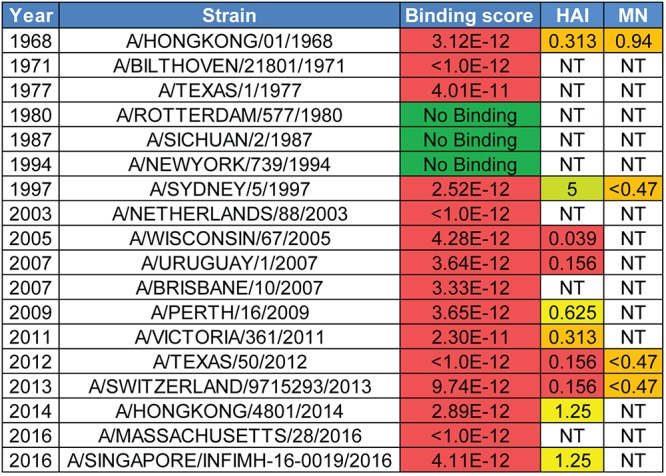
C585 IgG breadth of HA binding and virus neutralization for human H3N2 viruses. C585 IgG binding, hemagglutination inhibition, and microneutralization assay results against HAs from representative H3N2 strains. KD (dissociation constant [in molar]), hemagglutination inhibition (HAI) titers (in micrograms per milliliter), and microneutralization (MN) titers (in micrograms per milliliter) are shown. NT, not tested.
FIG 5.
Egg-derived HA sequence analysis. (A) Sequence alignment of vaccine strain Singapore INFIMH-16-0019 (SING16) and its egg-derived HA (SING16+egg). (B) Sequence alignment of vaccine strain A/HongKong/4801/2014 (HK14) and its egg-derived HA (HK14_egg). X at position 203 is mixed-sequence chromatograms that correlate to either T or I; X at position 225 is mixed-sequence chromatograms that correlate with either D or G. (C) Mapping the egg-derived mutations on the structure. Singapore INFIMH-16-0019 egg-derived mutations (red), A/HongKong/4801/2014 egg-derived mutation (salmon), and shared egg-derived mutations (purple) are indicated.
Recognition of the A/Sw/13 hemagglutinin by C585.
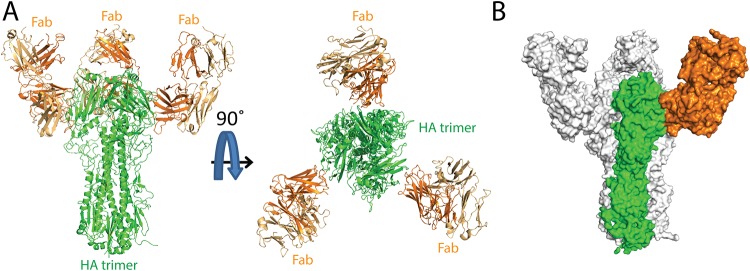
To elucidate the molecular basis for the broad H3 HA recognition by C585 MAb, the crystal structure of C585 Fab with A/Sw/13 HA was determined to 4.0-Å resolution (Table 1 and Fig. 6). Initial phases were obtained by molecular replacement using structures of H3 A/HongKong/1/1968 at 1.90-Å resolution (PDB code 4FNK) and a Fab model generated by MOE (molecular operating environment) (28) using the C585 sequence. One H3 HA trimer, bound by three C585 Fabs, is present in the crystal asymmetric unit. The C585 Fab and its interface with the HA head are well ordered with quality electron density. Additionally, the electron density of N-linked glycans at Asn38, Asn63, Asn133, Asn165, Asn246, Asn285, and Asn483 of the HA are visible. C585 Fab interacts with the HA1 head only with its heavy chain, including all three complementarity-determining regions (CDRs) (H1, H2, and H3), while the light chain makes no contact with the HA.
TABLE 1.
Data collection and refinement statistics
| Parametera | Value for Sw/13 HA-C585 Fab complexb |
|---|---|
| Data collection statistics | |
| Space group | P63 |
| Cell dimensions | |
| a, b, c (Å) | 287, 287, 182 |
| α, β, ɣ (°) | 90, 90, 120 |
| Resolution (Å) | 49.13–3.99 (4.07–3.99) |
| Rpim | 0.157 (0.920) |
| I/σI | 9.3 (1.4) |
| Completeness (%) | 99.5 (90.7) |
| Redundancy | 11.4 (9.7) |
| Refinement statistics | |
| Resolution (Å) | 47.9–4.0 |
| No. of reflections | 71,378 |
| Rwork/Rfree | 0.2092/0.2544 |
| No. of atoms | 21,597 |
| B-factors | 106.5 |
| RMSD | |
| Bond lengths (Å) | 0.014 |
| Bond angles (°) | 1.7586 |
| Ramachandran favored (%) | 91.87 |
| Ramachandran outliers (%) | 0.4 |
RMSD, root mean square deviation.
Values in parentheses are for the highest-resolution shell.
FIG 6.
Overall structure of Switzerland 2013 HA-C585 Fab complex. (A) The overall structure of the C585 Fab–Sw/13 HA complex is shown in cartoon representation from the side view and top view. The heavy chain (dark orange) and light chain (light chain) of C585 Fab and the Sw/13 HA trimer (green) are indicated. (B) Surface representation of HA-Fab interaction. One HA monomer is shown in green, with the other two monomers shown in light gray. C585 Fab is dark orange.
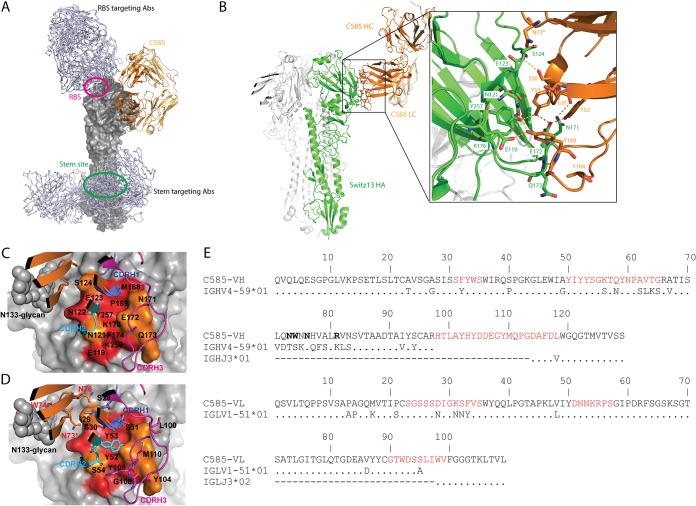
Notably, different from most other discovered H3 antibodies targeting either the RBS or the conserved stem region (Fig. 7A), C585 MAb engages an epitope on the side of the HA1 globular head domain, between the 2 β-sheets, which is close to but not overlapping with the RBS (Fig. 7B). The HA-MAb interface has extensive polar-polar interactions with a total solvent-accessible surface area of 622 Å2. The side chains of Ser30 and Ser31 from CDR-H1 form hydrogen bonds with the backbone amide of Ser124 and the carbonyl amide side chain of Asn171, respectively. A tyrosine cluster, including Tyr52, Tyr53 from CDR-H2, and Tyr109 from CDR-H3, forms an extensive hydrogen bond network with the side chains of Glu119, Asn121, Glu172, and Tyr257 from HA. The carbonyl group of Gly108 from CDR-H3 forms a hydrogen bond with the main chain amide of Gln173 from HA. By comparing C585 VH sequence with its germ line sequence, we found 14 amino acid SHMs on framework 3 (FR3) (Fig. 7E). Given this unusual feature, we examined whether these SHM residues play a role in the HA binding. Interestingly, besides Asn76, two SHM residues, Asn73 and Trp74, on FR3 directly contribute to the binding, the side chain of Asn73 forms a hydrogen bond with the hydroxyl group of Ser124 of HA, while the side chain of Trp74 contacts with Asn133 glycans (Fig. 7B to E).
FIG 7.
Recognition of the hemagglutinin off Switzerland 2013 (Sw/13) by C585 MAb. (A) C585 (shown as cartoon, colored dark orange and light orange) binds to H3 HA (shown as the surface of a monomer, colored gray) using a unique epitope comparing to other H3 HA antibodies. C05 (PDB 4FP8), S139/1 (PDB 4GMS), and F045-92 (PDB 4O58) antibodies targeting RBS and CR8020 (PDB 3SDY), CR8043 (PDB 4NM8), FI6V3 (PDB 3ZTJ), and CT149 (PDB 4UBD) targeting the stem are shown as gray lines. The RBS and stem region are circled in magenta and green, respectively. (B) Cartoon representation showing detailed interactions between C585 and Switzerland 13 HA. Interface residues are shown as sticks and colored as in Fig. 2. (C) Conservation of epitope residues in H3 HAs (1968 to 2018) is mapped on the structure of the Sw/13 HA-C585 Fab complex. Epitope residues are labeled. CDRs of heavy chain are shown as cartoons (CDRH1 in blue, CDRH2 in cyan, amd CDRH3 in magenta). Conservation of HA surface residues is indicated by color coding as follows: red, >94% conserved; orange, 74% to 94% conserved. (D) Same as panel C, but with paratope residues shown as sticks; N73, W74, and N76 are FR3 residues (shown in red); N73 and W74 are the SHM residues involved in binding (indicated by an additional asterisk). (E) Comparison of C585 heavy and light chains with their corresponding germ line segments. The different residues are shown. Identical amino acid residues are indicated by dots. The six complementarity-determining regions (CDRs) are shown in red, and the lowercase letters denote the insertions implied by the numbering scheme. C585 VH-FR3 residues that interact with the HA are shown in boldface type.
Sequence analysis of variation in the C585 epitope since 1968.
Antibodies targeting the HA head, including some antibodies that recognize dominant epitopes around the RBS, typically have a narrower breadth profile and strain specificity compared to antibodies targeting the more conserved HA2 stem. This is because the globular head region undergoes frequent antigenic variation, whereas the stem is typically not subject to immune pressure. Furthermore, the RBS region is smaller than a typical antibody footprint, and surrounding variable residues often limit the breadth of antibodies binding near the RBS. Our binding data showed that C585 recognizes most of the representative human H3N2 virus strains from 1968 to 2016, except for the strains evaluated between 1980 and 1994 (Fig. 4). To examine whether the binding correlates with conservation of this epitope, we performed two multisequence alignments using the ClustalW2 program: (i) 7,057 HA sequences from H3N2 influenza viruses circulating since 1968 available in the Influenza Virus Resource at the NCBI database, and (ii) 18 sequences of representative HAs from H3N2 influenza viruses circulating between 1968 and 2016 used for our binding assay. Residue conservation was calculated at each position of the epitope and mapped onto the A/Sw/13 HA surface (Fig. 7C and D). Of 14 C585 epitope residues, nine (Glu119, Asn122, Glu123, Met168, Pro169, Phe174, Lys176, Tyr257, and Lys259) have been highly conserved (>94%) among H3 HAs since 1968, and the remaining epitope residues are moderately conserved (74 to 94%) (Table 2). Alignment of the 18 HAs used in our binding assay showed good correlation of epitope conservation with the large-scale alignment result: Glu119, Glu123, Met168, Pro169, Asn171, Phe174, Lys176, Tyr257, and Lys259 are 100% conserved among the 18 selected HAs, while residues 121 (Asn/Ile), 122 (Asn/Thr/Asp), 124 (Ser/Gly/Asp), 172 (Glu/Asp/Gly), and 173 (Gln/Lys/Asn) are variable positions (Table 3). Therefore, the epitope variation among HAs from 1968 to 2016 is well represented by the HA sequences used in the binding assay.
TABLE 2.
Sequence conservation of the C585 epitope across the 7,057 influenza virus H3 HAs
|
HA residue no. |
% identitya |
|---|---|
| 119 | E (100) |
| 121 | N (82); K (11); I (5) |
| 122 | N (95); D (3) |
| 123 | E (100) |
| 124 | S (92); G (4); D (3) |
| 168 | M (99) |
| 169 | P (100) |
| 171 | N (87); K (12) |
| 172 | E (91); D (5); G (4) |
| 173 | Q (74); K (21) |
| 174 | F (100) |
| 176 | K (100) |
| 257 | Y (100) |
| 259 | K (100) |
The identity of the residue in HA (A/Sw/13) and the most common residue(s) at each position in the epitope are listed with percent identity for the most common residue(s).
TABLE 3.
A/Sw/13 HA contact residues with MAb C585 and conservation in selected influenza virus HA strains
| Yr | Strain | Binding scorea |
Contact residueb
in HA epitope |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 119 | 121 | 122 | 123 | 124 | 168 | 169 | 171 | 172 | 173 | 174 | 176 | 257 | 259 | |||
| 1968 | A/HongKong/8/1968 | 3 | . | I | T | . | G | . | . | . | D | N | . | . | . | . |
| 1971 | A/Bilthoven/21801/1971 | 3 | . | I | T | . | G | . | . | . | D | N | . | . | . | . |
| 1977 | A/Texas/1/1977 | 3 | . | I | . | . | G | . | . | . | D | N | . | . | . | . |
| 1980 | A/Rotterdam/577/1980 | 0 | . | I | . | . | G | . | . | . | G | N | . | . | . | . |
| 1987 | A/Sichuan/2/1987 | 0 | . | I | . | . | D | . | . | . | G | K | . | . | . | . |
| 1994 | A/NewYork/739/1994 | 0 | . | I | . | . | D | . | . | . | G | K | . | . | . | . |
| 1997 | A/Sydney/5/1997 | 3 | . | . | . | . | . | . | . | . | D | K | . | . | . | . |
| 2003 | A/Netherlands/88/2003 | 3 | . | . | . | . | . | . | . | . | . | K | . | . | . | . |
| 2005 | A/Wisconsin/67/2005 | 3 | . | . | D | . | . | . | . | . | . | K | . | . | . | . |
| 2007 | A/Uruguay/716/2007 | 3 | . | . | . | . | . | . | . | . | . | K | . | . | . | . |
| 2007 | A/Brisbane/10/2007 | 3 | . | . | . | . | . | . | . | . | . | K | . | . | . | . |
| 2009 | A/Perth/16/2009 | 3 | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 2011 | A/Victoria/361/2011 | 3 | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 2012 | A/Texas/50/2012 | 3 | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 2013 | A/Switzerland/9715293/2013 | 3 | E | N | N | E | S | M | P | N | E | Q | F | K | Y | K |
| 2014 | A/HongKong/4801/2014 | 3 | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 2016 | A/Massachusetts/28/2016 | 3 | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 2016 | A/Singapore/INFIMH-16-0019/2016 | 3 | . | K | . | . | . | . | . | K | . | . | . | . | . | . |
| 2016 | A/Sweden/96/2016 (3C.2a2) | NT | . | K | . | . | . | . | . | . | . | . | . | . | . | . |
| 2017 | A/British_Columbia/003/2017 (3C.2a1) | NT | . | K | . | . | . | . | . | K | . | . | . | . | . | . |
| 2017 | A/Newcastele/77/2017 (3C.3a) | NT | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
A KD of <1 nM was given a score of 3, and no detected binding is given a score of 0. NT, nor tested.
Residue variations from Sw/13 HA are shown in letters with conserved residues shown as dots.
Next, we examined epitope variations that abolished binding to MAb C585. Multiple residue variations compared to the A/Sw/13 HA were identified. Of these variations, Glu172 to Gly is the only common mutation shared by all HAs not binding to C585 in our test, indicating that Glu172 plays an important role in the C585-HA interaction (Table 3). This observation is consistent with the crystal structure in that the carboxyl group of Glu172 forms two hydrogen bonds with both Tyr52 and Tyr53 from CDRH2 of C585 (Fig. 7B). In contrast, mutation of Glu172 to the similar residue Asp can be tolerated for C585 binding, as Asp is likely to maintain the hydrogen bonds by its carboxyl group (Table 3).
Finally, we analyzed the tolerance of C585 for its epitope variation based on the structural information. In the A/Sw/13 HA, asparagine residues at positions 121 and 171 form hydrogen bonds with the hydroxyl group of Tyr109 and the carbonyl group of Ser31 from C585, respectively (Fig. 7B). The most frequent variants at the 121 and 171 sites are lysine mutations (Table 2), whose primary amine groups could potentially serve as proton donors to maintain the hydrogen bonds. For the Ile substitution at the 121 site (Table 3), the hydrophobic moiety of the side chain is likely to fit into the hydrophobic pocket formed by Tyr52, Tyr53, and Tyr109 of C585, therefore compensating for the loss of hydrogen bonding interactions. Moreover, main chain interactions at the antigen-antibody interface, such as where Ser30 and Gly108 of C585 interact with the amide groups of Ser124 and Glu173 from HA, are not likely to be sensitive to side chain mutations (Fig. 7B). A similar epitope tolerance mechanism was observed in a RBS-targeting antibody (7). Together, these structural features may provide a mechanism for C585 to tolerate most of the natural variation in its epitope on HAs from 1968 to 2016.
Moreover, sequence analysis of the C585 epitope residues from more recent viruses was performed (Table 3). The result showed that the C585 epitope residues are conserved in these recent strains, indicating potential neutralization activity against future viruses.
Glycosylation around the C585 binding site on H3 HA.
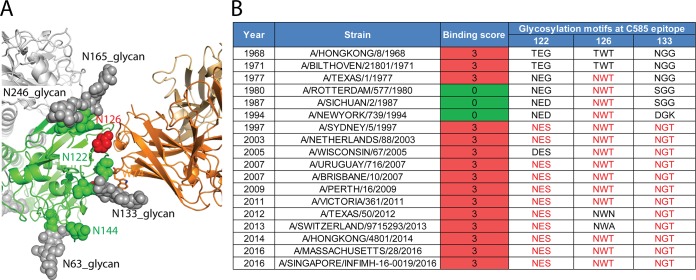
In addition to mechanisms of antigenic drift and shift due to sequence mutations, influenza virus HA can additionally evade immune detection by shielding antigenic sites with glycans. Sequence analysis of H3N2 HAs since 1968 indicate that up to eight N-glycosylation sites can be present in the head domain at positions 63, 122, 126, 133, 144, 158, 165, and 246 (Table 4). Among the six conserved sites in the A/Sw/13 HA head region (positions 63, 122, 133, 144, 165, and 246), electron density for glycans is visible at positions 63, 133, 165, and 246. Glycosylation at N133 is close to the epitope and makes contacts with Trp74 in the C585 heavy chain (Fig. 8A). Although the N126 glycosylation site is not present in the A/Sw/13 HA and N122 glycan density was not observed in the structure, these two potential glycosylation sites in recent seasonal H3 viruses are within the antibody footprint (Table 4). The N-glycans at these positions could either make direct contacts with C585 or result in steric hindrance. However, H3 HAs with different glycosylation states in our binding test showed a glycosylation-independent binding profile to C585, indicating that C585 is not sensitive to glycosylation differences at its epitope (Fig. 8B). Notably, it was observed that the orientation of HA glycans can adopt different conformations to accommodate antibody binding due to their flexibility (19, 20, 29). Similarly, the orientation of N-glycans at residues 122, 133, and 126 might be variable and adaptable for C585 binding in seasonal H3N2 viruses.
TABLE 4.
Conservation of the N-linked glycosylation sites on influenza virus H3 HAs from 1968 to 2018a
| HA residue numbering of N-glycosylation motif | N-linked glycosylation site (% conservation) | Comment |
|---|---|---|
| 63 x 65 | N(99) x T(100) | Clear glycan density |
| 122 x 124 | N(95)/D(3) x S(92)/G(4)/D(3) | No glycan density |
| 126 x 128 | N(97) x T(87)/A(12) | Not glycosylation site in Sw/13 HA |
| 133 x 135 | N(94)/D(4) x T(90) | Clear glycan density |
| 144 x 146 | N(48)/S(33) x S(99) | No glycan density |
| 158 x 160 | N(70)/K(22) x K(63)/T(34) | Not glycosylation site in Sw/13 HA |
| 165 x 167 | N(100) x T(100) | Clear glycan density |
| 246 x 248 | N(99) x T(98) | Clear glycan density |
Residues at each N-linked glycosylation sites are listed with percent identity for most common residue(s), where the N-glycosylation motif is Asn-x-Ser/Thr (x can be any residue other than proline).
FIG 8.
Glycosylation around C585 binding site on H3 HA. (A) Glycosylation sites on HA near the C585 binding interface. Glycans (gray spheres), predicted N-linked glycosylation sites (asparagine) (green spheres) and Asn126, a potential glycosylation site in seasonal flu virus, but not conserved in Switzerland 13 HA (red spheres) are indicated. (B) Comparisons of asparagine glycosylation in the globular head of Switzerland 13 HA with representative seasonal H3 HAs. Up to three potential N-glycosylation motifs are present around the epitope of MAb C585. Glycosylation motifs are listed in red letters; nonglycosylation motifs in the corresponding positions are listed in black letters. Measured binding scores are also presented for comparison. A KD of <1 nM is given a score of 3; no detected binding is given a score of 0.
DISCUSSION
Since the 1968 Hong Kong pandemic, influenza H3N2 viruses have circulated in humans for more than 50 years. A large number of mutations have accumulated, which makes the current H3 strain share only ∼80% sequence identity to the 1968 H3 strain. This highlights a major challenge for identifying a broadly protective or universal vaccine against seasonally drifting viruses, such as the H3 subtype. To date, most efforts in developing bnAbs or universal vaccines have been focusing on the stem region, which possesses highly conserved areas that can be targeted by human antibodies.
In this work, we investigate a monoclonal antibody isolated after B-cell repertoire profiling of a donor who exhibited serological breadth against a broad array of H3N2 seasonal viruses and report structural and functional studies of MAb C585, which showed breadth of neutralization against a broad range of H3 influenza viruses dating back to 1968 and recognizes a novel epitope on the globular head domain of the H3 HA. Altogether, our data suggested that the C585 epitope is a conserved, novel antigenic region that could potentially add value to broadly protective vaccines for eliciting breadth. The identification of an additional clonally unrelated MAb (C997) that competes for the C585 binding site also suggests that this domain is a target for eliciting broadly cross-protective immune responses. Interestingly, while a single MAb does have HAI activity against many strains across 50 years of antigenic drift, potency is not equivalent for all strains. The fact that vaccination yields this broadly neutralizing MAb with extended breadth and with its ability to neutralize drifting strains of the future supports “next-generation” influenza vaccination strategies targeting the HA globular head.
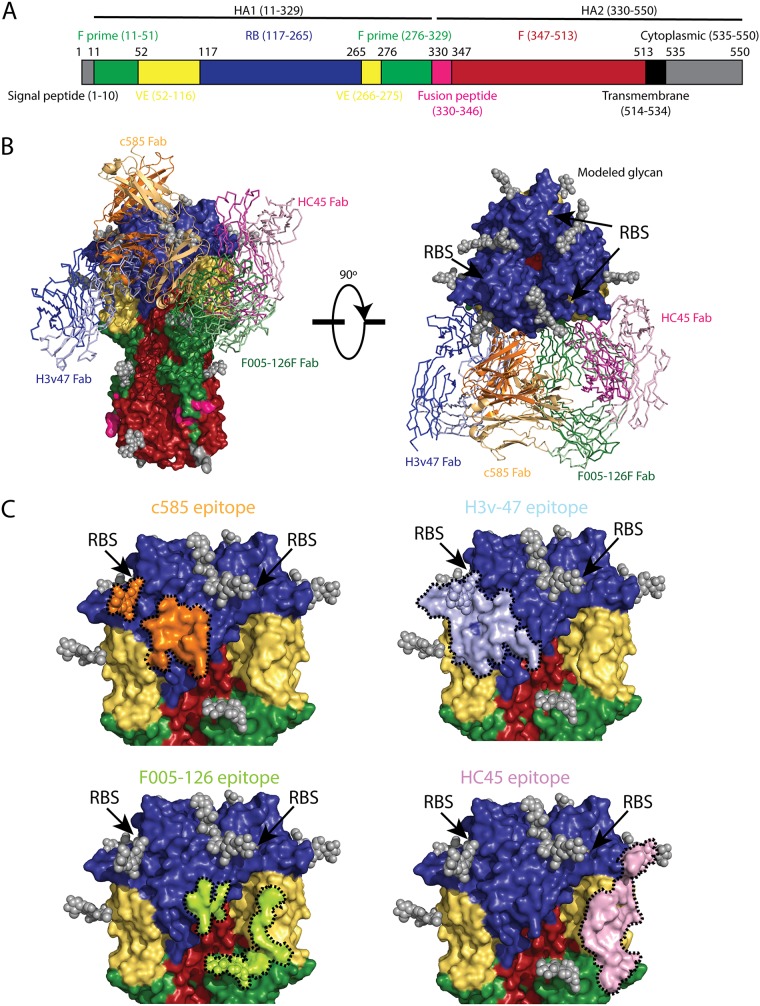
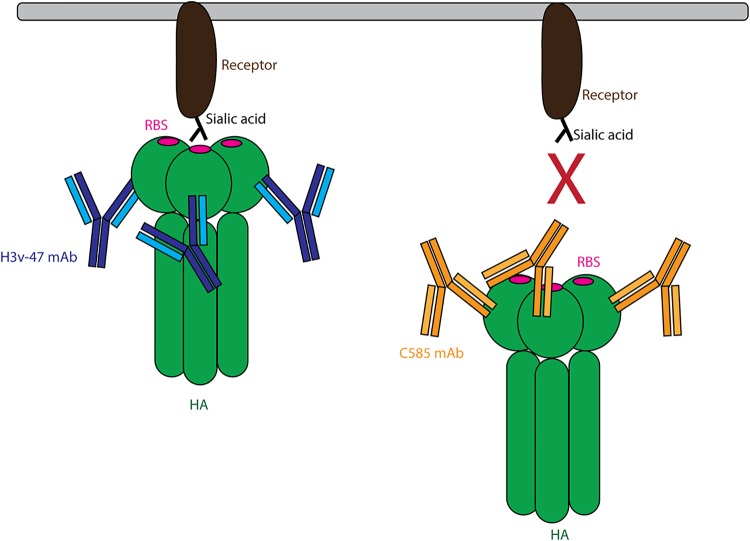
Most neutralizing antibodies that bind to the HA head of H3N2 viruses recognize epitopes in or surrounding the RBS, such as MAbs C05, F045-092, and S139 (Fig. 7A), which usually possess cross-subtype neutralizing activity (6, 7, 17). There is another class of MAbs that can neutralize diverse strains within a subtype but that do not cross-react with other subtypes (30, 31). MAbs in this class can target several subdomains of the H3N2 HA globular head, including the receptor-binding subdomain (RB), the vestigial esterase subdomain (VE), and F prime (F′) subdomain (Fig. 9A). To our knowledge, to date, four MAbs have been structurally characterized that recognize regions distal to the RBS within the H3N2 globular head: (i) H3v-47 targets a region spanning the RB and VE subdomains (32); (ii) F005-126 targets a cross section of the RB, VE, and F′ subdomains (33); (iii) HC45 targets mainly the VE subdomain and a small portion of the RB and F′ subdomains (34); (iv) BH151 targets a region very similar to HC45 epitope (35). Interestingly, C585 targets a unique epitope that is different from all previously characterized MAbs. The epitope of C585 is located only within the RB subdomain without overlap with other subdomains (Fig. 9C). By comparing the epitopes of C585 with the other MAbs with unique epitopes, it is noticed that, although H3v-47’s epitope is larger and covers more subdomains, it shares the most residues with C585’s epitope: Glu119, Asn121, Asn122, Glu172, Gln173, Phe174, and Tyr257. It was reported previously that H3v-47 exhibits MN activity by blocking viral egress from infected cells, but not HAI activity, which is consistent with the fact that its epitope is distant from the RBS (32, 36). In contrast, C585 showed both MN and HAI activities against various strains in the H3N2 subtype (Fig. 4). Comparison of the structures of C585 and H3v-47 in complex with HA showed that, although they share a large part of the epitope, the angles of their binding are different (Fig. 9B). This orientation difference could result in different effects on the receptor-binding ability of HA due to different steric hindrance outcomes caused by the antibodies. For instance, thanks to its unique angle of attack, C585’s Fc domain could block the RBS from interacting with the receptor. In contrast, H3v-47 binds to HA from a different direction that leaves HA free to bind its receptor (Fig. 10). This could be the reason why they behave differently in the HAI assay. Together with our functional assessment, the binding orientation of the C585 Fab with HA suggested a unique mechanism of neutralization through receptor binding inhibition.
FIG 9.
Comparison of C585 and other antibodies targeting HA head (non-RBS) showing that C585 binds a unique epitope in the HA head domain. (A) Schematic view of the H3 influenza hemagglutinin (HA) protein showing the different motifs present in HA. The stalk domain can be further divided into the F prime (green) or F (red) subdomains, whereas the globular head consists of the receptor-binding (RB) subdomain (blue) and vestigial esterase (VE) subdomain (yellow), followed by the fusion peptide (magenta), transmembrane domain (orange), and cytoplasmic domain (black). (B) Comparison of the binding site of C585 (orange, cartoon) with structurally characterized H3-binding antibodies binding to the HA head. The parts of HA are colored the same as in panel A. The HAs from all these HA-antibody complexes are aligned. These antibodies include H3v-47 (PDB 5W42) (blue), HC45 (PDB 1QFU) (pink) and F005-126 (PDB 3WHE) (green) binding to the globular head below the receptor-binding site (RBS) (location indicated by arrows). (C) Comparison of the detailed epitopes recognized by C585, H3v-47, F005-126, and HC45. The boundaries of the epitopes are outlined by black dotted lines. The RBS location is indicated by arrows.
FIG 10.
Schematic model of Mab C585 blocking substrate binding compared to MAb H3v47. H3v-47 binds to HA facing up, and therefore may not block substrate binding, as its Fc portion is pointing down. In contrast, C585 uses a facing down angle to bind HA, and therefore could indirectly block the RBS from interacting with the substrate by its Fc portion.
Additionally, MAb C585 appears to apply a biological solution similar to that used by MAb F005-126 (33) by making contact with HA via a heavily mutated FR3. C585 and F005-126 differ with respect to their point of contact between FR3 and HA and the degree of SHM within FR3, with C585 having 14 amino acid (AA) replacements compared to 5 AA mutations for F005-126. Indeed, high-throughput antibody sequencing analysis has identified this region as corresponding to the hypervariable region 4 (HV4) in T-cell receptors (37). This strategy has also been observed in anti-influenza stem-binding MAb CR6261 (20) and anti-HIV antibody 21c (38).
Although structure-based vaccine design strategy is still in its infancy, epitope-focused vaccinology (39) and B-cell lineage immunogen design (40) has shown some progress toward antibody-guided vaccine development. Specifically, while epitope-focused vaccinology would potentially involve the rational design of HAs able to induce C585-like antibodies, B-cell lineage immunogen design might involve generating C585-like antibodies by inducing their germ line or early progenitor B-cell clones with antigens presenting the epitope of interest. However, methods to induce expansion of epitope-specific clonal lineages in vivo are not well developed yet.
In summary, the combination of high potency and broad neutralizing activity exhibited by MAb C585 provides insight into mechanisms broadly neutralizing antibody responses targeting the influenza virus H3N2 subtype and can help characterize immune responses and potentially susceptible influenza virus strains. Additionally, the unique epitope targeted by C585 may induce production of broadly neutralizing antibodies against human H3 viruses, as observed following vaccination with A/Texas/50/2012, and together with other cross-neutralizing epitopes could contribute to the design of more broadly protective vaccines.
MATERIALS AND METHODS
BCR-SEQ analysis of donor 236.
We used the VH/VL paired sequencing approach using the 454 method and tagging all cDNA generated from an individual B cell with a unique DNA barcode before sequencing cDNA made from peripheral blood mononuclear cells (PBMCs) of donor 236 (27). We annotated heavy and light chain sequences with IMGT/HighV-QUEST (26) and performed immunogenetic analysis, including CDR analysis, somatic mutation, and clonotypes. Heavy and light chain sequences were concatenated and aligned for clonal lineage analysis and constructing phylogenetic tree using CLC Main Workbench 8.1. Statistical calculations were conducted using SAS JMP13 statistical software (SAS Institute, Cary, NC, USA).
Preparation of C585 Fab.
The gene fragments encoding the variable domains of the heavy chain (VH; positions 1 to 121) and light chain (VL; positions 1 to 107) of human MAb C585 were cloned into modified pTT5 vectors containing the IgG1 constant domain of CH1-His6 and CL, respectively. After sequencing confirmation, both plasmids were transiently cotransfected into Expi293F cells. After 5 days, the recombinant C585 Fab with a hexahistidine (His) tag at the C terminus was purified from Expi293F cell culture medium by HisTrap HP column (GE Healthcare), and the elution fraction was buffer exchanged to 1× phosphate-buffered saline (PBS). The amino acid sequence of the recombinant C585 Fab is shown below. The heavy chain sequence was QVQLQESGPGLVKPSETLSLTCAVSGASISSFYWSWIRQSPGKGLEWIAYIYYSGKTQYNPAVTGRATISLQNWNNHVALRVNSVTAADTAIYSCARHTLAYHYDDEGYMQPGDAFDLWGQGTMVTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTHHHHHH, and the light chain sequence was QSVLTQPPSVSAPAGQMVTIPCSGSSSDIGKSFVSWYQQLPGTAPKLVIYDNNKRPSGIPDRFSGSKSGTSATLGITGLQTGDEAVYYCGTWDSSLIWVFGGGTKLTVLGQPKAAPSVTLFPPSSEELQANKATLVCLISDFYPGAVTVAWKADSSPVKAGVETTTPSKQSNNKYAASSYLSLTPEQWKSHRSYSCQVTHEGSTVEKTVAPTECS.
Preparation of the recombinant A/Sw/13 hemagglutinin protein.
The gene fragments encoding the ectodomains of the HA proteins (residues 1 to 504 based on H3 numbering) from the A/Sw/13 (H3N2) strain was synthesized by Life Technologies (USA) and cloned into the baculovirus transfer vector, pAcGp67-C (BD Biosciences Pharmingen, San Jose, CA) by incorporating a GP67 signal peptide for HA secretion at the N terminus. The construct also incorporated a C-terminal thrombin cleavage site (LVPRGS), a “foldon” sequence (GYIPEAPRDGQAYVRKDGEWVLLSTFL) (41) and hexahistidine (6×His) tag at the extreme C terminus of the construct to enable purification. Transfection and virus amplification were conducted according to the baculovirus expression standard procedure (system manual, BD Biosciences Pharmingen) at BlueSky Lake Pharma (Worcester, MA). Small-scale expression scouting and optimization were conducted in Sf9, Sf21, and Trichoplusia ni cells with different dilutions (1:10,000 and 1:5,000) and harvest time points of 24, 48, and 72 h posttransfection to identify the conditions for the highest expression level of secreted protein. Sf21 cells at 1:10,000 of amplified virus stock dilution and harvest time of 72 h posttransfection were chosen for 25-liter scale-up expression. Clarified conditioned medium was 10× concentrated by tangential flow filtration (TFF) to 2.5 liters and buffer exchanged into immobilized metal ion affinity chromatography (IMAC) binding buffer (20 mM phosphate-buffered saline [pH 7.4], 0.5 M NaCl), followed by sterile filtration and transferring for purification on wet ice without freezing on the day of harvest. Recombinant HA was purified by IMAC capturing on HisTrap HP resin (GE Healthcare; 15-ml column volume [CV]). Imidazole was spiked into the loading sample up to 50 mM to minimize nonspecific binding and washed on column with 80 mM before linear gradient elution over 10 CVs with IMAC elution buffer (20 mM phosphate-buffered saline [pH 7.4], 0.5 M NaCl, 0.5 M imidazole). Elution fractions containing the purest and concentrated HA were pooled and dialyzed against 50 mM Tris HCl and 50 mM NaCl (pH 7.5) to polish by ion-exchange chromatography (IEC) in flowthrough mode on HiTrap Q HP resin (GE Healthcare; 10-ml CV). The polished HA pool was treated by dialysis against 20 mM Tris HCl and 150 mM NaCl (pH 7.5), followed by sterile filtration. The purified HA was subjected to thrombin digestion (GE Healthcare; a maximum of 50 U per mg of HA0) at room temperature for 16 h to remove the C-terminal Foldon domain and 6×His tag. The HA0 was further purified by size exclusion chromatography using a Superdex 200 10/300 column and then concentrated to approximately 10 mg/ml in storage buffer (20 mM Tris [pH 8.0] and 100 mM NaCl) for use in the structural study.
Biolayer interferometry.
Binding kinetics of C585 MAb to different recombinant hemagglutinins (rHAs) were determined by biolayer interferometry (BLI) using Octet RED96 system (Fortébio) and protein A biosensors. Before the start of the experiment, protein A biosensors were first hydrated for 20 min in 1× ForteBio kinetic buffer (PBS [pH 7.4] with 0.002% Tween 20, 0.01% albumin, and 0.005% sodium azide). ForteBio 1× kinetic buffer was also used for MAb and rHA dilutions and for baseline and dissociation phases. The kinetic measurement automated five-step procedure follows. In step 1, an initial reference baseline (baseline 1) of 120 s is performed in 1× kinetic buffer alone. In step 2, c585 MAb at a 10-μg/ml concentration is loaded onto a protein A biosensor at 25°C with 1,000 rpm agitation for 150 s. In step 3, a second baseline (baseline 2) of 120 s was determined. Once a stable baseline was obtained, a titration series of the antigen (concentration range from 0.3 to 10 μg/ml) is measured against an immobilized antibody with an association step (step 4) performed for 240 s, followed by monitoring the dissociation phase for 300 s (step 5). Acquired data were analyzed using ForteBio Data Analysis software (version 7.0) with global fitting and association rate (ka), disassociation rate (kd), and dissociation constant (KD) determination.
Antibody forensics.
An antibody forensics (AF) assay was used to determine the breadth of binding of MAbs to various rHAs (Protein Sciences Corp., Meriden, CT) coupled to magnetic beads (MagPlex microspheres; Luminex, Austin, TX) spanning the years 1968 to 2016. Briefly, antibody was fivefold serially diluted (starting from 10 μg/ml) and incubated with beads with constant shaking (600 rpm) for 2 h at 4°C. The plates were washed with PBS containing 0.05% Tween 20 (PBS-T), and biotinylated human-specific antibody against total IgG (Jackson Immunoresearch, West Grove, PA) was added to the plates. The plates were incubated for 40 min at 4°C, washed with PBS-T, and incubated with streptavidin, R-phycoerythrin conjugate (SAPE). The plates were washed again with PBS-T and read using a BioPlex-100/200 bead array reader. The strength of serially diluted antibody binding to influenza virus-specific rHAs was detected as a fluorescent intensity signal, and then the 50% effective concentration (EC50) (in micrograms per milliliter) was calculated using GraphPad Prism software.
HAI assay.
Influenza virus seed stocks used in hemagglutinin inhibition (HAI) assays were from the Centers for Disease Control and Prevention (Atlanta, GA, USA). The HAI assay was performed by following standard methodology established by the World Health Organization (WHO), which tests the quantity of MAb required to interfere with the agglutinating activity of 4 agglutinating units of virus. Briefly, 2× serial dilutions of MAb were mixed with 4 HA units of virus (HAU), followed by the addition of 0.5% turkey red blood cells after 1 h of incubation at room temperature. The HAI titer was determined as the lowest concentration of MAb resulting in complete inhibition of hemagglutination.
MN assay.
Microneutralization (MN) assay was performed by Bioqual, Inc., by following Standard Operating Procedure number BV-012 and the 2013 WHO Laboratory Procedures guidelines. Briefly, serial dilutions of MAb were incubated with influenza virus for 1 h at 37°C before being transferred to 96-well plates with MDCK cells. After 2 days of incubation, influenza virus was detected in an enzyme-linked immunosorbent assay (ELISA) to calculate the virus neutralization endpoint antibody titer. Reference serum, virus back-titration, and preimmune serum pools for each group were included as controls.
Crystallization and structure determination.
To prepare the C585 Fab-HA complex, the purified C585 Fab and HA were mixed in a 2:1 molar ratio and incubated on ice for 1 h. Then excess Fab was removed by size exclusion chromatography using a Superdex 200 increase 10/300GL column with 20 mM Tris (pH 8.0) and 100 mM NaCl as the mobile phase. Crystallization was performed by the sitting drop vapor diffusion method at 21°C by mixing 0.1 μl of the protein complex at 10 mg/ml and 0.1 μl of reservoir solution. Crystals of the C585 Fab–Sw/13 HA complex were grown from the drop with the reservoir solution containing 0.1 M sodium cacodylate (pH 6.5) and 1 M sodium citrate for a week. The crystal was cryoprotected in the reservoir solution supplemented with 25% (vol/vol) glycerol and then flash cooled in liquid nitrogen prior to data collection. The diffraction data were collected at SERCAT 22ID of Advanced Photon Source and processed with iMosflm (42) and Aimless from CCP4 package (43). The structure of the C585 Fab–Sw/13 HA complex was solved by the molecular replacement (MR) method using the Phaser program (44) with Hong Kong 1968 HA (PDB code 4FNK) and a Fab model generated by MOE (28) as the search models. Structural refinement was performed using Phenix (45), and the models were constructed using Coot (46). All structures were generated using PyMOL (http://www.pymol.org). The diffraction data and structural refinement statistics are summarized in Table 1.
Analysis of the H3 sequences.
All full-length H3N2 influenza HA sequences from 1968 to 2018 derived from human hosts were downloaded from the NCBI Influenza Virus Database (https://www.ncbi.nlm.nih.gov/genomes/FLU/Database/nph-select.cgi?go=database). The sequences were aligned using ClustalW (47).
Data availability.
The accession number for the atomic coordinates and structure factors for Fab C585 in complex with H3 HA (A/Switzerland/9715293/2013) reported in this paper is PDB 6PDX.
ACKNOWLEDGMENTS
This work was funded by Sanofi Pasteur and Sanofi Biologics.
We thank Samantha Cooper and Monica Lane for assistance with mass spectrometry.
The authors are current or former Sanofi employees except for Ted M. Ross, who is director of the Center for Vaccine and Immunology at the University of Georgia. Huawei Qiu is a former employee of Sanofi and is currently employed by Akrevia Therapeutics. Harry Kleanthous is a former employee of Sanofi and is currently employed by the Bill & Melinda Gates Foundation. Simon Delagrave is a former employee of Sanofi and is currently employed by Ring Therapeutics.
REFERENCES
- 1.Skehel JJ, Wiley DC. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem 69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 2.Berlanda Scorza F, Tsvetnitsky V, Donnelly JJ. 2016. Universal influenza vaccines: shifting to better vaccines. Vaccine 34:2926–2933. doi: 10.1016/j.vaccine.2016.03.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen JR, Ma C, Wong CH. 2011. Vaccine design of hemagglutinin glycoprotein against influenza. Trends Biotechnol 29:426–434. doi: 10.1016/j.tibtech.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Klenk HD, Garten W. 1994. Host cell proteases controlling virus pathogenicity. Trends Microbiol 2:39–43. doi: 10.1016/0966-842x(94)90123-6. [DOI] [PubMed] [Google Scholar]
- 5.Steinhauer DA. 1999. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology 258:1–20. doi: 10.1006/viro.1999.9716. [DOI] [PubMed] [Google Scholar]
- 6.Ekiert DC, Kashyap AK, Steel J, Rubrum A, Bhabha G, Khayat R, Lee JH, Dillon MA, O’Neil RE, Faynboym AM, Horowitz M, Horowitz L, Ward AB, Palese P, Webby R, Lerner RA, Bhatt RR, Wilson IA. 2012. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 489:526–532. doi: 10.1038/nature11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee PS, Ohshima N, Stanfield RL, Yu W, Iba Y, Okuno Y, Kurosawa Y, Wilson IA. 2014. Receptor mimicry by antibody F045-092 facilitates universal binding to the H3 subtype of influenza virus. Nat Commun 5:3614. doi: 10.1038/ncomms4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu NC, Wilson IA. 2018. Structural insights into the design of novel anti-influenza therapies. Nat Struct Mol Biol 25:115–121. doi: 10.1038/s41594-018-0025-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sparrow E, Friede M, Sheikh M, Torvaldsen S, Newall AT. 2016. Passive immunization for influenza through antibody therapies, a review of the pipeline, challenges and potential applications. Vaccine 34:5442–5448. doi: 10.1016/j.vaccine.2016.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neu KE, Henry Dunand CJ, Wilson PC. 2016. Heads, stalks and everything else: how can antibodies eradicate influenza as a human disease? Curr Opin Immunol 42:48–55. doi: 10.1016/j.coi.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corti D, Cameroni E, Guarino B, Kallewaard NL, Zhu Q, Lanzavecchia A. 2017. Tackling influenza with broadly neutralizing antibodies. Curr Opin Virol 24:60–69. doi: 10.1016/j.coviro.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lang S, Xie J, Zhu X, Wu NC, Lerner RA, Wilson IA. 2017. Antibody 27F3 broadly targets influenza A group 1 and 2 hemagglutinins through a further variation in VH1-69 antibody orientation on the HA stem. Cell Rep 20:2935–2943. doi: 10.1016/j.celrep.2017.08.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whittle JR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, Kepler TB, Liao HX, Harrison SC. 2011. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci U S A 108:14216–14221. doi: 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt AG, Xu H, Khan AR, O’Donnell T, Khurana S, King LR, Manischewitz J, Golding H, Suphaphiphat P, Carfi A, Settembre EC, Dormitzer PR, Kepler TB, Zhang R, Moody MA, Haynes BF, Liao H-X, Shaw DE, Harrison SC. 2013. Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc Natl Acad Sci U S A 110:264–269. doi: 10.1073/pnas.1218256109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu R, Krause JC, McBride R, Paulson JC, Crowe JE Jr, Wilson IA. 2013. A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin. Nat Struct Mol Biol 20:363–370. doi: 10.1038/nsmb.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong M, Lee PS, Hoffman RM, Zhu X, Krause JC, Laursen NS, Yoon SI, Song L, Tussey L, Crowe JE Jr, Ward AB, Wilson IA. 2013. Antibody recognition of the pandemic H1N1 influenza virus hemagglutinin receptor binding site. J Virol 87:12471–12480. doi: 10.1128/JVI.01388-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee PS, Yoshida R, Ekiert DC, Sakai N, Suzuki Y, Takada A, Wilson IA. 2012. Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity. Proc Natl Acad Sci U S A 109:17040–17045. doi: 10.1073/pnas.1212371109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, Silacci C, Fernandez-Rodriguez BM, Agatic G, Bianchi S, Giacchetto-Sasselli I, Calder L, Sallusto F, Collins P, Haire LF, Temperton N, Langedijk JP, Skehel JJ, Lanzavecchia A. 2011. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 19.Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, van der Vlugt R, Lamrani M, Korse HJ, Geelen E, Sahin O, Sieuwerts M, Brakenhoff JP, Vogels R, Li OT, Poon LL, Peiris M, Koudstaal W, Ward AB, Wilson IA, Goudsmit J, Friesen RH. 2012. Highly conserved protective epitopes on influenza B viruses. Science 337:1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA. 2009. Antibody recognition of a highly conserved influenza virus epitope. Science 324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakamura G, Chai N, Park S, Chiang N, Lin Z, Chiu H, Fong R, Yan D, Kim J, Zhang J, Lee WP, Estevez A, Coons M, Xu M, Lupardus P, Balazs M, Swem LR. 2013. An in vivo human-plasmablast enrichment technique allows rapid identification of therapeutic influenza A antibodies. Cell Host Microbe 14:93–103. doi: 10.1016/j.chom.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 22.Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, Santelli E, Stec B, Cadwell G, Ali M, Wan H, Murakami A, Yammanuru A, Han T, Cox NJ, Bankston LA, Donis RO, Liddington RC, Marasco WA. 2009. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol 16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wyrzucki A, Dreyfus C, Kohler I, Steck M, Wilson IA, Hangartner L. 2014. Alternative recognition of the conserved stem epitope in influenza A virus hemagglutinin by a VH3-30-encoded heterosubtypic antibody. J Virol 88:7083–7092. doi: 10.1128/JVI.00178-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DiLillo DJ, Tan GS, Palese P, Ravetch JV. 2014. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcgammaR interactions for protection against influenza virus in vivo. Nat Med 20:143–151. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nunez IA, Carlock MA, Allen JD, Owino SO, Moehling KK, Nowalk P, Susick M, Diagle K, Sweeney K, Mundle S, Vogel TU, Delagrave S, Ramgopal M, Zimmerman RK, Kleanthous H, Ross TM. 2017. Impact of age and pre-existing influenza immune responses in humans receiving split inactivated influenza vaccine on the induction of the breadth of antibodies to influenza A strains. PLoS One 12:e0185666. doi: 10.1371/journal.pone.0185666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giudicelli V, Duroux P, Kossida S, Lefranc MP. 2017. IG and TR single chain fragment variable (scFv) sequence analysis: a new advanced functionality of IMGT/V-QUEST and IMGT/HighV-QUEST. BMC Immunol 18:35. doi: 10.1186/s12865-017-0218-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan YC, Blum LK, Kongpachith S, Ju CH, Cai X, Lindstrom TM, Sokolove J, Robinson WH. 2014. High-throughput sequencing of natively paired antibody chains provides evidence for original antigenic sin shaping the antibody response to influenza vaccination. Clin Immunol 151:55–65. doi: 10.1016/j.clim.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vilar S, Cozza G, Moro S. 2008. Medicinal chemistry and the molecular operating environment (MOE): application of QSAR and molecular docking to drug discovery. Curr Top Med Chem 8:1555–1572. doi: 10.2174/156802608786786624. [DOI] [PubMed] [Google Scholar]
- 29.Brandenburg B, Koudstaal W, Goudsmit J, Klaren V, Tang C, Bujny MV, Korse HJ, Kwaks T, Otterstrom JJ, Juraszek J, van Oijen AM, Vogels R, Friesen RH. 2013. Mechanisms of hemagglutinin targeted influenza virus neutralization. PLoS One 8:e80034. doi: 10.1371/journal.pone.0080034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Velkov T, Ong C, Baker MA, Kim H, Li J, Nation RL, Huang JX, Cooper MA, Rockman S. 2013. The antigenic architecture of the hemagglutinin of influenza H5N1 viruses. Mol Immunol 56:705–719. doi: 10.1016/j.molimm.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 31.Ren H, Zhou P. 2016. Epitope-focused vaccine design against influenza A and B viruses. Curr Opin Immunol 42:83–90. doi: 10.1016/j.coi.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 32.Bangaru S, Zhang H, Gilchuk IM, Voss TG, Irving RP, Gilchuk P, Matta P, Zhu X, Lang S, Nieusma T, Richt JA, Albrecht RA, Vanderven HA, Bombardi R, Kent SJ, Ward AB, Wilson IA, Crowe JE Jr. 2018. A multifunctional human monoclonal neutralizing antibody that targets a unique conserved epitope on influenza HA. Nat Commun 9:2669. doi: 10.1038/s41467-018-04704-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iba Y, Fujii Y, Ohshima N, Sumida T, Kubota-Koketsu R, Ikeda M, Wakiyama M, Shirouzu M, Okada J, Okuno Y, Kurosawa Y, Yokoyama S. 2014. Conserved neutralizing epitope at globular head of hemagglutinin in H3N2 influenza viruses. J Virol 88:7130–7144. doi: 10.1128/JVI.00420-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fleury D, Barrere B, Bizebard T, Daniels RS, Skehel JJ, Knossow M. 1999. A complex of influenza hemagglutinin with a neutralizing antibody that binds outside the virus receptor binding site. Nat Struct Biol 6:530–534. doi: 10.1038/9299. [DOI] [PubMed] [Google Scholar]
- 35.Fleury D, Daniels RS, Skehel JJ, Knossow M, Bizebard T. 2000. Structural evidence for recognition of a single epitope by two distinct antibodies. Proteins 40:572–578. doi:. [DOI] [PubMed] [Google Scholar]
- 36.Bangaru S, Nieusma T, Kose N, Thornburg NJ, Finn JA, Kaplan BS, King HG, Singh V, Lampley RM, Sapparapu G, Cisneros A III, Edwards KM, Slaughter JC, Edupuganti S, Lai L, Richt JA, Webby RJ, Ward AB, Crowe JE Jr. 2016. Recognition of influenza H3N2 variant virus by human neutralizing antibodies. JCI Insight 1:e86673. doi: 10.1172/jci.insight.86673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Briney BS, Willis JR, Crowe JE Jr. 2012. Location and length distribution of somatic hypermutation-associated DNA insertions and deletions reveals regions of antibody structural plasticity. Genes Immun 13:523–529. doi: 10.1038/gene.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Diskin R, Marcovecchio PM, Bjorkman PJ. 2010. Structure of a clade C HIV-1 gp120 bound to CD4 and CD4-induced antibody reveals anti-CD4 polyreactivity. Nat Struct Mol Biol 17:608–613. doi: 10.1038/nsmb.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lanzavecchia A, Fruhwirth A, Perez L, Corti D. 2016. Antibody-guided vaccine design: identification of protective epitopes. Curr Opin Immunol 41:62–67. doi: 10.1016/j.coi.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 40.Haynes BF, Kelsoe G, Harrison SC, Kepler TB. 2012. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat Biotechnol 30:423–433. doi: 10.1038/nbt.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Letarov AV, Londer YY, Boudko SP, Mesyanzhinov VV. 1999. The carboxy-terminal domain initiates trimerization of bacteriophage T4 fibritin. Biochemistry (Mosc) 64:817–823. [PubMed] [Google Scholar]
- 42.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. 2011. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr 67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. 2010. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res 38:W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The accession number for the atomic coordinates and structure factors for Fab C585 in complex with H3 HA (A/Switzerland/9715293/2013) reported in this paper is PDB 6PDX.