Some RNA virus genomes are suppressed in the nucleotide combination of a cytosine followed by a guanosine (CpG), indicating that they are detrimental to the virus. The antiviral protein ZAP binds viral RNA containing CpGs and prevents the virus from multiplying. However, it remains unknown how the number and position of CpGs in viral genomes affect restriction by ZAP and whether CpGs have other antiviral mechanisms. Importantly, manipulating the CpG content in viral genomes could help create new vaccines. HIV-1 shows marked CpG suppression, and by introducing CpGs into its genome, we show that ZAP efficiently targets a specific region of the viral genome, that the number of CpGs does not predict the magnitude of antiviral activity, and that CpGs can inhibit HIV-1 gene expression through a ZAP-independent mechanism. Overall, the position of CpGs in the HIV-1 genome determines the magnitude and mechanism through which they inhibit the virus.
KEYWORDS: CpG, KHNYN, RNA, ZAP, human immunodeficiency virus, innate immunity, virus
ABSTRACT
CpG dinucleotides are suppressed in the genomes of many vertebrate RNA viruses, including HIV-1. The cellular antiviral protein ZAP (zinc finger antiviral protein) binds CpGs and inhibits HIV-1 replication when CpGs are introduced into the viral genome. However, it is not known if ZAP-mediated restriction is the only mechanism driving CpG suppression. To determine how CpG dinucleotides affect HIV-1 replication, we increased their abundance in multiple regions of the viral genome and analyzed the effect on RNA expression, protein abundance, and infectious-virus production. We found that the antiviral effect of CpGs was not correlated with their abundance. Interestingly, CpGs inserted into some regions of the genome sensitize the virus to ZAP antiviral activity more efficiently than insertions into other regions, and this sensitivity can be modulated by interferon treatment or ZAP overexpression. Furthermore, the sensitivity of the virus to endogenous ZAP was correlated with its sensitivity to the ZAP cofactor KHNYN. Finally, we show that CpGs in some contexts can also inhibit HIV-1 replication by ZAP-independent mechanisms, and one of these is the activation of a cryptic splice site at the expense of a canonical splice site. Overall, we show that the location and sequence context of the CpG in the viral genome determines its antiviral activity.
IMPORTANCE Some RNA virus genomes are suppressed in the nucleotide combination of a cytosine followed by a guanosine (CpG), indicating that they are detrimental to the virus. The antiviral protein ZAP binds viral RNA containing CpGs and prevents the virus from multiplying. However, it remains unknown how the number and position of CpGs in viral genomes affect restriction by ZAP and whether CpGs have other antiviral mechanisms. Importantly, manipulating the CpG content in viral genomes could help create new vaccines. HIV-1 shows marked CpG suppression, and by introducing CpGs into its genome, we show that ZAP efficiently targets a specific region of the viral genome, that the number of CpGs does not predict the magnitude of antiviral activity, and that CpGs can inhibit HIV-1 gene expression through a ZAP-independent mechanism. Overall, the position of CpGs in the HIV-1 genome determines the magnitude and mechanism through which they inhibit the virus.
INTRODUCTION
The frequency of CpG dinucleotides is suppressed in many vertebrate RNA viruses, indicating that they may be deleterious (1–5). Supporting this hypothesis, increasing the CpG content in picornaviruses and influenza A virus inhibits their replication (6–10). However, the mechanisms by which CpG dinucleotides attenuate viral replication remain unclear. Importantly, introduction of CpG dinucleotides into viral genomes using synthetic biology techniques may be a new way to develop live attenuated virus vaccines, and a full understanding of how CpG dinucleotides inhibit viral replication is necessary to develop this approach (9, 10).
HIV-1 encodes the three polyproteins found in all retroviruses (Gag, Pol, and Env), two regulatory proteins (Tat and Rev), and four accessory proteins (Vif, Vpr, Vpu, and Nef) (11). CpG dinucleotides are suppressed throughout the HIV-1 genomic RNA (gRNA), and introducing CpGs into gag or env inhibits viral replication (12–16). Furthermore, analysis of clinical HIV-1 samples found that mutations that create new CpG dinucleotides in HIV-1 are twice as costly as those that do not and that increased CpG dinucleotide abundance in env may predict disease progression (17, 18). There are at least four mechanisms by which CpG dinucleotides could inhibit HIV-1. First, CpG DNA methylation-induced transcriptional silencing could repress viral gene expression (12, 13, 19). Second, introduction of CpGs into cis-acting elements or structures required for viral replication may render them nonfunctional. Third, CpGs could create deleterious cis-acting elements or structures. Fourth, they could act as a pathogen-associated molecular pattern (PAMP) that is recognized by the innate immune system. Supporting the hypothesis that CpGs in viral RNA could be a PAMP, the antiviral protein ZAP (zinc finger antiviral protein) has recently been shown to bind regions of HIV-1 RNA containing CpG dinucleotides and to inhibit HIV-1 with increased CpG content in env (16).
ZAP (encoded by the gene ZC3HAV1) was initially discovered as a cellular factor inhibiting murine leukemia virus (MLV) gene expression (20). Subsequent studies have shown that ZAP also inhibits alphaviruses, filoviruses, and hepadnaviruses, as well as some flaviviruses and picornaviruses (21–25). ZAP inhibits viral replication by binding viral RNA and targeting it for degradation and/or inhibiting its translation (20, 21, 26–28). It may also have other mechanisms of antiviral activity. However, ZAP does not restrict all viruses, and yellow fever virus, Zika virus, dengue virus, herpes simplex virus 1, vesicular stomatitis virus, and poliovirus are resistant to its antiviral activity (21, 24). ZAP does not have enzymatic activity and interacts with other cellular proteins, such as TRIM25 and KHNYN, to restrict viral replication (29–31). Why some viruses are sensitive to ZAP and others are resistant, and whether the CpG abundance and context in viruses determines this, remains unknown. It is also unclear whether the deleterious effect of CpG dinucleotides on viral replication is mediated exclusively by ZAP or through additional mechanisms.
Overall, the specific mechanisms by which CpG dinucleotides inhibit viral replication are not well understood for any virus. Because CpGs are highly suppressed in HIV-1, it is an excellent model virus to study the antiviral effects of the dinucleotide. In this study, we introduced CpGs into different contexts and regions of the viral genome and analyzed how they restricted viral gene expression and replication. First, we determined whether there was a position-dependent effect of CpGs on viral replication. Second, we analyzed whether there was a correlation between endogenous ZAP activity and the abundance of CpG dinucleotides. Third, we tested whether increasing ZAP abundance increased its antiviral effect. In sum, we found that CpGs in different contexts and locations inhibited viral gene expression through ZAP-dependent inhibition of gene expression and ZAP-independent changes in pre-mRNA splicing. Importantly, the number of introduced CpG dinucleotides did not predict the magnitude of their antiviral activity or inhibition by endogenous ZAP. ZAP appears to target a specific region in env containing introduced CpGs more efficiently than other regions in the viral genome, though high levels of ZAP can target most regions of the genome containing CpGs. Our results indicate that the context and position of CpG dinucleotides in the HIV-1 genome determine how they inhibit viral replication through ZAP-dependent and -independent mechanisms.
RESULTS
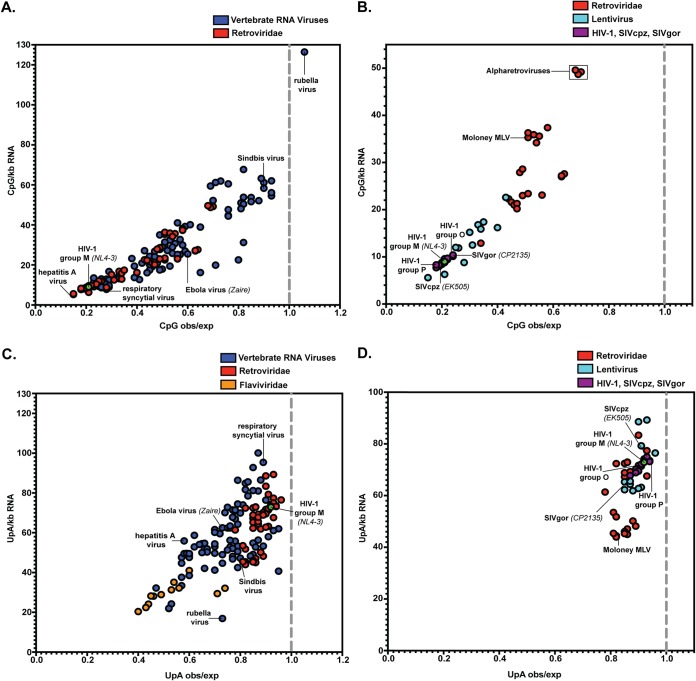
To determine how CpG suppression in HIV-1 compared with that in other viruses that infect vertebrates, we compared the CpG frequencies in a panel of viruses (Fig. 1A; see Data Set S1 in the supplemental material). Because RNA viruses have large variations in the frequencies of A, C, G, and U in their genomes, we calculated the number of CpGs per kilobase of RNA and the observed/expected ratio of CpGs. This analysis showed that there is a broad range of CpG suppression in RNA viruses. As previously shown, togaviruses show little CpG suppression, with >40 CpGs/kb and an observed/expected ratio of >0.75 (1–5). Many viruses show moderate CpG suppression, with an observed/expected CpG ratio of ∼0.5. However, there are some viruses in which CpG abundance is highly suppressed, including hepatitis A virus, respiratory syncytial virus, and HIV-1 (Fig. 1A; see Data Set S1). Within the retrovirus family, lentiviruses have high levels of CpG suppression (6 to 23 CpGs/kb; observed/expected ratio, 0.2 to 0.4) and alpharetroviruses have low levels of suppression (∼50 CpGs/kb; observed/expected ratio, ∼0.7) (Fig. 1B; see Data Set S1). Viruses closely related to HIV-1 have ∼10 CpGs/kb and an observed/expected ratio of ∼0.2.
FIG 1.
CpG abundance is highly suppressed and UpA abundance is not substantially suppressed in HIV-1 compared to many viruses that infect vertebrate cells. (A) The number of CpGs per kilobase and the CpG observed (obs)/expected (exp) ratio were calculated for each virus. (B) The numbers of CpGs per kilobase and CpG observed/expected ratio were plotted for the family Retroviridae. Members of the genus Lentivirus and the other retroviruses are also shown. The viruses naturally found in humans, chimpanzees, and gorillas are shown in purple. (C) The number of UpAs per kilobase and the UpA observed/expected ratio were calculated for each virus. (D) The numbers of UpAs per kilobase and UpA observed/expected ratio were plotted for the family Retroviridae. The viruses naturally found in humans, chimpanzees, and gorillas are shown in purple. See Data Set S1 for the value for each virus.
In addition to CpG dinucleotides, UpA dinucleotides are suppressed in many RNA viruses (1, 3, 4). Recently, it was reported that ZAP interacts with viral RNA containing UpAs and restricts echovirus 7 containing introduced UpAs (32). Therefore, we analyzed the UpA abundances in our panel of viruses (Fig. 1C; see Data Set S1). While some vertebrate RNA viruses, such as flaviviruses, show marked UpA suppression (<30 UpAs/kb; observed/expected ratio, <0.5) (Fig. 1C), UpA frequency in retroviruses is not substantially suppressed (Fig. 1D). Specifically, viruses closely related to HIV-1 have ∼70 UpAs/kb and observed/expected ratios of ∼0.9. In sum, CpGs, but not UpAs, appear to be potently suppressed in HIV-1.
To better understand the potential evolutionary pressures that have led to CpG suppression in HIV-1, we explored the effects of introducing CpGs into different regions of the viral genome using synonymous mutations. However, HIV-1 contains several overlapping reading frames that constrain where CpGs can be introduced (Fig. 2A). Furthermore, it is important that the synonymous mutations introducing CpGs do not disrupt RNA elements that regulate viral replication. The HIV-1 open reading frames (ORFs) contain multiple cis-acting regulatory elements, including the programmed ribosomal frameshift sequence in gag (33); the Rev response element (RRE) in env (34); polypurine tracts in pol and nef (35); and splicing signals in pol, vif, vpr, tat, rev, and env (36). In addition, there could be uncharacterized elements. Therefore, we identified sequences in the HIV-1 open reading frames that contain reduced variability at synonymous sites, which could indicate the presence of functional RNA elements (37). This analysis identified many of the known HIV-1 linear and structural RNA regulatory elements, including the region at the 5′ end of gag required for dimerization and encapsidation, the ribosomal frameshift sequence required for Pol translation, the polypurine tracts required for reverse transcription, several splicing regulatory sequences, and the RRE (Fig. 2B to E; see Data Set S2 in the supplemental material). We synonymously introduced CpG dinucleotides into gag, pol, and env sequences that the analysis revealed were unlikely to contain cis-acting elements (see Data Set S2).
FIG 2.
Overlapping functional elements are localized to specific regions of the HIV-1 genome. (A) Schematic representation of the HIV-1 provirus with the open reading frames indicated. (B) Analysis of conservation at synonymous sites. The brown line depicts the ratio of the observed number of substitutions to the number expected under a null model of neutral evolution at synonymous sites for a 25-codon sliding window. (C to E) The red lines show the corresponding P values for 25-, 15-, and 9-codon sliding-window analyses. The gray dashed lines indicate approximate false-positive thresholds with a P value of 0.05 after correcting for multiple tests in each plot. See Data Set S2 for the z-score for each codon in the HIV-1 open reading frames in the 9-, 15-, or 25-codon sliding windows. The regions with known cis-acting elements are identified: FS, programmed ribosomal frameshift sequence (33); cPPT, central polypurine tract (35); SRS, splicing regulatory sequences (36); RRE, Rev response element (34); PPT, polypurine tract (35).
The number of CpGs introduced into the HIV-1 genome is not correlated with the antiviral effect or ZAP sensitivity.
An important experimental consideration when studying how CpGs regulate HIV-1 is that CpG DNA methylation-induced transcriptional silencing could potentially inhibit HIV-1 gene expression. However, CpGs in plasmids amplified in bacteria are not methylated when the plasmid is transiently transfected into mammalian cells (38–40), and we have therefore used an experimental approach in which HIV-1 proviral DNA plasmids are transfected into HeLa or 293T cells. A region in env immediately after vpu does not contain any detectable cis-acting RNA elements, which makes it a good region to analyze the effect of introducing CpGs (Fig. 2; see Data Set S2). It has previously been shown that introducing 36 CpGs into env nucleotides (nt) 86 to 561 (HIV-1env86–561CpG) inhibits HIV-1 genomic-RNA abundance, Gag expression, Env expression, and infectious-virus production (Fig. 3A, C, and E and Table 1) (16, 31). Importantly, this restriction is eliminated in ZAP knockout cells (Fig. 3B, C, and E), indicating that it is due to the recognition of the CpG dinucleotides by ZAP. To further characterize how CpGs affect HIV-1 replication, we inserted 48 CpGs into env nt 611 to 1014 (HIV-1env611–1014CpG) (Fig. 3A and Table 1) and analyzed their effect on HIV-1 genomic-RNA abundance, Env expression, Gag expression, and infectious-virus production. These CpGs inhibited HIV-1 infectious-virus production more potently than the 36 CpGs in HIV-1env86–561CpG in the control CRISPR cells (Fig. 3C). However, in the ZAP knockout cells, Gag expression, Env expression, and virion production were only partially increased for HIV-1env611–1014CpG, and infectious-virus production was rescued only ∼2-fold (Fig. 3C). This indicates that the 48 CpGs introduced into this region of env caused both ZAP-dependent and ZAP-independent suppression of infectious-virus production. Nef is expressed from fully spliced mRNAs that do not contain the env region with the introduced CpGs (36). As expected, the CpGs introduced into HIV-1env86–561CpG did not decrease Nef expression (Fig. 3C). However, there was decreased Nef expression for HIV-1env611–1014CpG, and this was not rescued in the ZAP CRISPR cells. This suggests that another mechanism, such as altered splicing, contributes to the CpG-mediated decrease in infectious-virus production for HIV-1env611–1014CpG. Furthermore, genomic-RNA abundance for HIV-1env611–1014CpG was not fully restored to wild-type levels in ZAP knockout cells, indicating that the CpGs in HIV-1env611–1014CpG inhibit genomic-RNA abundance through ZAP-dependent and ZAP-independent mechanisms (Fig. 3E).
FIG 3.
Introduction of CpGs into env inhibits HIV-1 infectious-virus production through ZAP-dependent and -independent mechanisms. (A) Schematic representation of the HIV-1 env region with the regions of synonymously introduced CpGs in HIV-1env86–561CpG, HIV-1env611–1014CpG, and HIV-1env86–1014CpG highlighted. (B) ZAP expression in HeLa untransduced, control, ZAP-ex4, and ZAP-ex6 CRISPR cells was detected using immunoblotting. The untransduced cells are the parental HeLa cells that were not transduced with a CRISPR-Cas9 lentiviral vector. (C and D) HeLa control, ZAP-ex4, and ZAP-ex6 CRISPR cells were transfected with pHIV-1, pHIV-1env86–561CpG, pHIV-1env611–1014CpG, or pHIV-1env86–1014CpG plus pGFP (C) or pVSV-G (D). The culture supernatants were used to infect TZM-bl reporter cells to measure infectious-virus production. Gag expression in the media, as well as Gag, Hsp90, Env, actin, and Nef expression in the cell lysates, was detected using immunoblotting. The bar charts show the averages of the results of four independent experiments normalized to wild-type HIV-1 in HeLa control CRISPR cells. (E) Genomic-RNA abundance was quantified by qRT-PCR in cell lysates. The bar charts show the averages of the results of three independent experiments normalized to wild-type HIV-1 in HeLa control CRISPR cells. The error bars represent standard deviations. *, P < 0.05 as determined by a two-tailed unpaired t test; ns, not significant. The black asterisks/ns compare the virus containing introduced CpGs in the control CRISPR cells to wild-type HIV-1 in the control CRISPR cells. The red asterisks/ns compare the virus containing introduced CpGs between the ZAP CRISPR cells (red bars) and the control CRISPR cells (black bars).
TABLE 1.
CpGs and mutations introduced by codon modification
| Virus | Total no. within the modified regiona
|
||
|---|---|---|---|
| CpGs in wild-type virus | CpGs introduced | Mutations introduced | |
| HIV-1gag22–165CM | 4 | 11 | 49 |
| HIV-1gag22–261CM | 4 | 18 | 80 |
| HIV-1gag22–378CM | 4 | 26 | 109 |
| HIV-1gag22–378CM–no-CpG | 4 | 0 | 79 |
| HIV-1gag22–378CpG | 4 | 26 | 30 |
| HIV-1gag22–378DC | 4 | 0 | 23 |
| HIV-1gag22–378CM–5nt-CpG | 4 | 26 | 71 |
| HIV-1gag22–651CpG | 4 | 30 | 37 |
| HIV-1gag660–1185CpG | 5 | 32 | 36 |
| HIV-1gag22–1185CpG | 9 | 62 | 73 |
| HIV-1gag694–1206CpG | 5 | 60 | 72 |
| HIV-1pol795–1386CpG | 1 | 53 | 61 |
| HIV-1env86–561CpG | 1 | 36 | 43 |
| HIV-1env611–1014CpG | 4 | 48 | 52 |
| HIV-1env86–1014CpG | 5 | 84 | 95 |
| HIV-1–IRES-GFP | NA | 96 | NA |
| HIV-1–IRES-Renilla | NA | 96 | NA |
| HIV-1Gag-Venus | NA | 64 | NA |
| HIV-1Gag-STOP-Venus | NA | 64 | NA |
NA, not applicable.
We also combined the two regions in env containing 36 and 48 CpGs for a total of 84 CpGs (HIV-1env86–1014CpG) (Fig. 3A and Table 1). This inhibited HIV-1 infectious-virus production in an approximately additive manner compared to the two regions’ individual effects through both ZAP-dependent and ZAP-independent effects on genomic-RNA abundance and viral protein expression (Fig. 3C and E). To determine the contribution of decreased Env expression to CpG-mediated inhibition, we pseudotyped the viruses with the vesicular stomatitis virus glycoprotein (VSV-G) and found that the ZAP-dependent and -independent defects in infectious-virus production were still present (Fig. 3D).
We then introduced 53 CpGs into a region of pol that did not contain any known or detectable cis-acting elements (HIV-1pol795–1386CpG) (Fig. 2 and 4A and Table 1; see Data Set S2). Surprisingly, this large number of CpGs caused only a small (∼2-fold) reduction in Gag expression and infectious-virus production in control CRISPR cells, and this effect was eliminated in the ZAP knockout cells (Fig. 4B). This suggests that the magnitude of ZAP-dependent restriction is not simply proportional to the absolute number of CpGs added to the viral genome.
FIG 4.
Introduction of 53 CpGs into pol only moderately sensitizes the virus for ZAP- and KHNYN-dependent inhibition of infectious-virus production. (A) Schematic representation of HIV-1 pol with the region of synonymously introduced CpGs in HIV-1pol795–1386CpG highlighted. (B) HeLa control, ZAP-ex4, and ZAP-ex6 CRISPR cells were transfected with pHIV-1 or pHIV-1pol795–1386CpG plus pGFP. Viral infectivity was measured using TZM-bl reporter cells infected with cell culture supernatants. Gag expression in the media, as well as Gag and Hsp90 expression in the cell lysates, was detected by immunoblotting. The bar charts show the average values of the results of four independent experiments normalized to the values obtained for wild-type HIV-1 in the HeLa control CRISPR cells. The black asterisk compares the virus containing introduced CpGs in the control CRISPR cells to wild-type HIV-1 in the control CRISPR cells. The red asterisk/ns compares the virus containing introduced CpGs between the ZAP CRISPR cells (red bars) and the control CRISPR cells (black bars). (C and D) HeLa cells were transfected with 500 ng pHIV-1, pHIV-1EnvCpG86–561, or pHIV-1pol795–1386CpG and 500 ng of pGFP-FLAG or 31.25 ng, 62.5 ng, 125 ng, 250 ng, or 500 ng pKHNYN-1-FLAG plus the amount of pGFP-FLAG required to make 500 ng total. (C) Viral infectivity was measured using TZM-bl reporter cells infected with cell culture supernatants. Each point shows the average value of the results of three independent experiments normalized to the value obtained for wild-type HIV-1 in HeLa cells. *, P < 0.05; ns, not significant, as determined by a two-tailed unpaired t test. The black asterisks/ns compare the virus containing introduced CpGs to wild-type HIV-1 with 0 ng of KHNYN. The red asterisks/ns compare the virus containing introduced CpGs between the points of KHNYN overexpression to 0 ng of KHNYN. (D) Gag expression in the media, as well as Gag, Hsp90, Env, actin, and KHNYN-FLAG expression in the cell lysates, was detected using immunoblotting. (E) HeLa control and KHNYN-ex3 CRISPR cells were transfected with pHIV-1 or pHIV-1env86–561CpG, pHIV-1env611–1014CpG, pHIV-1env86–1014CpG, or pHIV-1pol795–1386CpG plus pGFP. Viral infectivity was measured using TZM-bl reporter cells infected with cell culture supernatants. Gag expression in the media, as well as Gag, Hsp90, Env. and actin expression in the cell lysates, was detected using immunoblotting. The bar charts show the average values of the results of three independent experiments normalized to the values obtained for wild-type HIV-1 in the HeLa control CRISPR cells. The error bars represent standard deviations. *, P < 0.05 as determined by a two-tailed unpaired t test. The black asterisks compare the virus containing introduced CpGs in the control CRISPR cells to wild-type HIV-1 in the control CRISPR cells. The red asterisks compare the virus containing introduced CpGs between the KHNYN CRISPR cells (red bars) and the control CRISPR cells (black bars).
We have recently identified KHNYN as an essential ZAP cofactor for CpGs to inhibit HIV-1env86–561CpG gene expression and infectious-virus production (31). Our previous work showed that KHNYN overexpression inhibited HIV-1env86–561CpG much more potently than wild-type HIV-1, indicating that the introduced CpGs were required for KHNYN to inhibit HIV-1 (31). To determine if inhibition by KHNYN correlated with CpG abundance or ZAP sensitivity, we overexpressed KHNYN on wild-type HIV-1, HIV-1env86–561CpG, or HIV-1pol795–1386CpG (Fig. 4C and D). KHNYN antiviral activity was correlated with the sensitivity of the virus to endogenous ZAP, with HIV-1env86–561CpG inhibited much more potently than HIV-1pol795–1386CpG or wild-type HIV-1. We also tested whether endogenous KHNYN was required to restrict HIV-1env611–1014CpG, HIV-1env86–1014CpG, and HIV-1pol795–1386CpG using the KHNYN CRISPR cells that we previously characterized (31). Wild-type HIV-1 infectious-virus production was not affected by depleting KHNYN, while HIV-1env86–561CpG infectious-virus production and gene expression were substantially increased (31) (Fig. 4E). The CpG-mediated inhibition of HIV-1env611–1014CpG and HIV-1env86–1014CpG infectious-virus production was partially rescued in KHNYN CRISPR cells, which was correlated with their restriction by ZAP (Fig. 3C and 4E). The small decrease in infectious-virus production for HIV-1pol795–1386CpG was rescued in the KHNYN CRISPR cells, indicating that the 53 introduced CpGs in pol moderately inhibited HIV-1 through ZAP and KHNYN (Fig. 4E).
HIV-1 strains containing reporter genes, such as those encoding green fluorescent protein (GFP) or luciferase, are important experimental tools and have large numbers of CpGs in the reporter gene. Therefore, it was important to determine whether ZAP inhibits these viruses, because this could confound the interpretation of results obtained under some experimental conditions. Specifically, we analyzed whether ZAP could restrict HIV-1 containing the encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES) followed by enhanced GFP (eGFP) (IRES-GFP) or Renilla luciferase (IRES-Renilla) (Fig. 5A and Table 1) (41, 42). Both IRES-GFP and IRES-Renilla introduced 96 CpGs into the viral genome. While both reporter viruses produced less Gag and infectious virus than wild-type HIV-1, ZAP depletion did not increase this production (Fig. 5B). We also analyzed the effect of a Venus fluorescent protein-plus-inker sequence that introduced 64 CpGs as a fusion protein with Gag (Fig. 5A and Table 1) (43). Interestingly, this sequence did not sensitize Gag abundance or virus-like particle (VLP) production to ZAP (Fig. 5C). Because a ribosome could displace ZAP bound to CpGs in an open reading frame as it moves along the mRNA, it is possible that CpGs in a 3′ untranslated region (UTR) could inhibit HIV-1 gene expression in a ZAP-dependent manner more effectively than CpGs in a coding region. Therefore, we inserted stop codons between Gag and Venus in both the Gag and Pol reading frames (Fig. 5A and Table 1). However, the CpGs in the context of the 3′ UTR also did not promote ZAP-mediated inhibition of Gag expression or VLP production (Fig. 5C). In sum, the total number of CpGs introduced into the HIV-1 genome does not correlate with their antiviral activity in the context of endogenous ZAP levels in HeLa cells.
FIG 5.
HIV-1–IRES-GFP, HIV-1–IRES-Renilla, and HIV-1 containing a Gag-Venus fusion protein are not targeted by endogenous levels of ZAP in HeLa cells. (A) Schematic representation of the HIV-1 nef/long terminal repeat (LTR) region with the introduced GFP and Renilla reporter genes and HIV-1 Gag-Venus fusion constructs. The nucleotide sequence shows the insertion between the gag and venus open reading frames that introduces stop codons (highlighted in red). (B) Control, ZAP-ex4, and ZAP-ex6 CRISPR cells were transfected with pHIV-1, HIV-1env86–561CpG, pHIV-1-IRES-GFP, or pHIV-1-IRES-Renilla plus pGFP. Infectious-virus production was measured using TZM-bl reporter cells infected with cell culture supernatants. Gag expression in the media, as well as Gag and Hsp90 expression in the cell lysates, was detected using immunoblotting. The bar chart shows the average values of the results of three independent experiments normalized to the values obtained for wild-type HIV-1 in HeLa control CRISPR cells. (C) Control and ZAP-ex6 CRISPR cells were transfected with pHIV-1Gag-Venus or pHIV-1Gag-STOP-Venus. Gag expression in the media, as well as Gag and Hsp90 expression in the cell lysates, was detected using immunoblotting. The bar chart shows the average values of the results of three independent experiments normalized to the virus in control CRISPR cells. The error bars represent standard deviations. *, P < 0.05; ns, not significant, as determined by a two-tailed unpaired t test. The black asterisks/ns compare the virus containing introduced CpGs in the control CRISPR cells to wild-type HIV-1 in the control CRISPR cells. The red asterisks/ns compare the virus containing introduced CpGs between the ZAP CRISPR cells (red bars) and the control CRISPR cells (black bars).
Increasing ZAP abundance inhibits CpG-containing HIV-1.
We then analyzed whether increasing ZAP abundance further inhibited HIV-1 containing introduced CpGs. First, we treated control and ZAP knockout HEK293T cells with type I interferon (IFN-I) (Fig. 6). Similar to previous reports, this consistently increased expression of the short isoform of ZAP (ZAP-S) by ∼2-fold and had no substantial effect on expression of the long isoform of ZAP (ZAP-L) (Fig. 6C) (44, 45). IFN-I inhibited all of the viruses. For wild-type HIV-1, the magnitudes of inhibition were similar in the control and ZAP knockout cells (Fig. 6A), which is consistent with the observation that endogenous ZAP does not target wild-type HIV-1 (16, 31). The magnitude of IFN-I inhibition of HIV-1env86–561CpG was reduced when ZAP was depleted, indicating that ZAP contributes to the antiviral effect of IFN-I on the virus. Importantly, IFN-I treatment augmented the ZAP-dependent inhibition of HIV-1pol795–1386CpG. This suggests that CpG-rich sequences in an HIV open reading frame that are only weakly restricted by endogenous ZAP levels can be further sensitized by IFN-I. However, even in the presence of IFN-I, the magnitude of inhibition by the 53 introduced CpGs in pol was lower than that of the inhibition mediated by the 36 introduced CpGs in env. For HIV-1–IRES-GFP and HIV-1–IRES-Renilla, the magnitude of IFN-I inhibition was not decreased upon ZAP depletion (Fig. 6B). This indicates that endogenous ZAP does not target these viruses, despite the large numbers of CpGs that have been introduced into the viral genome.
FIG 6.
Type I interferon inhibits HIV-1 containing introduced CpG dinucleotides using ZAP-dependent and ZAP-independent mechanisms and induces an ∼2-fold increase in ZAP-S expression in 293T cells. (A and B) HEK293T control and ZAP-ex6 CRISPR cells were transfected with pHIV-1, pHIV-1env86–561CpG, pHIV-1pol795–1386CpG, pHIV-1-IRES-GFP, or pHIV-1-IRES-Renilla plus pGFP with (+) or without (−) 1,000 U/ml of type I interferon treatment. Infectious-virus production was measured using TZM-bl reporter cells infected with cell culture supernatants. Gag expression in the media, as well as Gag and Hsp90 expression in the cell lysates, was detected using immunoblotting. The bar charts show the average values of the results of four independent experiments normalized to wild-type HIV-1 in HEK293T control CRISPR cells. The numbers above the bars represent the fold decrease in relative infectious-virus production due to the type I interferon treatment. The error bars represent standard deviations. *, P < 0.05; ns, not significant, as determined by a two-tailed unpaired t test. The black asterisks/ns compare the virus containing introduced CpGs in the control CRISPR cells to wild-type HIV-1 in the control CRISPR cells. The red asterisks/ns compare results without and with IFN-I treatment. The black bars are samples in control CRISPR cells and the red bars are samples in ZAP CRISPR cells. (C) Bar chart showing the average values of the results of 15 independent 1,000-U/ml interferon treatments for ZAP-S and ZAP-L expression with a representative Western blot for ZAP expression in HEK293T control and ZAP-ex6 CRISPR cells with and without type I interferon treatment. *, P < 0.05; ns, not significant, as determined by a two-tailed unpaired t test.
Because IFN-I only moderately upregulated ZAP-S, we also overexpressed ZAP-S or ZAP-L in HeLa cells. This increased ZAP-S expression ∼5-fold and ZAP-L expression ∼20-fold (Fig. 7C). Both ZAP isoforms inhibited all of the viruses tested at least 2-fold (Fig. 7). Interestingly, at high levels of ZAP-S or ZAP-L, the 36 introduced CpGs in env and the 53 CpGs in pol inhibited HIV-1 infectious-virus production to similar levels (Fig. 7A). Both HIV-1–IRES-GFP and HIV-1–IRES-Renilla were potently inhibited by overexpressed ZAP, indicating that when ZAP abundance is high enough, the CpGs introduced into these viruses can be targeted (Fig. 7B). Therefore, the inhibition observed for HIV-1–IRES-GFP and HIV-1–IRES-Renilla at high ZAP levels indicates that CpGs in contexts that are not targeted by the endogenous ZAP levels in HeLa cells (Fig. 4) can be targeted if ZAP abundance is substantially increased (Fig. 7B).
FIG 7.
ZAP overexpression inhibits wild-type HIV-1 and HIV-1 containing introduced CpG dinucleotides. (A and B) HeLa cells were transfected with pHIV-1, pHIV-1env86–561CpG, pHIV-1pol795–1386CpG, pHIV-1-IRES-GFP, or pHIV-1-IRES-Renilla plus either pHA-Renilla, pHA-ZAP-S, or pHA-ZAP-L. Infectious-virus production was measured using TZM-bl reporter cells infected with cell culture supernatants. Gag expression in the media, as well as Gag and Hsp90 expression in the cell lysates, was detected using immunoblotting. The bar charts show the average values of the results of four independent experiments normalized to wild-type HIV-1 plus HA-Renilla. The numbers above the bars represent the fold decrease in relative infectious-virus production due to ZAP-S or ZAP-L overexpression. (C) Bar chart and representative Western blot for ZAP expression showing the average ZAP abundance for 15 independent transfections of p-HA-ZAP-S or pHA-ZAP-L in HeLa cells. The error bars represent standard deviations. *, P < 0.05 as determined by a two-tailed unpaired t test. The black asterisks compare the virus containing introduced CpGs in the control CRISPR cells to wild-type HIV-1 in the control CRISPR cells. The red asterisks compare the virus with HA-Renilla overexpression (black bars) to HA-ZAP overexpression (red bars).
CpG dinucleotides introduced into the 5′ end of gag inhibit HIV-1 replication in a ZAP-independent manner.
We previously introduced CpG dinucleotides into nt 22 to 378 of gag in two different contexts (Fig. 8A and Table 1) and found that they inhibited viral replication with different phenotypes on Gag expression (15). For HIV-1gag22–378CM, the codon modified (CM) sequence was derived from a codon-optimized Gag-Pol construct and introduced 109 synonymous nucleotide changes and 26 CpGs (Table 1) (15, 46). In the context of a single-cycle infectivity assay, HIV-1gag22–378CM Gag expression and infectious-virus production were decreased to the limit of detection (Fig. 8B). This correlated with a large decrease in genomic-RNA abundance in the cell lysate and medium (Fig. 8C and D). The CpG dinucleotides were necessary to inhibit the virus, because when they were removed (while leaving the 79 mutations that did not introduce a CpG dinucleotide) to create HIV-1gag22–378CM–no-CpG (Fig. 8A and Table 1), gRNA abundance, Gag expression, and infectious-virion production were substantially increased (Fig. 8B). However, when the 26 CpG dinucleotides were introduced without the additional mutations in the codon-optimized Gag sequence to create HIV-1gag22–378CpG (Fig. 8A and Table 1), infectious-virus production was decreased by >95%, even though there was no substantial decrease in Gag expression (Fig. 8B).
FIG 8.
Introduction of CpG dinucleotides into gag nt 22 to 378 inhibits HIV-1 infectious-virus production. (A) Schematic representation of HIV-1 gag with introduced synonymous mutations in HIV-1gag22–378CM, HIV-1gag22–378CpG, HIV-1gag22–378CM–no-CpG, HIV-1gag22–378DC, and HIV-1gag22–378CM–5nt-CpG highlighted. (B to E) HeLa cells were transfected with pHIV-1, pHIV-1gag22–378CM, pHIV-1gag22–378CM–no-CpG, pHIV-1gag22–378CpG, or pHIV-1gag22–378DC (B to D) or pHIV-1, pHIV-1gag22–378CM, pHIV-1gag22–378CpG, or pHIV-1gag22–378CM–5nt-CpG and pGFP (E). The culture supernatants were used to infect TZM-bl reporter cells to measure the amount of infectious-virus production. (B and E) Gag expression in the media, as well as Gag, Hsp90, Env, and actin expression in the cell lysates, was detected using immunoblotting. (C and D) Genomic-RNA abundance was quantified by qRT-PCR in cell lysates (C) and media (D). The bar charts show the average values of the results of three (B and E) or four (C and D) independent experiments normalized to the value obtained for wild-type HIV-1. The error bars represent standard deviations. *, P < 0.05 as determined by a two-tailed unpaired t test comparing the virus containing synonymous mutations to wild-type HIV-1.
Deletion of nt 22 to 378 in gag does not substantially decrease infectious-virus production (15, 47), indicating that there are no essential cis-acting elements in the region. However, altering the RNA sequence could modulate the local RNA structure in ways that a large deletion did not, and the 5′ region of gag has been shown to indirectly regulate gRNA packaging by regulating the structure of the 5′ UTR (48, 49). To determine if the CpGs in HIV-1gag22–378CpG specifically decreased infectious-virus production or if a cis-acting regulatory element in the region had been mutated, we changed the codons previously mutated to introduce CpGs into a different codon (DC) that was not the wild-type HIV-1 codon and did not introduce a CpG where possible to produce pHIV-1gag22–378DC (Fig. 8A and Table 1). HIV-1gag22–378DC produced levels of infectious virus similar to those of wild-type HIV-1 (Fig. 8B). We also measured gRNA abundance in the cell lysate and medium. HIV-1gag22–378CpG had ∼60% and ∼80% decreases in gRNA in the lysate and medium, respectively, while HIV-1gag22–378DC had levels of gRNA similar to those of wild-type HIV-1 (Fig. 8C and D). This indicates that the introduced CpGs are necessary for the reduction in infectious-virus production and not a result of mutating essential cis-acting elements in the region that modulate gRNA packaging or other steps of HIV-1 replication. In sum, while HIV-1gag22–378CM and HIV-1gag22–378CpG have the same 26 introduced CpGs, there is a much larger decrease in Gag expression and intracellular gRNA abundance for HIV-1gag22–378CM, and this is due to the introduced CpGs. This suggests that the sequence surrounding the CpG dinucleotides modulates their inhibitory effect.
To test the effect of the sequence proximal to the CpG, we changed all of the mutations in HIV-1gag22–378CM that were not within 5 nucleotides of an introduced CpG back to the wild-type HIV-1 sequence to produce HIV-1gag22–378CM–5nt-CpG (Fig. 8A and Table 1). Interestingly, HIV-1gag22–378CM–5nt-CpG Gag expression and infectious-virus production were very similar to those of HIV-1gag22–378CM (Fig. 8E). This indicates that the sequence immediately surrounding the CpG dinucleotides influences the degree to which they inhibit HIV-1 gene expression. We also analyzed intracellular Env abundance and found that Env expression was reduced to undetectable levels for HIV-1gag22–378CM and HIV-1gag22–378CM–5nt-CpG (Fig. 8E). Env expression was also decreased for HIV-1gag22–378CpG, which likely accounts for the decreased infectious-virus production, in addition to the reduced gRNA levels present in virions (Fig. 8D). The decrease in Env expression was unexpected, because the region in the HIV-1 genome containing the introduced CpGs in these viruses is present only in the unspliced transcript encoding Gag and Gag-Pol and not in the singly spliced env mRNAs. Overall, the local sequence context of the CpGs in gag nt 22 to 378 determines the magnitude of inhibition for both Gag and Env expression.
We then analyzed whether ZAP was necessary for the CpGs in HIV-1gag22–378CM and HIV-1gag22–378CpG to inhibit Gag expression and infectious-virus production. In contrast to HIV-1env86–561CpG, ZAP depletion did not increase infectious-virus production for HIV-1gag22–378CM and HIV-1gag22–378CpG (Fig. 9A). This shows that the CpGs introduced into the 5′ region of gag inhibit HIV-1 gene expression and infectious-virus production through a ZAP-independent mechanism. To determine if introducing a larger number of CpGs into gag could make it ZAP sensitive, we cloned pHIV-1gag22–1188CpG, which contains 62 CpG dinucleotides distributed across ∼1,100 nt (Fig. 9B and Table 1). In addition, we produced pHIV-1gag22–654CpG and HIV-1gag658–1188CpG, which contain 30 and 32 CpGs, respectively. Introducing CpGs into nt 22 to 654 of gag led to large decreases in Gag expression, Env expression, and infectious-virus production (Fig. 9C). Interestingly, Gag expression for HIV-1gag22–1188CpG and HIV-1gag22–654CpG was increased in the ZAP knockout cells, though this did not affect Env expression or infectious-virus production. In contrast, introduction of 32 CpGs into nt 658 to 1188 of gag had no effect on Gag expression, Env expression, or infectious-virus production (Fig. 9C). Therefore, we introduced 60 CpGs into this 3′ region of gag (HIV-1gag694–1206CpG) (Fig. 9B and Table 1) and analyzed the effect on infectious-virus production in control and ZAP CRISPR cells (Fig. 9D). The 60 CpGs in the 3′ region of gag inhibited HIV-1 infectious-virus production about 2-fold, which is similar to the effect that 53 CpGs had in pol and less than the 36 CpGs in env. Overall, in some contexts, CpGs introduced into gag allow ZAP to inhibit Gag expression, but the number of CpGs is not correlated with the magnitude of the inhibitory effect. Furthermore, the CpGs introduced into the 5′ region of gag inhibit infectious-virus production in a ZAP-independent manner.
FIG 9.
CpG dinucleotides in gag have ZAP-dependent and ZAP-independent effects, but the number of CpGs is not correlated with antiviral activity. (A, C, and D) HeLa control, ZAP-ex4, and ZAP-ex6 CRISPR cells were transfected with pHIV-1, pHIV-1env88-561CpG, pHIV-1gag22–378CM, or pHIV-1gag22–378CpG (A); pHIV-1, pHIV-1gag22–1185CpG, pHIV-1gag22–651CpG, or pHIV-1gag660-1185CpG (C); or pHIV-1, pHIV-1env88-561CpG, pHIV-1pol795–1386CpG, or pHIV-1gag694–1206CpG plus pGFP (D). The culture supernatants were used to infect TZM-bl reporter cells to measure infectious-virus production. Gag expression in the media, as well as Gag, Hsp90, Env, and actin expression in the cell lysates, was detected using immunoblotting. The bar charts show the average values of the results of three (C and D) or four (A) independent experiments normalized to HIV-1 in HeLa control CRISPR cells. The error bars represent standard deviations. *, P < 0.05; ns, not significant, as determined by a two-tailed unpaired t test. The black asterisks/ns compare the virus containing introduced CpGs in the control CRISPR cells to wild-type HIV-1 in the control CRISPR cells. The red asterisks/ns compare the virus containing introduced CpGs between the ZAP CRISPR cells (red bars) and the control CRISPR cells (black bars). (B) Schematic representation of HIV-1 gag with the regions of synonymously introduced CpGs in HIV-1gag22–1185CpG, HIV-1gag22–651CpG, HIV-1gag660-1185CpG, and HIV-1gag694–1206CpG highlighted.
CpG dinucleotides introduced into gag can inhibit HIV-1 gene expression by modulating pre-mRNA splicing.
The HIV-1 genomic RNA undergoes extensive alternative splicing to mediate expression of all of the viral genes, and synonymous mutations in gag have previously been shown to disrupt HIV-1 splicing (36, 50). Since the CpGs in the 5′ region of gag reduced Env expression in all sequence contexts tested (Fig. 8E and 9C), we speculated that they could affect splicing. Therefore, we analyzed RNA abundance when progressively longer regions of codon-optimized gag sequence were added to the virus, which introduced 11, 18, or 26 CpGs (HIV-1gag22–165CM, HIV-1gag22–261CM, and HIV-1gag22–378CM) (Fig. 10A and Table 1). We have previously analyzed these viruses and have shown that they are deficient for genomic-RNA abundance and Gag expression (15). A comparison of the total RNA and the genomic RNA indicated that the abundance of both was decreased by the synonymous mutations (Fig. 10B). To determine whether the decrease in viral RNA abundance is due to altered splicing, we used RNA-seq to sequence the transcriptome in each sample and determine which splice sites were used to produce the mRNAs for each virus (Fig. 10C and Table 2; see Data Set S3 in the supplemental material). This analysis showed that a preexisting cryptic splice donor (CD1) was activated. Importantly, this donor is outside the region into which the CpGs were introduced (Fig. 10D). The frequency of use of the cryptic splice donor increased with the length of the codon-optimized gag sequence and coincided with a decrease in the utilization of canonical splice donor 1 (SD1). Activation of the cryptic splice donor increased the length of the first exon incorporated into all of the spliced viral RNAs to include the gag sequence prior to CD1. This led to the incorporation of the Gag initiation codon in every transcript upstream of the canonical start codon for the encoded protein (Fig. 10C and D), which is predicted to result in inefficient translation of all HIV-1 proteins encoded by a singly or fully spliced mRNA, including Tat and Rev. This could account for the decrease in total RNA levels, genomic-RNA levels, and Gag and Env expression that we observed for HIV-1gag22–378CM (Fig. 8 and 10) (15).
FIG 10.
Codon modification in the 5′ end of gag activates a cryptic splice donor. (A) Schematic representation of the HIV-1 gag region with introduced synonymous mutations in HIV-1gag22–165CM, HIV-1gag22–261CM, or HIV-1gag22–378CM highlighted. (B) HeLa cells were transfected with pHIV-1, pHIV-1gag22–165CM, pHIV-1gag22–261CM, or pHIV-1gag22–378CM. Total and genomic-RNA abundances were quantified by qRT-PCR in the cell lysates. The bar charts show the average values of the results of two independent experiments normalized to wild-type HIV-1. (C) Relative usage of splice donor 1 and cryptic splice donor 1 upon codon modification of HIV-1. The 9,173-nt HIV-1 genomic RNA and features are depicted in the “HIV-1 genome” track. Canonical donors (D1 to D4) and acceptors (A1 to A7), a codon modification-induced cryptic donor (CD1), and the gag start codon (AUG) are shown in the “Splice Sites and Gag AUG” track. The number of reads supporting use of D1 (nt 291) or CD1 (nt 718) paired with a given splice acceptor is depicted by line width (y axis). The splice data line height is arbitrary. The read numbers shown are the sums across duplicate samples per thousand HIV-mapping reads. (D) Alignment of gag sequences for HIV-1, HIV-1gag22–165CM, HIV-1gag22–261CM, and HIV-1gag22–378CM. Introduced CpGs are highlighted in red, and CpGs found in the wild-type HIV-1 gag sequence are highlighted in yellow. The cryptic splice donor (CD1) activated by the synonymous mutations in gag nt 22 to 378 is highlighted in green.
TABLE 2.
Splicing events from splice donor 1 or cryptic splice donor 1 to the canonical HIV-1 splice acceptors as a percentage of total HIV-1 splicing events
| Start (nt) | End (nt) | Splice sitesa | % of total events for HIV-1 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| WT.1 | WT.2 | CM22–165.1 | CM22–165.2 | CM22–261.1 | CM22–261.2 | CM22–378.1 | CM22–378.2 | |||
| 291 | 4460 | D1-A1 | 11.9 | 11.9 | 3.0 | 3.4 | 2.5 | 1.5 | 0.2 | 0.0 |
| 291 | 4937 | D1-A2 | 4.4 | 4.4 | 3.2 | 3.3 | 2.9 | 2.1 | 0.0 | 0.0 |
| 291 | 5324 | D1-A3 | 3.0 | 3.1 | 3.4 | 3.5 | 4.0 | 3.1 | 0.0 | 0.0 |
| 291 | 5483 | D1-A4c | 1.2 | 1.4 | 0.6 | 1.1 | 0.1 | 0.4 | 0.0 | 0.0 |
| 291 | 5501 | D1-A4a | 4.1 | 3.7 | 1.9 | 2.0 | 1.6 | 0.9 | 0.0 | 0.0 |
| 291 | 5507 | D1-A4b | 2.2 | 2.2 | 1.7 | 1.6 | 1.1 | 1.1 | 0.3 | 0.1 |
| 291 | 5523 | D1-A5 | 25.7 | 26.2 | 30.4 | 33.3 | 29.7 | 26.2 | 0.7 | 0.9 |
| 291 | 7916 | D1-A7 | 0.9 | 1.0 | 0.9 | 0.8 | 0.0 | 0.4 | 0.0 | 0.0 |
| 718 | 4460 | CD1-A1 | 0.0 | 0.0 | 0.6 | 0.4 | 2.0 | 1.5 | 7.8 | 7.4 |
| 718 | 4937 | CD1-A2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.5 | 0.8 | 1.7 | 1.4 |
| 718 | 5324 | CD1-A3 | 0.0 | 0.0 | 0.1 | 0.1 | 1.4 | 1.1 | 1.6 | 1.8 |
| 718 | 5483 | D1-A4c | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 718 | 5501 | CD1-A4a | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 1.0 | 1.2 |
| 718 | 5507 | CD1-A4b | 0.0 | 0.0 | 0.0 | 0.0 | 0.4 | 0.4 | 3.2 | 3.0 |
| 718 | 5523 | CD1-A5 | 0.0 | 0.0 | 0.2 | 0.4 | 4.7 | 4.4 | 24.9 | 23.1 |
| 718 | 7916 | CD1-A7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
D1, splice donor 1; CD1, cryptic splice donor 1.
DISCUSSION
There is selection against CpG dinucleotides in many vertebrate RNA viruses, and introducing them into viral genomes may allow novel vaccines to be developed (1–5, 9, 10). However, to attenuate viral replication, it is unclear how many CpGs are required, whether there is an optimal location to insert CpGs, and whether all CpGs inhibit via ZAP. Due to the profound suppression of CpG abundance in HIV-1, we used it as a model system to analyze how CpG dinucleotides inhibit viral replication.
Our results show that CpGs can inhibit HIV-1 replication through at least two independent, but not mutually exclusive, mechanisms. First, they can recruit ZAP and target the viral RNA for degradation. Second, they can inhibit replication by altering pre-mRNA splicing. In addition, CpGs could silence HIV-1 transcription through DNA methylation. The multiple mechanisms by which CpGs inhibit HIV-1 infectious-virus production may explain why they are strongly suppressed in the virus, even compared to other RNA viruses, and may also explain why small changes in the number of CpGs in env are linked to disease progression (18). Introducing CpGs into env nt 86 to 561 potently inhibits genomic-RNA abundance, Env expression, Gag expression, and infectious-virus production in a ZAP-dependent manner. However, ZAP depletion does not fully rescue infectious-virus production when CpGs are introduced into several other regions of the HIV-1 genome, highlighting the ZAP-independent effects of CpGs, as well as the sensitivity of the 5′ region of env for CpGs that mediate ZAP antiviral activity.
We have characterized the ZAP-independent effect for CpGs introduced into the 5′ end of gag. These CpGs have a dramatic effect on genomic-RNA levels, Gag expression, and Env expression by promoting the use of a cryptic 5′ splice donor at the expense of SD1. Interestingly, the magnitude of this effect is modulated by the sequence identity immediately surrounding the CpGs. It should be noted that these experiments were done in the context of transiently transfected proviral constructs in HeLa cells, but previous studies have shown that the splicing patterns for HIV-1 are similar in transfected cells and infected T cells (51, 52). The introduced CpGs do not directly enhance the strength of this splice site because they are upstream of the splice donor and do not affect the sequence itself. A previous report has identified that introducing synonymous mutations into the 5′ end of gag promotes splicing at this cryptic donor, but the role of CpGs in this was not characterized (50). We found that CpGs introduced into this region have multiple effects on viral replication, including decreases in genomic-RNA stability, Gag expression, virion production, and infectivity per genome (15). All of these phenotypes are likely due to the decreased use of SD1 and the corresponding increase in splicing from the cryptic splice donor in gag. The CpGs introduced into the 5′ end of gag could inhibit a preexisting exonic splicing silencer (ESS) or introduce an exonic splicing enhancer (ESE). We favor the hypothesis that the CpGs introduced an ESE, because the synonymous mutations promote splicing at the cryptic splice donor in a length-dependent manner and the sequence within 5 nucleotides surrounding the CpG modulates the magnitude of the decrease in Env and Gag expression. However, further experiments will be required in order to characterize how CpGs modulate splicing. There are approximately 1,500 RNA binding proteins in the human genome, most of which do not have a well-characterized recognition sequence, though several have been reported to bind sequences that contain CpGs (53, 54). Therefore, an unknown number of RNA binding proteins bind CpGs, and we do not yet know which protein regulates HIV-1 splicing in a CpG-dependent manner. In addition, introducing CpGs into the HIV-1 genome may affect its local or long-range RNA structures; posttranscriptional modifications, such as cytosine methylation; or other aspects of RNA biology (55–57).
Surprisingly, the magnitude of ZAP-mediated inhibition was not correlated with the number of CpGs introduced into the viral genome, and some regions of the genome can tolerate substantial numbers of CpGs. Thirty-six CpGs inserted into the 5′ region of env had the greatest ZAP-dependent inhibitory phenotype. This corresponds to the observations by Takata et al., who first identified this region in a panel of viruses with large numbers of synonymous mutations in different regions of the HIV-1 genome (16, 50). The introduction of 53 CpGs into pol or 60 CpGs into the 3′ end of gag produced only a small amount of ZAP-dependent inhibition of infectious-virus production. KHNYN overexpression also produced less inhibition of infectious-virus production when 53 CpGs were introduced into pol than when 36 CpGs were added to env. This supports our hypothesis that KHNYN antiviral activity is controlled by ZAP’s ability to target the viral RNA (31). Only when ZAP levels were very high due to overexpression from a cDNA plasmid did the 53 CpGs in pol mediate a level of repression similar to that mediated by the 36 CpGs introduced into env nt 86 to 561. This highlights the facts that this region in env is very sensitive to the endogenous levels of ZAP in HeLa cells and that the position or local context of the CpG is important for ZAP to inhibit the virus. Interestingly, the weak inhibition of infectious-virus production by the 53 CpGs in pol mediated by endogenous ZAP levels could be substantially enhanced by IFN-I treatment. Therefore, part of the anti-HIV activity mediated by IFN-I (58) may be to promote ZAP targeting CpGs in contexts where it normally does so inefficiently. The fact that there is only a small increase in ZAP-S abundance upon IFN-I treatment raises the question of whether this induction of ZAP is sufficient to explain the phenotype or whether increased abundance or activity of ZAP cofactors, such as TRIM25 or KHNYN, may contribute (29–31, 59).
Because HiV-1 strains containing reporter genes are commonly used research tools, we investigated whether the large numbers of CpGs in the EMCV IRES, GFP, or Renilla luciferase could sensitize these viruses to ZAP. Adding 96 CpGs to the 3′ end of the genome in the context of IRES-GFP or IRES-Renilla luciferase did not sensitize the virus to the endogenous levels of ZAP in HeLa or HEK293T cells. Similarly, HIV-1 with GFP in place of nef has previously been shown not to be targeted by endogenous levels of ZAP in HeLa or MT4 cells (16). While HIV-1–IRES-GFP and HIV-1–IRES-Renilla were not inhibited by ZAP after IFN-I treatment, they were inhibited when high levels of ZAP were present due to overexpression from a cDNA plasmid. Therefore, ZAP abundance can determine whether CpG-containing viral genomes are targeted, though it is unclear whether the ZAP levels produced from plasmid-based overexpression can be achieved in a relevant in vivo context. This suggests that these reporter viruses are useful tools that may not be affected by ZAP under many experimental conditions.
An important area of future research is to determine why CpGs in some contexts or regions are efficiently targeted by ZAP and others are not. To date, the primary evidence that ZAP directly binds CpGs comes from PAR-CLIP experiments (16). The advantage of this technique is that it captures ZAP binding to CpGs in a living cell. However, other cellular factors present could modulate ZAP’s binding specificity. Several groups have shown that the ZAP cofactor TRIM25 can bind cellular and viral RNA, and it has been reported to regulate ZAP binding to Sindbis virus RNA (29, 30, 60–65). Therefore, TRIM25 or other ZAP cofactors could bind specific motifs in viral RNA to determine the sensitivity of the RNA to ZAP-mediated antiviral activity. In addition, it is not known how many ZAP molecules are required to bind RNA to mediate antiviral activity or if they have to be clustered in a specific way. While structural and mutagenesis studies of the RNA binding domain in ZAP have shown that it is a dimer that may have two RNA binding cavities within a large RNA binding cleft, how it binds CpG dinucleotides remains unknown (66, 67). To fully understand how specific CpGs mediate ZAP-dependent antiviral activity, it will be essential to understand how ZAP binds CpGs in specific RNA contexts and structures and the role its cofactors play in modulating its RNA binding activity.
It will be interesting to compare how CpGs inhibit HIV-1 to how they inhibit other RNA viruses and if they do so by targeting viral RNA for degradation, by inhibiting its translation, or through other mechanisms. CpGs directly or indirectly introduced into coding and noncoding regions of picornaviruses have shown that they can potently attenuate viral replication and create strains that protect animals from challenge with the wild-type virus (6–8, 10, 68). ZAP has recently been shown to be necessary for introduced CpGs to inhibit the picornavirus echovirus 7 (32). CpGs have also been shown to attenuate influenza A virus and to protect animals from lethal challenge by the wild-type virus (9). However, the molecular mechanism of attenuation remains unclear, and the CpGs could inhibit via ZAP, altered RNA splicing similar to what we have observed in HIV-1, or other mechanisms. This highlights both a challenge and an opportunity for introducing CpG dinucleotides to create live attenuated vaccines. The multiple mechanisms of action, as well as the position and context dependence of CpG-mediated viral inhibition, pose a challenge to determining the engineering principles for attenuating viruses with a predicted magnitude and mechanism. The opportunity is that CpGs can be used to attenuate viruses through multiple and potentially additive or synergistic mechanisms, which may enhance the utility of this approach.
MATERIALS AND METHODS
Sequence analysis of viral genomes.
The “analyze base composition” tool in MacVector (MacVector Inc.) was used to calculate the CpG and UpA observed/expected ratios for the viral sequences. The observed/expected ratio was calculated using the following formula: number of dinucleotide occurrences/(frequency of first nucleotide × frequency of second nucleotide), where the frequency of the base is the number of occurrences of the base divided by the total number of bases in the sequence. For the retroviral sequences, the sequence representing the packaged genome was used, i.e., the sequence encompassing the 5′ repeat (R) to the 3′ R.
Synonymous site conservation analysis.
Synonymous site conservation was calculated with synplot2 (37) for codons in a gag-pol-vif-vpr-5′ tat-vpu-env-nef in-frame fusion. The 16-nt noncoding sequence between tat and vpu was deleted, the frameshift heptanucleotide was changed from UUUUUUA to UUUUUUAA to fuse gag and pol, and single-nucleotide insertions (N) or deletions were applied as necessary to fuse ORFs in frame. Sequences were translated into amino acid sequences, which were aligned with CLUSTAL W (69). The amino acid alignment was then converted back to a nucleotide alignment with tranalign from the EMBOSS package (70). The alignment was then mapped to the coordinates of a specific reference sequence (viz. EU541617) by removing alignment positions that contained a gap character in the reference sequence. After calculating the synonymous site conservation statistics with synplot2, they were mapped to EU541617 genome coordinates (i.e., by reinserting noncoding regions) for display (Fig. 2). The following GenBank sequences were used in the alignment: AB023804, AF321523, EU861977, JN944905, JX236669, M38429, AB287379, AY805330, FJ670516, JN944907, JX236670, AB485648, FJ771006, JN944941, JX236671, AB485649, FJ771007, JN944942, JX236672, AF004394, FJ771008, JN944943, JX236673, U26942, AF004885, FJ771009, JN944944, JX236676, U34603, AF005494, FJ771010, JN944945, JX236677, U34604, AF005496, GQ290462, JN944946, JX236678, U39362, AF077336, GU237072, JN944947, JX236679, U63632, AF082394, DQ676872, GU362882, JN944948, K03454, U88824, AF082395, DQ979025, JN106043, JX236668, M27323, AF286237, EU541617, KC156212, KC156213, KC156214, KC156215, KC156218, and KC156221. We also used the WARO, MCST, RHGA, STCOr1, and CH457 sequences, which have previously been described (71, 72). The z-scores for 9-, 15-, and 25-codon windows were as follows: 9-codon window, z ≥ 3.603808; 15-codon window, z ≥ 3.468872; 25-codon window, z ≥ 3.329161. These equate to a P value of 0.05, with a correction for multiple testing based on the number of independent (i.e., nonoverlapping) windows of length x codons in the 2,870-codon alignment, i.e., 0.05/(2,870/x). The scores correspond to the whole x-codon window centered on a given codon.
Plasmids.
All primers were ordered from Eurofins, and the PCRs were performed with Phusion high-fidelity polymerase (New England Biolabs). Diagnostic restriction enzyme digestion and DNA sequencing (Eurofins Genomics) were used to ensure the correct identity of modified sequences inserted into plasmids. The pHIV-1NL4-3 constructs used in this study contain the provirus sequence from pHIV-1NL4-3 (73) cloned into pGL4.10 (Promega) (15). The sequences for HIV-1gag22–378DC, pHIV-1gag22–654CpG, pHIV-1gag654–1188CpG, pHIV-1gag22–1188CpG, pHIV-1gag694–1204CpG, pHIVenv611–1014CpG, and pHIV-1pol795–1386CpG were synthesized by Life Technologies and cloned into pHIV-1NL4-3. CpG dinucleotides were introduced through synonymous mutations without altering cryptic splice sites predicted by the MaxEnt tool in Human Splicing Finder (74, 75) in pHIV-1gag22–654CpG, pHIV-1gag654–1188CpG, pHIV-1gag22–1188CpG, pHIV-1gag694–1204CpG, pHIVenv611–1014CpG, and pHIV-1pol795–1386CpG. To generate pHIV-1 Gag-Venus, the SphI and XmaI fragment of pGag-Venus (43) was amplified via PCR and subcloned into pHIV-1NL4-3 in pGL4.10. pHIV-1 Gag-STOP-Venus was generated by PCR amplification of the SphI and XmaI sites in pGag-Venus using a primer that introduced the 3 stop codons, which was then subcloned into pHIV-1Gag-Venus. Hemagglutinin (HA)-Renilla, HA–ZAP-S, and HA–ZAP-L sequences were amplified by PCR and cloned into pcDNA3.1(+) (Invitrogen). pHIV-1gag22–165CM, pHIV-1gag22–261CM, pHIV-1gag22–378CM, HIV-1gag22–378CM–no-CpG, pHIV-1gag22–378CpG, pHIV-1-IRES-GFP, pHIV-1-IRES-Renilla, pHIV-1env 86–561CpG, pKHNYN-1, pGFP-FLAG, pGFP, and pVSV-G have been previously described (15, 31, 41, 42, 76, 77). Of note, pHIV-1gag22–165CM, pHIV-1gag22–261CM, and pHIV-1gag22–378CM were previously called pHIV-1 CM22–165, pHIV-1 CM22–261, and pHIV-1 CM22–378, respectively (15). pHIV-1gag22–378CM–no-CpG was previously called pHIV-1 CM22–378lowCpG, and pHIV-1gag22–378CpG was called pHIV-1 CpG22–378 (15). pHIV-1env86–561CpG was previously called pHIV-1EnvCpG86–561 (31).
Cell culture and cell lines.
HeLa, TZM-bl, and HEK293T cells were cultured in Dulbecco’s modified Eagle medium (Gibco) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. All the cell lines were grown at 37°C in a humidified atmosphere with 5% CO2. For the production of ZAP and KHNYN knockout cell lines by CRISPR-Cas9, ZAP- and KHNYN-targeting guide sequences were inserted into a lentivirus-based CRISPR plasmid (LentiCRISPRv2-puro) from Addgene (no. 52961) (78). The CRISPR guide sequences were as follows: Luciferase-G1 (control), 5′-CTT TAC CGA CGC ACA TAT CG-3′; ZAP-ex4, 5′-TCT GGT AGA AGT TAT ATC TG-3′; ZAP-ex6, 5′-ACT TCC ATC TGC CTT ACC GG-3′; and KHNYN-ex3, 5′-GGG GGT GAG CGT CCT TCC GA-3. LentiCRISPR vectors encoding guide RNAs targeting luciferase or ZAP were produced in HEK293T cells seeded in 6-well plates using 10 μl polyethylenimine (PEI) with 0.5 μg pVSV-G (76), 1.0 μg pCMVΔR8.91 (79), and 1.0 μg LentiCRISPRv2 (78). Virus-containing supernatant was harvested 48 h after transfection, rendered cell free via filtration through 0.45-μm filters (Millipore), and used to transduce HeLa or HEK293T cells, followed by selection in 1 μg/ml puromycin (Sigma-Aldrich). ZAP CRISPR HeLa cell clones were generated by limiting dilution. The ZAP-ex6 and KHNYN-ex3 CRISPR clones have previously been described and were called ZAP-G1 CRISPR clone and KHNYN-G1 CRISPR clone B (31). Loss of ZAP protein expression was confirmed by Western blotting.
Transfections.
HeLa and HEK293T cells were seeded in 6-well plates and transfected at a confluence of 70%. HeLa cells were transfected using TransIT-LT1 (Mirus) according to the manufacturer’s instructions at a ratio of 3 μl TransIT-LT1 to 1 μg DNA. HEK293T cells were transfected according to the manufacturer’s instructions using PEI (1 mg/ml; Sigma-Aldrich) at a ratio of 4 μl PEI to 1 μg DNA. For each transfection, 0.5 μg pHIV-1 and 0.5 μg pGFP, pGFP-FLAG, pKHNYN-1, pVSVG, pHA-ZAP-S, pHA-ZAP-L, or pHA-Renilla was used, for a total of 1 μg DNA. The transfection medium was replaced with fresh medium 6 h (HEK293T) or 24 h (HeLa) posttransfection. The cells were lysed 48 h posttransfection, and the medium was recovered. In experiments performed with type I interferon, 1,000 U/ml of IFN-I (Universal Type I Interferon; PBL Assay Science) was added to the cells upon medium change 6 h posttransfection. The medium was filtered through a 0.45-μm filter, and the virions were pelleted for 2 h at 20,000 × g through a 20% sucrose cushion in phosphate-buffered saline (PBS) solution.
TZM-bl cell infectivity assay.
Supernatant was recovered 48 h posttransfection and filtered as previously described. TZM-bl cells were seeded at 70% confluence in 24-well plates and infected by overnight incubation with filtered virus stocks. Forty-eight hours postinfection, the cells were lysed, and the amount of infectious-virus production was measured by induction of β-galactosidase using the Galacto-Star system (Applied Biosystems) following the manufacturer’s instructions. β-Galactosidase activity was quantified as relative light units per second using a PerkinElmer luminometer.
Analysis of protein expression by immunoblotting.
Approximately 48-h posttransfection, HeLa or HEK293T cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1% sodium dodecyl sulfate [SDS], 1% Triton X-100, 1% sodium deoxycholate). The medium was filtered through a 0.45-μm filter, and the virions were pelleted through a 20% sucrose cushion in PBS solution for 2 h at 20,000 × g. The pellet was resuspended in 2× loading buffer (60 mM Tris-HCl, pH 6.8, 10% β-mercaptoethanol, 10% glycerol, 2% SDS, 0.1% bromophenol blue). Cell lysates and virions were resolved by SDS-polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane. The antibodies used in the study were 1:50 anti-HIV-1 p24Gag (mouse; NIH AIDS Reagent Program; catalog no. 1513) (80), 1:3,000 anti-HIV-1 gp160/120 rabbit (ADP421; Centralized Facility for AIDS Reagents [CFAR]), 1:1,000 anti-Hsp90 (sc7947; Santa Cruz Biotechnology), 1:5,000 β-actin (ac-15; Sigma), 1:5,000 anti-ZAP (Abcam; ab154680), 1:2,500 anti-HA (Biolegend; 901514), 1:2,500 anti-DYKDDDDK (rabbit; Cell Signaling; 14793), 1:1,000 anti-HIV-1 Nef (mouse; NIH AIDS Reagent Program; catalog no. 1539) (81). 1:10,000 Dylight 800-conjugated anti-mouse/rabbit secondary antibodies (Cell Signaling Technology; 5151S and 5257S), 1:5,000 anti-mouse IRDye 680RD (Li-CoR; 926-68070), 1:5,000 anti-rabbit horseradish peroxidase (HRP) (Cell Signaling Technology; 7074), and 1:5,000 anti-mouse HRP (Cell Signaling Technology; 7076). Bound primary antibodies were detected via Li-CoR infrared imaging (Li-CoR UK Ltd.) or using the Amersham ECL Prime Western blotting detection reagent (GE Life Sciences) for HRP-linked antibodies using an ImageQuant (LAS4000 Mini).
Quantitative reverse transcription (qRT)-PCR.
HeLa cells were transfected at a confluence of 70% in a 6-well plate and after 48 h were washed with 1× PBS and lysed. The RNA was extracted using an RNeasy kit (Qiagen) following the manufacturer’s instructions. The supernatant was also collected and treated for 3 h at 37°C with RQ1 DNase (Invitrogen) to decrease plasmid DNA contamination. cDNA was synthesized using a high-capacity cDNA archive kit (Applied Biosystems), and 1 μg of RNA from virions was isolated using a QIAamp viral RNA minikit following the manufacturer’s instructions. Because carrier RNA was added to the lysis buffer, the total RNA isolated was quantified using a Qubit 3.0 fluorometer (ThermoFisher) and normalized so that 20 ng of RNA from each sample was reverse transcribed using the high-capacity cDNA archive kit (Applied Biosystems). Quantitative PCRs (qPCRs) were performed in triplicate with TaqMan universal PCR mix using an Applied Biosystems 7500 real-time PCR system. The HIV-1NL4-3 gRNA primers were GGCCAGGGAATTTTCTTCAGA/TTGTCTCTTCCCCAAACCTGA (forward/reverse), and the probe was 6-carboxyfluorescein (FAM)-ACCAGAGCCAACAGCCCCACCAGA-6-carboxytetramethylrhodamine (TAMRA). The HIV-1NL4-3 total RNA primers were TAACTAGGGAACCCACTGC/GCTAGAGATTTTCCACACTG (forward/reverse), and the probe was FAM-ACACAACAGACGGGCACACACTA-TAMRA. The absolute number of copies was determined using the slope of the standard curve at a qPCR efficiency between 95% and 105%.
Analysis of HIV-1 splicing.
HeLa cells were transfected at a confluence of 70% in a 6-well plate and after ∼40 h were washed with 1× PBS and lysed. The RNA was extracted using an RNeasy kit (Qiagen) following the manufacturer’s instructions. The RNA samples were quantified using a Qubit 2.0 fluorometer (Life Technologies, Carlsbad, CA, USA), and RNA integrity was checked with RNA screen tape on an Agilent 2200 TapeStation (Agilent Technologies, Palo Alto, CA, USA). The multiplexed RNA-sequencing library was prepared using an Illumina TruSeq stranded mRNA library preparation kit following the manufacturer’s protocol. Sequencing libraries were validated using DNA analysis screen tape on an Agilent 2200 TapeStation and quantified by using a Qubit 2.0 fluorometer, as well as by quantitative PCR (Applied Biosystems, Carlsbad, CA, USA). The sequencing libraries were multiplexed and clustered on two flow cell lanes. After clustering, the flow cell was loaded on the Illumina HiSeq instrument according to the manufacturer’s instructions. The samples were sequenced, using a 2-by-150 paired-end (PE) high-output configuration, by Genewiz. Image analysis and base calling were conducted with HiSeq control software (HCS) on the HiSeq instrument. The raw sequence data (.bcl files) generated from the Illumina HiSeq were converted into fastq files and demultiplexed using the Illumina bcl2fastq program version 2.17. Adapter trimming was performed using BBDuk (https://jgi.doe.gov/data-andtools/bbtools/), and read pairs were merged with BBmerge (https://jgi.doe.gov/data-and-tools/bbtools/) in order to increase base call quality and generate long, whole-fragment reads. The reads were then aligned with the human genome (hg38) and the HIVNL4-3 genomic-RNA sequence simultaneously using Hisat2 (82). HIV-mapping junction-spanning reads were isolated using regtools (https://github.com/griffithlab/regtools) to allow per-junction read counting. To visualize junctions of interest, data from replicates were first merged using the Picard (http://broadinstitute.github.io/picard) MergeSamFiles function, followed by generation of sashimi plots using Gviz (83). The percentage of HIV-1 junction-spanning reads was calculated by dividing the number of reads for each junction by the total number of junction-spanning reads in the library.
Statistical analysis.
Statistical significance was determined using unpaired two-tailed t tests calculated using Microsoft Excel software. Data are represented as means ± standard deviations. Significance was ascribed to P values of <0.05.
Data availability.
The RNA-seq data are available in the NCBI Gene Expression Omnibus (GEO) under accession number GSE134159. The GenBank accession numbers for the HIV-1 genomic-RNA nucleotide sequences are as follows: HIV-1 (strain NL4-3), MN685337; HIV-1gag22–165CM, MN685338; HIV-1gag22–261CM, MN685339; HIV-1gag22–378CM, MN685340; HIV-1gag22–378CM–no-CpG, MN685341; HIV-1gag22–378CpG, MN685342; HIV-1gag22–378DC, MN685343; HIV-1gag22–378CM–5nt-CpG, MN685344; HIV-1gag22–651CpG, MN685345; HIV-1gag660-1185CpG, MN685346; HIV-1gag22–1185CpG, MN685347; HIV-1gag694–1206CpG, MN685348; HIV-1pol795–1386CpG, MN685349; HIV-1env86–561CpG, MN685350; HIV-1env611–1014CpG, MN685351; HIV-1env86–1014CpG, MN685352.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Swanson and Neil laboratories for helpful discussions. The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: TZM-bl from John C. Kappes, Xiaoyun Wu, and Tranzyme Inc.; HIV-1 p24 hybridoma (183-H12-5C; catalog no. 1513) from Bruce Chesebro; and anti-HIV-1 NefMonoclonal (6.2) from Kai Krohn and Vladimir Ovod. The antiserum to HIV-1 gp120 no. 20 (ARP421) was obtained from the NIBSC Center for AIDS Reagents. We thank Akira Ono and Harmit Malik for generously providing reagents. We also thank Robert Gifford for providing advice and sequence information on retroviral phylogeny.
These studies were funded by MRC Discovery award MC/PC/15068 and a Wellcome Trust Senior Research Fellowship (WT098049AIA) to S.J.D.N., a Wellcome Trust Senior Research Fellowship (106207) to A.E.F., Medical Research Council grant MR/M019756/1 to R.S. and C.M.S., and Medical Research Council grant MR/S000844/1 to S.J.D.N. and C.M.S. Medical Research Council grant MR/S000844/1 is part of the EDCTP2 programme supported by the European Union. M.F. is supported by the United Kingdom Medical Research Council (MR/R50225X/1) and is a King’s College London member of the MRC Doctoral Training Partnership in Biomedical Sciences. I.A.-A. and R.H.-W. are supported by the King’s Bioscience Institute and the Guy’s and St Thomas’ Charity Prize Ph.D. Program in Biomedical and Translational Science. This work was also supported by the Department of Health via a National Institute for Health Research Comprehensive Biomedical Research Center award to Guy’s and St. Thomas’ NHS Foundation Trust in partnership with King’s College London and King’s College Hospital NHS Foundation Trust.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Greenbaum BD, Levine AJ, Bhanot G, Rabadan R. 2008. Patterns of evolution and host gene mimicry in influenza and other RNA viruses. PLoS Pathog 4:e1000079. doi: 10.1371/journal.ppat.1000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karlin S, Doerfler W, Cardon LR. 1994. Why is CpG suppressed in the genomes of virtually all small eukaryotic viruses but not in those of large eukaryotic viruses? J Virol 68:2889–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simmonds P, Xia W, Baillie JK, McKinnon K. 2013. Modelling mutational and selection pressures on dinucleotides in eukaryotic phyla—selection against CpG and UpA in cytoplasmically expressed RNA and in RNA viruses. BMC Genomics 14:610. doi: 10.1186/1471-2164-14-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rima BK, McFerran NV. 1997. Dinucleotide and stop codon frequencies in single-stranded RNA viruses. J Gen Virol 78:2859–2870. doi: 10.1099/0022-1317-78-11-2859. [DOI] [PubMed] [Google Scholar]
- 5.Kunec D, Osterrieder N. 2016. Codon pair bias is a direct consequence of dinucleotide bias. Cell Rep 14:55–67. doi: 10.1016/j.celrep.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 6.Atkinson NJ, Witteveldt J, Evans DJ, Simmonds P. 2014. The influence of CpG and UpA dinucleotide frequencies on RNA virus replication and characterization of the innate cellular pathways underlying virus attenuation and enhanced replication. Nucleic Acids Res 42:4527–4545. doi: 10.1093/nar/gku075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burns CC, Campagnoli R, Shaw J, Vincent A, Jorba J, Kew O. 2009. Genetic inactivation of poliovirus infectivity by increasing the frequencies of CpG and UpA dinucleotides within and across synonymous capsid region codons. J Virol 83:9957–9969. doi: 10.1128/JVI.00508-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fros JJ, Dietrich I, Alshaikhahmed K, Passchier TC, Evans DJ, Simmonds P. 2017. CpG and UpA dinucleotides in both coding and non-coding regions of echovirus 7 inhibit replication initiation post-entry. Elife 6:e29112. doi: 10.7554/eLife.29112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaunt E, Wise HM, Zhang H, Lee LN, Atkinson NJ, Nicol MQ, Highton AJ, Klenerman P, Beard PM, Dutia BM, Digard P, Simmonds P. 2016. Elevation of CpG frequencies in influenza A genome attenuates pathogenicity but enhances host response to infection. Elife 5:e12735. doi: 10.7554/eLife.12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tulloch F, Atkinson NJ, Evans DJ, Ryan MD, Simmonds P. 2014. RNA virus attenuation by codon pair deoptimisation is an artefact of increases in CpG/UpA dinucleotide frequencies. Elife 3:e04531. doi: 10.7554/eLife.04531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frankel AD, Young JA. 1998. HIV-1: fifteen proteins and an RNA. Annu Rev Biochem 67:1–25. doi: 10.1146/annurev.biochem.67.1.1. [DOI] [PubMed] [Google Scholar]
- 12.Kypr J, Mrazek J, Reich J. 1989. Nucleotide composition bias and CpG dinucleotide content in the genomes of HIV and HTLV 1/2. Biochim Biophys Acta 1009:280–282. doi: 10.1016/0167-4781(89)90114-0. [DOI] [PubMed] [Google Scholar]
- 13.Shpaer EG, Mullins JI. 1990. Selection against CpG dinucleotides in lentiviral genes: a possible role of methylation in regulation of viral expression. Nucleic Acids Res 18:5793–5797. doi: 10.1093/nar/18.19.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohno S, Yomo T. 1990. Various regulatory sequences are deprived of their uniqueness by the universal rule of TA/CG deficiency and TG/CT excess. Proc Natl Acad Sci U S A 87:1218–1222. doi: 10.1073/pnas.87.3.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antzin-Anduetza I, Mahiet C, Granger LA, Odendall C, Swanson CM. 2017. Increasing the CpG dinucleotide abundance in the HIV-1 genomic RNA inhibits viral replication. Retrovirology 14:49. doi: 10.1186/s12977-017-0374-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takata MA, Goncalves-Carneiro D, Zang TM, Soll SJ, York A, Blanco-Melo D, Bieniasz PD. 2017. CG dinucleotide suppression enables antiviral defence targeting non-self RNA. Nature 550:124–127. doi: 10.1038/nature24039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Theys K, Feder AF, Gelbart M, Hartl M, Stern A, Pennings PS. 2018. Within-patient mutation frequencies reveal fitness costs of CpG dinucleotides and drastic amino acid changes in HIV. PLoS Genet 14:e1007420. doi: 10.1371/journal.pgen.1007420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wasson MK, Borkakoti J, Kumar A, Biswas B, Vivekanandan P. 2017. The CpG dinucleotide content of the HIV-1 envelope gene may predict disease progression. Sci Rep 7:8162. doi: 10.1038/s41598-017-08716-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alinejad-Rokny H, Anwar F, Waters SA, Davenport MP, Ebrahimi D. 2016. Source of CpG depletion in the HIV-1 genome. Mol Biol Evol 33:3205–3212. doi: 10.1093/molbev/msw205. [DOI] [PubMed] [Google Scholar]
- 20.Gao G, Guo X, Goff SP. 2002. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 297:1703–1706. doi: 10.1126/science.1074276. [DOI] [PubMed] [Google Scholar]
- 21.Bick MJ, Carroll JW, Gao G, Goff SP, Rice CM, MacDonald MR. 2003. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J Virol 77:11555–11562. doi: 10.1128/jvi.77.21.11555-11562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muller S, Moller P, Bick MJ, Wurr S, Becker S, Gunther S, Kummerer BM. 2007. Inhibition of filovirus replication by the zinc finger antiviral protein. J Virol 81:2391–2400. doi: 10.1128/JVI.01601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao R, Nie H, Cai D, Zhang J, Liu H, Yan R, Cuconati A, Block TM, Guo JT, Guo H. 2013. Inhibition of hepatitis B virus replication by the host zinc finger antiviral protein. PLoS Pathog 9:e1003494. doi: 10.1371/journal.ppat.1003494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiu HP, Chiu H, Yang CF, Lee YL, Chiu FL, Kuo HC, Lin RJ, Lin YL. 2018. Inhibition of Japanese encephalitis virus infection by the host zinc-finger antiviral protein. PLoS Pathog 14:e1007166. doi: 10.1371/journal.ppat.1007166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li M, Yan K, Wei L, Yang J, Lu C, Xiong F, Zheng C, Xu W. 2015. Zinc finger antiviral protein inhibits coxsackievirus B3 virus replication and protects against viral myocarditis. Antiviral Res 123:50–61. doi: 10.1016/j.antiviral.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 26.Guo X, Carroll JW, Macdonald MR, Goff SP, Gao G. 2004. The zinc finger antiviral protein directly binds to specific viral mRNAs through the CCCH zinc finger motifs. J Virol 78:12781–12787. doi: 10.1128/JVI.78.23.12781-12787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo X, Ma J, Sun J, Gao G. 2007. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc Natl Acad Sci U S A 104:151–156. doi: 10.1073/pnas.0607063104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu Y, Wang X, Goff SP, Gao G. 2012. Translational repression precedes and is required for ZAP-mediated mRNA decay. EMBO J 31:4236–4246. doi: 10.1038/emboj.2012.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li MM, Lau Z, Cheung P, Aguilar EG, Schneider WM, Bozzacco L, Molina H, Buehler E, Takaoka A, Rice CM, Felsenfeld DP, MacDonald MR. 2017. TRIM25 enhances the antiviral action of zinc-finger antiviral protein (ZAP). PLoS Pathog 13:e1006145. doi: 10.1371/journal.ppat.1006145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng X, Wang X, Tu F, Wang Q, Fan Z, Gao G. 2017. TRIM25 is required for the antiviral activity of zinc finger antiviral protein. J Virol 91:e00088-17. doi: 10.1128/JVI.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ficarelli M, Wilson H, Pedro Galao R, Mazzon M, Antzin-Anduetza I, Marsh M, Neil SJ, Swanson CM. 2019. KHNYN is essential for the zinc finger antiviral protein (ZAP) to restrict HIV-1 containing clustered CpG dinucleotides. Elife 8:e46767. doi: 10.7554/eLife.46767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Odon V, Fros JJ, Goonawardane N, Dietrich I, Ibrahim A, Alshaikhahmed K, Nguyen D, Simmonds P. 2019. The role of ZAP and OAS3/RNAseL pathways in the attenuation of an RNA virus with elevated frequencies of CpG and UpA dinucleotides. Nucleic Acids Res 47:8061–8083. doi: 10.1093/nar/gkz581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brierley I, Dos Ramos FJ. 2006. Programmed ribosomal frameshifting in HIV-1 and the SARS-CoV. Virus Res 119:29–42. doi: 10.1016/j.virusres.2005.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pollard VW, Malim MH. 1998. The HIV-1 Rev protein. Annu Rev Microbiol 52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- 35.Le Grice SF. 2012. Human immunodeficiency virus reverse transcriptase: 25 years of research, drug discovery, and promise. J Biol Chem 287:40850–40857. doi: 10.1074/jbc.R112.389056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sertznig H, Hillebrand F, Erkelenz S, Schaal H, Widera M. 2018. Behind the scenes of HIV-1 replication: alternative splicing as the dependency factor on the quiet. Virology 516:176–188. doi: 10.1016/j.virol.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 37.Firth AE. 2014. Mapping overlapping functional elements embedded within the protein-coding regions of RNA viruses. Nucleic Acids Res 42:12425–12439. doi: 10.1093/nar/gku981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kass SU, Goddard JP, Adams RL. 1993. Inactive chromatin spreads from a focus of methylation. Mol Cell Biol 13:7372–7379. doi: 10.1128/mcb.13.12.7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsieh CL. 1994. Dependence of transcriptional repression on CpG methylation density. Mol Cell Biol 14:5487–5494. doi: 10.1128/mcb.14.8.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsieh CL. 1997. Stability of patch methylation and its impact in regions of transcriptional initiation and elongation. Mol Cell Biol 17:5897–5904. doi: 10.1128/mcb.17.10.5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Munch J, Rajan D, Schindler M, Specht A, Rucker E, Novembre FJ, Nerrienet E, Muller-Trutwin MC, Peeters M, Hahn BH, Kirchhoff F. 2007. Nef-mediated enhancement of virion infectivity and stimulation of viral replication are fundamental properties of primate lentiviruses. J Virol 81:13852–13864. doi: 10.1128/JVI.00904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manganaro L, de Castro E, Maestre AM, Olivieri K, Garcia-Sastre A, Fernandez-Sesma A, Simon V. 2015. HIV Vpu interferes with NF-kappaB activity but not with interferon regulatory factor 3. J Virol 89:9781–9790. doi: 10.1128/JVI.01596-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chukkapalli V, Hogue IB, Boyko V, Hu WS, Ono A. 2008. Interaction between the human immunodeficiency virus type 1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphosphate is essential for efficient gag membrane binding. J Virol 82:2405–2417. doi: 10.1128/JVI.01614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goodier JL, Pereira GC, Cheung LE, Rose RJ, Kazazian HH Jr. 2015. The broad-spectrum antiviral protein ZAP restricts human retrotransposition. PLoS Genet 11:e1005252. doi: 10.1371/journal.pgen.1005252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hayakawa S, Shiratori S, Yamato H, Kameyama T, Kitatsuji C, Kashigi F, Goto S, Kameoka S, Fujikura D, Yamada T, Mizutani T, Kazumata M, Sato M, Tanaka J, Asaka M, Ohba Y, Miyazaki T, Imamura M, Takaoka A. 2011. ZAPS is a potent stimulator of signaling mediated by the RNA helicase RIG-I during antiviral responses. Nat Immunol 12:37–44. doi: 10.1038/ni.1963. [DOI] [PubMed] [Google Scholar]
- 46.Mostoslavsky G, Kotton DN, Fabian AJ, Gray JT, Lee JS, Mulligan RC. 2005. Efficiency of transduction of highly purified murine hematopoietic stem cells by lentiviral and oncoretroviral vectors under conditions of minimal in vitro manipulation. Mol Ther 11:932–940. doi: 10.1016/j.ymthe.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 47.Reil H, Bukovsky AA, Gelderblom HR, Gottlinger HG. 1998. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J 17:2699–2708. doi: 10.1093/emboj/17.9.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y, Nikolaitchik OA, Rahman SA, Chen J, Pathak VK, Hu WS. 2017. HIV-1 sequence necessary and sufficient to package non-viral RNAs into HIV-1 particles. J Mol Biol 429:2542–2555. doi: 10.1016/j.jmb.2017.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kharytonchyk S, Brown JD, Stilger K, Yasin S, Iyer AS, Collins J, Summers MF, Telesnitsky A. 2018. Influence of gag and RRE sequences on HIV-1 RNA packaging signal structure and function. J Mol Biol 430:2066–2079. doi: 10.1016/j.jmb.2018.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takata MA, Soll SJ, Emery A, Blanco-Melo D, Swanstrom R, Bieniasz PD. 2018. Global synonymous mutagenesis identifies cis-acting RNA elements that regulate HIV-1 splicing and replication. PLoS Pathog 14:e1006824. doi: 10.1371/journal.ppat.1006824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emery A, Zhou S, Pollom E, Swanstrom R. 2017. Characterizing HIV-1 splicing by using next-generation sequencing. J Virol 91:e02515-16. doi: 10.1128/JVI.02515-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Purcell DF, Martin MA. 1993. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J Virol 67:6365–6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gerstberger S, Hafner M, Tuschl T. 2014. A census of human RNA-binding proteins. Nat Rev Genet 15:829–845. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, Na H, Irimia M, Matzat LH, Dale RK, Smith SA, Yarosh CA, Kelly SM, Nabet B, Mecenas D, Li W, Laishram RS, Qiao M, Lipshitz HD, Piano F, Corbett AH, Carstens RP, Frey BJ, Anderson RA, Lynch KW, Penalva LO, Lei EP, Fraser AG, Blencowe BJ, Morris QD, Hughes TR. 2013. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499:172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McIntyre W, Netzband R, Bonenfant G, Biegel JM, Miller C, Fuchs G, Henderson E, Arra M, Canki M, Fabris D, Pager CT. 2018. Positive-sense RNA viruses reveal the complexity and dynamics of the cellular and viral epitranscriptomes during infection. Nucleic Acids Res 46:5776–5791. doi: 10.1093/nar/gky029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lavender CA, Gorelick RJ, Weeks KM. 2015. Structure-based alignment and consensus secondary structures for three HIV-related RNA genomes. PLoS Comput Biol 11:e1004230. doi: 10.1371/journal.pcbi.1004230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Courtney DG, Tsai K, Bogerd HP, Kennedy EM, Law BA, Emery A, Swanstrom R, Holley CL, Cullen BR. 2019. Epitranscriptomic addition of m(5)C to HIV-1 transcripts regulates viral gene expression. Cell Host Microbe 26:217–227. doi: 10.1016/j.chom.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Doyle T, Goujon C, Malim MH. 2015. HIV-1 and interferons: who’s interfering with whom? Nat Rev Microbiol 13:403–413. doi: 10.1038/nrmicro3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shaw AE, Hughes J, Gu Q, Behdenna A, Singer JB, Dennis T, Orton RJ, Varela M, Gifford RJ, Wilson SJ, Palmarini M. 2017. Fundamental properties of the mammalian innate immune system revealed by multispecies comparison of type I interferon responses. PLoS Biol 15:e2004086. doi: 10.1371/journal.pbio.2004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choudhury NR, Heikel G, Trubitsyna M, Kubik P, Nowak JS, Webb S, Granneman S, Spanos C, Rappsilber J, Castello A, Michlewski G. 2017. RNA-binding activity of TRIM25 is mediated by its PRY/SPRY domain and is required for ubiquitination. BMC Biol 15:105. doi: 10.1186/s12915-017-0444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kwon SC, Yi H, Eichelbaum K, Fohr S, Fischer B, You KT, Castello A, Krijgsveld J, Hentze MW, Kim VN. 2013. The RNA-binding protein repertoire of embryonic stem cells. Nat Struct Mol Biol 20:1122–1130. doi: 10.1038/nsmb.2638. [DOI] [PubMed] [Google Scholar]
- 62.Manokaran G, Finol E, Wang C, Gunaratne J, Bahl J, Ong EZ, Tan HC, Sessions OM, Ward AM, Gubler DJ, Harris E, Garcia-Blanco MA, Ooi EE. 2015. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science 350:217–221. doi: 10.1126/science.aab3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanchez JG, Sparrer KMJ, Chiang C, Reis RA, Chiang JJ, Zurenski MA, Wan Y, Gack MU, Pornillos O. 2018. TRIM25 binds RNA to modulate cellular anti-viral defense. J Mol Biol 430:5280–5293. doi: 10.1016/j.jmb.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Choudhury NR, Nowak JS, Zuo J, Rappsilber J, Spoel SH, Michlewski G. 2014. Trim25 is an RNA-specific activator of Lin28a/TuT4-mediated uridylation. Cell Rep 9:1265–1272. doi: 10.1016/j.celrep.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, Krijgsveld J, Hentze MW. 2012. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 66.Law LM, Albin OR, Carroll JW, Jones CT, Rice CM, Macdonald MR. 2010. Identification of a dominant negative inhibitor of human zinc finger antiviral protein reveals a functional endogenous pool and critical homotypic interactions. J Virol 84:4504–4512. doi: 10.1128/JVI.02018-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen S, Xu Y, Zhang K, Wang X, Sun J, Gao G, Liu Y. 2012. Structure of N-terminal domain of ZAP indicates how a zinc-finger protein recognizes complex RNA. Nat Struct Mol Biol 19:430–435. doi: 10.1038/nsmb.2243. [DOI] [PubMed] [Google Scholar]
- 68.Coleman JR, Papamichail D, Skiena S, Futcher B, Wimmer E, Mueller S. 2008. Virus attenuation by genome-scale changes in codon pair bias. Science 320:1784–1787. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 71.Parrish NF, Gao F, Li H, Giorgi EE, Barbian HJ, Parrish EH, Zajic L, Iyer SS, Decker JM, Kumar A, Hora B, Berg A, Cai F, Hopper J, Denny TN, Ding H, Ochsenbauer C, Kappes JC, Galimidi RP, West AP, Bjorkman PJ, Wilen CB, Doms RW, O'Brien M, Bhardwaj N, Borrow P, Haynes BF, Muldoon M, Theiler JP, Korber B, Shaw GM, Hahn BH. 2013. Phenotypic properties of transmitted founder HIV-1. Proc Natl Acad Sci U S A 110:6626–6633. doi: 10.1073/pnas.1304288110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Parrish NF, Wilen CB, Banks LB, Iyer SS, Pfaff JM, Salazar-Gonzalez JF, Salazar MG, Decker JM, Parrish EH, Berg A, Hopper J, Hora B, Kumar A, Mahlokozera T, Yuan S, Coleman C, Vermeulen M, Ding H, Ochsenbauer C, Tilton JC, Permar SR, Kappes JC, Betts MR, Busch MP, Gao F, Montefiori D, Haynes BF, Shaw GM, Hahn BH, Doms RW. 2012. Transmitted/founder and chronic subtype C HIV-1 use CD4 and CCR5 receptors with equal efficiency and are not inhibited by blocking the integrin alpha4beta7. PLoS Pathog 8:e1002686. doi: 10.1371/journal.ppat.1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol 59:284–291. doi: 10.1128/JVI.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yeo G, Burge CB. 2004. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 75.Desmet F-O, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C. 2009. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fouchier RA, Meyer BE, Simon JH, Fischer U, Malim MH. 1997. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J 16:4531–4539. doi: 10.1093/emboj/16.15.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Swanson CM, Sherer NM, Malim MH. 2010. SRp40 and SRp55 promote the translation of unspliced human immunodeficiency virus type 1 RNA. J Virol 84:6748–6759. doi: 10.1128/JVI.02526-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sanjana NE, Shalem O, Zhang F. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. 1997. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol 15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- 80.Chesebro B, Wehrly K, Nishio J, Perryman S. 1992. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J Virol 66:6547–6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ovod V, Lagerstedt A, Ranki A, Gombert FO, Spohn R, Tahtinen M, Jung G, Krohn KJ. 1992. Immunological variation and immunohistochemical localization of HIV-1 Nef demonstrated with monoclonal antibodies. AIDS 6:25–34. doi: 10.1097/00002030-199201000-00003. [DOI] [PubMed] [Google Scholar]
- 82.Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hahne F, Ivanek R. 2016. Visualizing genomic data using Gviz and bioconductor. Methods Mol Biol 1418:335–351. doi: 10.1007/978-1-4939-3578-9_16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data are available in the NCBI Gene Expression Omnibus (GEO) under accession number GSE134159. The GenBank accession numbers for the HIV-1 genomic-RNA nucleotide sequences are as follows: HIV-1 (strain NL4-3), MN685337; HIV-1gag22–165CM, MN685338; HIV-1gag22–261CM, MN685339; HIV-1gag22–378CM, MN685340; HIV-1gag22–378CM–no-CpG, MN685341; HIV-1gag22–378CpG, MN685342; HIV-1gag22–378DC, MN685343; HIV-1gag22–378CM–5nt-CpG, MN685344; HIV-1gag22–651CpG, MN685345; HIV-1gag660-1185CpG, MN685346; HIV-1gag22–1185CpG, MN685347; HIV-1gag694–1206CpG, MN685348; HIV-1pol795–1386CpG, MN685349; HIV-1env86–561CpG, MN685350; HIV-1env611–1014CpG, MN685351; HIV-1env86–1014CpG, MN685352.