Cytomegalovirus (CMV) infections result in morbidity and mortality in immunocompromised individuals, and the virus is also a major cause of birth defects in newborns. Currently, because of the unavailability of vaccines against this virus and restricted antiviral therapies with low toxicity, as well as the emergency of resistant strain of this virus, the understanding of viral late gene regulation may provide clues to study new antiviral drugs or vaccines. In this study, we report that MCMV protein pM49 is critical for viral late gene transcription, based on its interaction with pM95. This finding reveals the important role of pM49-pM95 interaction in the regulation of viral late gene expression and that it could be a future potential target for therapeutic intervention in CMV diseases.
KEYWORDS: cytomegalovirus, late gene, pM49, pM95, transactivation factors
ABSTRACT
Late gene expression of betaherpesviruses and gammaherpesviruses is tightly controlled by virus-encoded transactivation factors (vTFs). We recently proved that the 6 vTFs of murine cytomegalovirus (MCMV) form a complex to regulate late gene transcription. pM49, one of the vTFs that has not been studied before, was identified to be a component of the complex that interacts with pM95. In this study, we began to investigate the potential role of pM49 in viral late gene expression. A recombinant MCMV expressing C-terminal FLAG-tagged pM49 was constructed to study the expression kinetics and localization of pM49. pM49 was expressed at the late time of virus infection. Inhibition of viral DNA synthesis by phosphonate sodium phosphonic acid (PAA) abolished pM49 expression, indicating that it is a late protein. pM49 colocalized with pM44 at the viral replication compartment, similarly to other viral vTFs that have been reported. Mutant virus lacking full-length pM49 expression failed to express viral late genes, leading to nonproductive infection. The expression of immediate early and early genes was not affected, and viral DNA synthesis was only minimally affected during pM49-deficient virus infection. All of these data support the role of pM49 in viral late gene expression. After a series of mutagenesis analyses, two key residues, K325 and C326, were identified as required for pM49-pM95 interaction. Cells expressing pM49 with either single mutation of these two residues failed to rescue the late gene expression and support the replication of pM49-deficient virus. Our results indicated that pM49-pM95 interaction is essential for viral late gene expression.
IMPORTANCE Cytomegalovirus (CMV) infections result in morbidity and mortality in immunocompromised individuals, and the virus is also a major cause of birth defects in newborns. Currently, because of the unavailability of vaccines against this virus and restricted antiviral therapies with low toxicity, as well as the emergency of resistant strain of this virus, the understanding of viral late gene regulation may provide clues to study new antiviral drugs or vaccines. In this study, we report that MCMV protein pM49 is critical for viral late gene transcription, based on its interaction with pM95. This finding reveals the important role of pM49-pM95 interaction in the regulation of viral late gene expression and that it could be a future potential target for therapeutic intervention in CMV diseases.
INTRODUCTION
Human cytomegalovirus (HCMV), a prototypical member of the betaherpesviruses, is a ubiquitous pathogen in humans and usually presents with asymptomatic infections in healthy individuals (1, 2). However, HCMV is also an opportunistic pathogen and may result in significant morbidity and mortality in immunocompromised populations, such as HIV/AIDS or transplant patients (3, 4). HCMV is also a contributing factor to birth defects, including deafness, blindness and mental retardation. Because HCMV is limited to human hosts by virtue of its species specificity, murine cytomegalovirus (MCMV) is used as its small-animal model due to its conserved genome organization, gene expression, virion structure, and tissue tropism (5, 6). Studying the functions of conserved genes in MCMV will shed light on the life cycle of HCMV.
All herpesviruses, including CMV, establish lifelong latent infection in the host (7). Under appropriate stimuli, the lytic stage is activated, followed by virus production and cell damage. During this lytic period, viral genes are expressed in a temporal, highly ordered cascade, which can be divided into immediate early (IE), early (E), and late (L) phases (8, 9). IE genes are expressed immediately after virus entry, requiring only incoming virion-associated proteins and host cell factors (10). IE genes products are responsible for activating early gene transcription and remolding of the cellular microenvironment to support viral replication (11). Early proteins are necessary for viral replication compartment formation and DNA synthesis. Following DNA synthesis, viral late genes begin to transcribe; these main encoding structural proteins associated with virion assembly, maturation, and release. Although IE and E gene expression is independent of DNA synthesis, L gene expression can be extremely enhanced by DNA synthesis. While there is a wealth of research and analysis on IE genes and E genes, little was known about mechanisms in late gene regulation until R. Sun’s group reported that four proteins encoded by open reading frame 18 (ORF18), ORF30, ORF31, and ORF34 of murine gammaherpesvirus 68 (MHV68) were essential for viral late gene expression (12–15). Depletion of any of these proteins abrogated late gene transcription, but had little effect on DNA replication and immediate early or early gene expression. Besides these four proteins, ORF24 from Kaposi’s sarcoma-associated herpesvirus (KSHV) and BFRF2 from Epstein-Barr virus (EBV) were also identified as indispensable for late gene transcription (15, 16). These six proteins are conserved among betaherpesviruses and gammaherpesviruses (Table 1) and are also known as viral transactivation factors (vTFs) (10, 17–24). These vTFs form the viral preinitiation complex (vPIC), which assembles, along with other viral and host transcription factors, at the viral late gene promoter (23, 25). Among these, EBV BcRF1 have been identified as a TATA box protein (TBP)-like protein thanks to its structural similarity to the TBP protein and its capacity to specifically bind with viral noncanonical TATT boxes (26, 27); pUL79 from HCMV has been shown to facilitate transcription elongation (28), but little is known about the functions and mechanisms of other vTFs.
TABLE 1.
Conserved transactivator genes between betaherpesviruses and gammaherpesviruses
| Viral subfamily | Virus | Gene name (function) | |||||
|---|---|---|---|---|---|---|---|
| Gammaherpesviruses | EBV | BcRF1 (TBP-like) | BFRF2 | BGLF3 | BVLF1 | BDLF4 | BDLF3.5 |
| KSHV | ORF24 | ORF66 | ORF34 | ORF18 | ORF31 | ORF30 | |
| MHV68 | ORF24 | ORF66 | ORF34 | ORF18 | ORF31 | ORF30 | |
| Betaherpesviruses | HCMV | UL87 | UL49 | UL95 | UL79 (elongation) | UL92 | UL91 |
| MCMV | M87 | M49 | M95 | M79 | M92 | M91 | |
Our previous study dissected the relationships among six vTFs (pM49, pM95, pM91, pM79, pM87, and pM92) from MCMV, using immunoprecipitation (IP) approaches (20). We showed that these six vTFs formed a protein complex, comprising three strong protein-protein pair interactions (pM49-pM95, pM91-pM79, and pM87-pM95) and one weak protein-protein pair interaction (pM87-pM92). We also showed that this complex consisted of two components that may perform different functions. One component included the four proteins pM49, pM95, pM79, and pM91. We showed that these four proteins appeared to bind directly or indirectly to one another, forming a core within the complex. The other component appeared to just contains pM92. These two components were found to interact with each other via the introduction of pM87, and then the whole complex contributed to vPIC assembly. Furthermore, our analyses revealed that the core could be disrupted by the lack of pM49 or pM79, suggesting that pM49-pM95 and pM91-pM79 interactions were essential for the core complex formation (20).
In this study, we focused on pM49 and its interaction with pM95. Our results revealed that pM49 plays an important role in viral late gene expression. Our point mutation analyses suggested that pM49-pM95 interaction is essential for the production of infectious virus particles and indicated that pM49 serves as a vPIC component and participates in viral late gene transcription.
RESULTS
pM49 is a late protein localizing to the viral replication compartment.
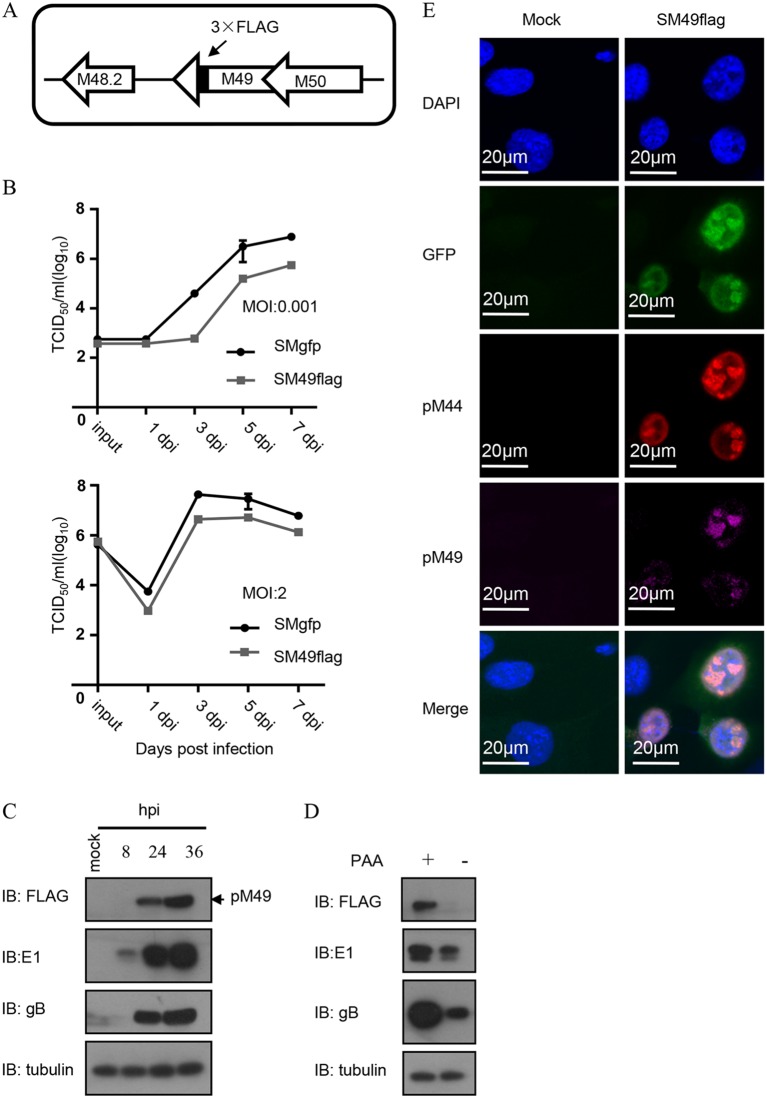
To study the expression and localization of pM49 during viral infection, we engineered a recombinant SM49flag virus with a 3×FLAG tag inserted at the pM49 C terminus. Growth curve analysis showed that replication of the recombinant virus (SM49flag) was slightly impaired (less than 10-fold) compared to that of the wild-type virus (SMgfp) at a low multiplicity of infection (MOI) of 0.001. SM49flag viral growth was less affected when performed at a high multiplicity of infection (MOI = 2); therefore, we performed subsequent experiments at an MOI of 2, where the C-terminal FLAG tag minimally disturbed pM49 function. To determine pM49 expression level, we infected mouse embryonic fibroblast 10.1 (MEF10.1) cells with SM49flag and analyzed the accumulation of FLAG-tagged pM49 by immunoblotting using antibody against the FLAG tag. pM49 protein levels were only detected at 24 h postinfection (hpi); however, the expression levels increased at 36 hpi. The expression of pM49 was completely disrupted after infected cells were treated with the DNA synthesis inhibitor phosphonate sodium phosphonic acid (PAA). This suggested that pM49 expression was late and that its expression depends on viral DNA replication.
Furthermore, we examined the intracellular localization of pM49 using immunofluorescence (IF). Our results showed that pM49 localized to the nuclei of infected cells, where it formed a puncta structure that colocalized with pM44, a marker of this viral replication compartment (Fig. 1E). Thus, similarly to other vTFs, pM49 was a nuclear protein that localized to the viral replication compartment during MCMV infection.
FIG 1.
Expression kinetics and localization of pM49. (A) Schematic of the genome structure of the pSM49flag bacterial artificial chromosome (BAC) generated from wild-type BAC (pSMgfp). (B) Growth curve analyses. MEF10.1 cells were infected with SMgfp or SM49flag at an MOI of 0.001 and at an MOI of 2. Cell-free viruses were collected at indicated time points postinfection, and virus titers were determined by 50% tissue culture infective dose (TCID50) assay. (C) Viral protein profile. MEF10.1 cells were infected with SM49flag at an MOI of 2 and harvested at the indicated time points postinfection. The accumulation of host and viral protein was analyzed by immunoblot (IB) assay. (D) pM49 expression and viral DNA synthesis. MEF10.1 cells were infected with SM49flag virus at an MOI of 2, in the presence or absence of PAA (200 μg/ml). At 24 hpi, cells were harvested and analyzed by IB assay. (E) pM49 localization analysis by immunofluorescence (IF) assay. MEF10.1 were mock infected or infected with SM49flag. At 24 hpi, cells were fixed and subjected to immunostaining with a rabbit anti-FLAG antibody (purple) and a mouse anti-pM44 antibody (red). Cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) to visualize the nuclei (blue).
pM49 is essential for MCMV replication.
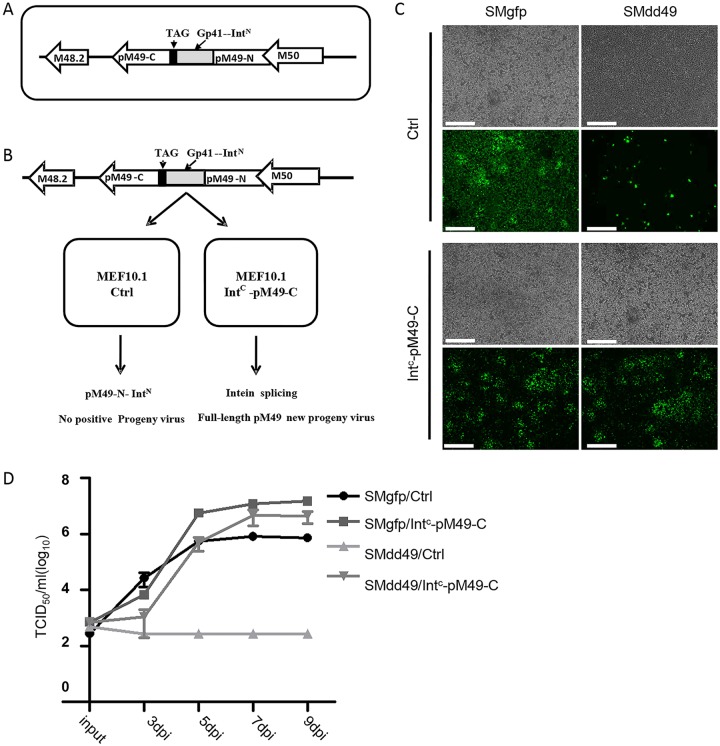
To investigate the role of pM49 in the MCMV life cycle, we constructed a pM49-deficient recombinant virus using the intein-mediated modulation of protein stability (imPS) system previously developed in our laboratory (29).There are many examples in CMV research where expressing a deleted gene in trans failed to rescue the gene-deficient recombinant virus (20, 30). The imPS system is useful and convenient for overcoming such issues. Briefly, a protein destabilization domain and one half of the split intein is fused to the N or C terminus of the gene of interest. The protein products of the fusion gene will be quickly degraded and lose function. However, expressing the other half of split intein in trans reconstitutes the enzymatically active intein, thereby mediating cleavage of the protein destabilization domain, rescuing the gene product and virus replication. In this work, we inserted the split intein into the middle of pM49 coding sequence because its N-terminal coding sequence overlaps with M50, whereas 3×FLAG tag C-terminal insertion slightly affects functional output. As shown in Fig. 2A, the coding sequence of gp41-1 N-terminal split intein (IntN; gray box) and a stop codon (TAG; black box) were inserted at 793 nucleotides downstream of the pM49 start codon. The recombinant pM49-deficient virus SMdd49 expressed a truncated pM49 containing the N-terminal 264 amino acids of the full-length protein, with IntN fused to its C terminus (pM49-N-IntN).To reconstitute the full-length pM49, we constructed a lentiviral vector that expressed the gp41-1 C-terminal split intein (IntC) fused with the C-terminal half of the pM49 coding sequence (IntC-pM49-C). MEF10.1 cells were transduced with this lentiviral vector to generate complementary cells expressing the IntC-pM49-C fusion protein (Fig. 2B). When the pM49-deficient bacterial artificial chromosome (BAC) pSMdd49 was transfected into these complementary cells, pM49-N-IntN expressed from the BAC genome and IntC-pM49-C expressed from the lentiviral vector interacted to reconstitute the full-length pM49 via protein splicing (Fig. 2B and C). As shown in Fig. 2C, transfection of pSMdd49 into MEF10.1control cells led to green fluorescent protein (GFP) expression in scattered individual cells at 5 days postinfection. In contrast, transfection of pSMdd49 into IntC-pM49-C-expressing complementary cells led to the spread of GFP expression in all cells, similar to that of wild-type BAC pSMgfp transfection into control or complementary cells (Fig. 2C). As expected, growth curve analyses showed that no SMdd49 virus was recovered from MEF10.1 control cells, while SMdd49 virus collected from IntC-pM49-C-expressing complementary cells grew to wild-type virus levels (Fig. 2D). These data suggest that full-length pM49 was essential for MCMV replication.
FIG 2.
pM49 is essential for MCMV replication. (A) Schematic depicting the structure of pSMdd49 BAC used this study. pM49 coding sequence was inserted with a gp41-1 split intein N (IntN, gray box) coding sequence and a stop codon (TAG, black box) at 793 nt downstream of the start codon, resulting in an interrupted protein translation and the production of pM49-N-IntN. (B) Complementary model of the pM49-deficient virus (SMdd49) using the intein-mediated modulation of protein stability (imPS) system. Without Intc-pM49-C, cells infected with SMdd49 only expressed the fusion protein of pM49-N-IntN. However, intein splicing that occurred in complementary cells produced full-length pM49 to support viral growth. (C and D) MEF10.1Ctrl and MEF10.1 Intc-pM49-C cells were infected with SMgfp or SMdd49 at an MOI of 0.001. The green fluorescent protein (GFP) signals were observed at 5 days postinfection (dpi) using fluorescence microscope. Bar, 500 μm. Cell-free virus was collected at indicated time points postinfection, and virus titers were determined by TCID50 assay.
pM49 is indispensable for viral late gene transcription.
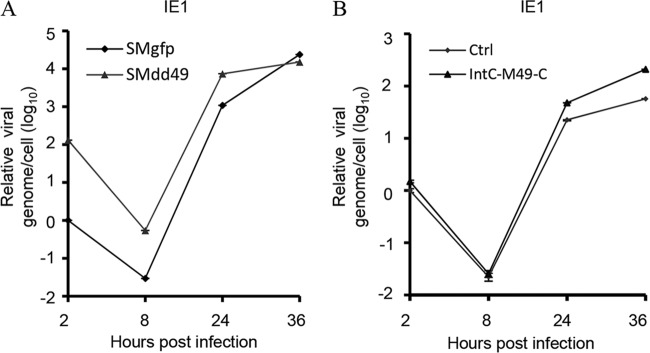
After proposing the critical role of pM49 in viral growth, we were keen to ascertain if pM49 was required for viral DNA synthesis due to its localization signal in the viral replication compartment. To test this hypothesis, we infected MEF10.1 cells with SMgfp or SMdd49 at an MOI of 2 and extracted DNA at specific time points postinfection. DNA quantities were analyzed using quantitative PCR (qPCR), and values were normalized to actin DNA levels. Interestingly, we found that the amount of input DNA for SMdd49 was greater than that for SMgfp (Fig. 3A), indicating that SMdd49 virus particles were less infectious than those of the wild-type virus, since more mutant viral genomes were required to reach an equivalent infectious unit. When quantifying newly produced viral DNA at 24 and 36 hpi, we found no significant differences in viral DNA synthesis in the presence or absence of M49 (Fig. 3A). The slightly higher levels of viral DNA accumulation in SMdd49-infected cells at 24 hpi may be caused by higher levels of input DNA. To address this observation, we infected MEF10.1 control and complementary cells with the same pM49-deficient virus stock at an MOI of 2. We then extracted and analyzed intracellular DNA as described above. Our results showed that viral DNA accumulation was similar at 24 hpi in both cell types, and levels were slightly lower in control cells at 36 hpi compared to those in complementary cells (Fig. 3B). Therefore, we concluded that pM49 minimally affected viral DNA replication.
FIG 3.
pM49 has a minimal effect on viral DNA replication. (A) MEF10.1 cells were infected with SMgfp or SMdd49 at an MOI of 2, and intracellular DNA was extracted from infected cells at the indicated time points postinfection. DNA levels were analyzed with quantitative PCR (qPCR), and the values of IE1 were normalized to actin DNA levels. The quantity of DNA from SMgfp-infected cells at 2 hours postinfection (hpi) was set to 1. (B) MEF10.1Ctrl and MEF10.1 Intc-pM49-C cells were infected with SMdd49 at an MOI of 2. Extracted DNA was analyzed as described in panel A. The quantity of DNA from MEF10.1Ctrl cells at 2 hpi was set to 1.
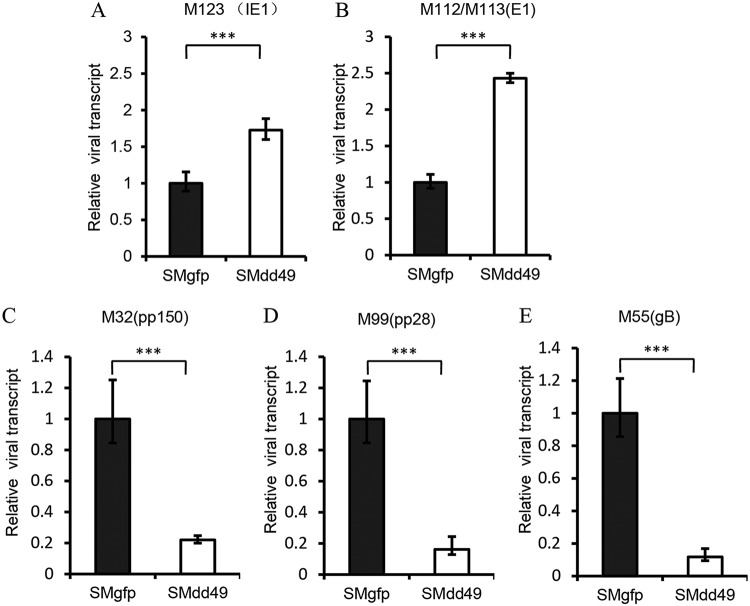
We next tested if pM49 played a role in viral late gene transcription, since its EBV homolog, BFRF2, was reported to be essential for viral late gene transcription (16). After SMgfp or SMdd49 infection, we measured immediate early, early, and late gene transcription levels in MEF10.1 cells using qPCR. At the early time point (8 hpi), transcript accumulation of the immediate early gene M123 (IE1) and the early gene M112/113 (E1) was higher in the absence of pM49 (Fig. 4A and B). As shown (Fig. 3A), the viral genome of SMdd49 was larger than that of SMgfp at the same MOI. Therefore, we concluded that these subtle differences in immediate early and early gene transcript accumulation between SMgfp and SMdd49 infection probably resulted from the different initial input of viral genome copies. At a late time point (36 hpi), the transcript accumulation of three late genes, pM55 (gB), pM99 (pp28), and pM32 (pp150), was 5- to 10-fold lower during SMdd49 infection compared to that during SMgfp infection (Fig. 4C to E), indicating that pM49 was essential for viral late gene transcription.
FIG 4.
pM49 is indispensable for viral late gene transcription. MEF10.1 cells were infected with SMgfp or SMdd49 at an MOI of 2. At 8 hpi (A and B) or 36 hpi (C, D, and E), total RNA was harvested and quantified by reverse transcription-quantitative PCR (RT-qPCR) analysis using the primers listed in Table 3. Viral gene transcript levels were normalized to mouse Gapdh gene transcript levels, and the normalized transcript levels of SMgfp-infected cells were set to 1. Statistical analysis was performed using Student’s t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
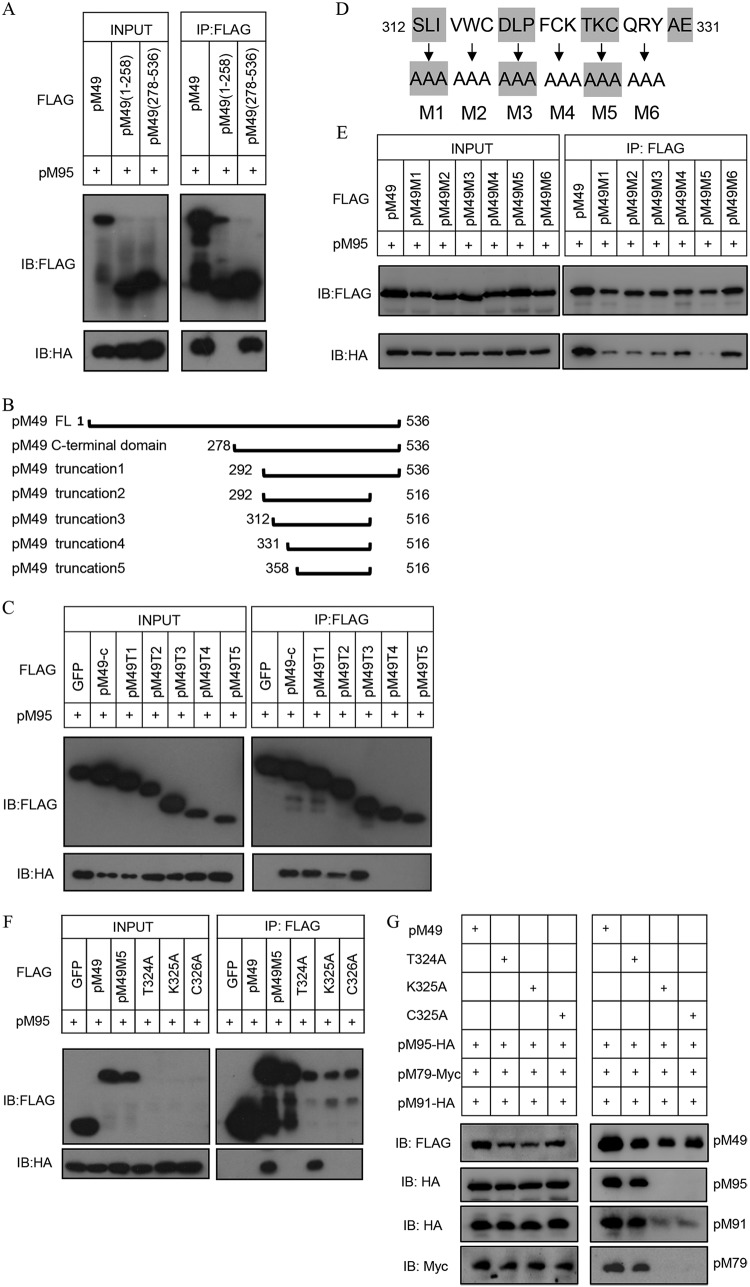
The K325 and C326 residues of pM49 are required for pM49-M95 interaction.
In our previous study, we showed that pM49 interacted with pM95, facilitating assembly of the vTF core complex in HEK293T cells (20). Protein sequence alignments of pM49 and pUL49 revealed that their C-terminal domains were highly conserved. We first divided the 536-amino acid pM49 coding sequence in half. After confirming the C-terminal domain was responsible for pM49-pM95 interaction using an IP approach (Fig. 5A), we constructed five truncations where the C-terminal regions were progressively deleted, based on the predicted secondary structures (Fig. 5B). Our IP results showed that residues 312 to 331 were important for pM49-pM95 interaction (Fig. 5C). We then performed a 3-alanine screening on residues 312 to 329 (Fig. 5D). Almost all pM49 alanine mutants reduced the interaction, and the mutant (residues 324 to 326 to A) abrogated pM49-pM95 binding (Fig. 5E). Next, the three residues T324, K325, and C326 in pM49 were replaced with alanine to perform single-site mutagenesis. Strikingly, the single mutants K325A and C326A abolished the pM49-pM95 interaction (Fig. 5F), whereas the single mutant T324A had no effect. To further investigate whether K325A and C326A disrupt the core complex formation or not, HEK293T cells were transfected with FLAG-tagged wild-type or mutant pM49 together with expression plasmids of other three vTFs of the core complex. After 48 h, cells were harvested and lysed for IP experiments. Consistent with the results presented above, the interaction among pM49, pM79 and pM91 was completely disrupted by K325A and C326A but not by T324A (Fig. 5G), suggesting that K325 and C326 were important for the core complex formation.
FIG 5.
Mapping the key residues required for pM49 and pM95 interaction. (A, C, E, and F) HEK293T cells were cotransfected with hemagglutinin (HA)-tagged pM95 and either FLAG-tagged GFP or pM49 (wild type, truncations, or mutations). Cell lysates were used for immunoprecipitation (IP) assay with anti-FLAG M2 beads, followed by IB analysis with indicated antibodies. T1 to T5 are pM49 truncation mutants. M1 to M6 are pM49 alanine scanning mutants. T324A, K325A, and C326A indicate pM49 point mutants. (B) The scheme of pM49 serial truncation mutant construction analyses. The start and terminal amino acid residues for each mutant are indicated. (D) The scheme of pM49 alanine scanning mutant construction analyses. M1, M2, M3, M4, M5, and M6 represent residues 312 to 314, 315 to 317, 318 to 320, 321 to 323, 324 to 326, and 327 to 329, respectively, that are mutated to alanine. (G) HEK293T cells were transfected with FLAG-tagged wild-type or mutant pM49 together with HA-tagged pM91, pM95, and Myc-tagged pM79. Cell lysates were treated as described in panel A.
The pM49-M95 interaction is important for viral late gene expression.
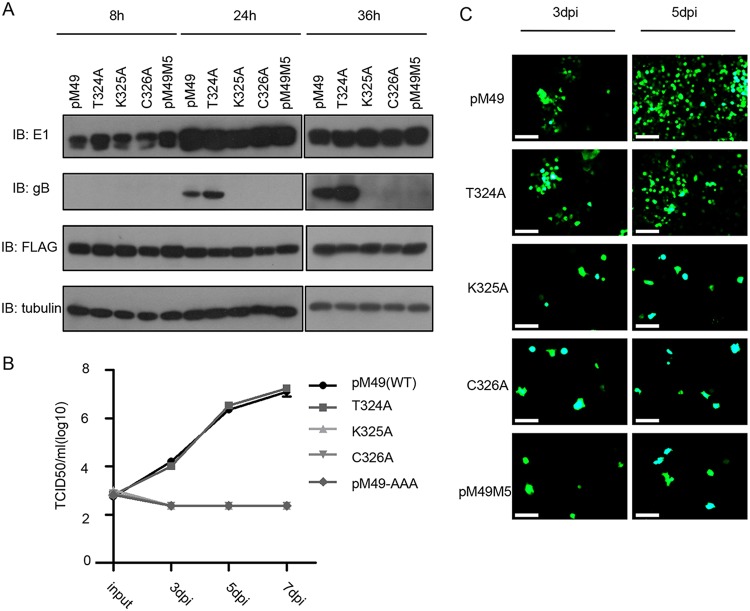
To confirm the importance of pM49 and pM95 interactions, we generated a MEF10.1 cell line stably expressing wild-type pM49 and pM49 mutants by retrovirus transduction. After SMdd49 infection, the phenotype of cells expressing the T324A mutant was similar to that of cells expressing wild-type pM49, while the growth of this deficient virus could not be reconstituted with cells expressing pM49 mutants that abrogated pM49 and pM95 interaction (Fig. 6B). Consistently, growth curve analyses showed similar results, suggesting that pM49 and pM95 interaction was critical for viral production, probably through the regulation of vPIC assembly (Fig. 6C).
FIG 6.
The pM49-pM95 interaction is important for MCMV growth. (A) MEF10.1 cells expressing wild-type pM49 or pM49 mutants were infected with SMdd49 at an MOI of 2 and harvested at the indicated time points postinfection. Host and viral protein expression were analyzed by IB assay. (B and C) MEF10.1 cells expressing wild-type pM49 or pM49 mutants were infected with SMdd49 at an MOI of 0.001, and the GFP signals were observed using fluorescence microscopy (bar, 100 μm). Cell-free virus was collected at the indicated time points postinfection, and virus titers were determined by TCID50 assay.
To determine the basis of viral growth defects caused by pM49-pM95 interaction disruption, our engineered cell lines were harvested at specific time points post SMdd49 infection for immunoblot (IB) analyses. Data from these studies showed the loss of the late gene marker gB in cells expressing the pM49 mutants K325A and C326A compared to cells expressing wild-type pM49 and the T324A mutant (Fig. 6A). Taken together, these data suggested that the pM49-M95 interaction was important for MCMV late gene transcription and viral replication.
DISCUSSION
In this study, we demonstrated the essential role of pM49 in viral late gene expression during MCMV infection; pM49 accumulated at a late time and localized to the viral replication compartment. Without pM49, late gene transcripts were markedly reduced, while early gene products and DNA synthesis were slightly affected. We also mapped the key residues within pM49 that are critical for its interaction with pM95. Mutation of two conserved residues in pM49, K325 and C326, abolished the pM49-pM95 interaction and the core complex formation, resulting in reduced virus production. These data demonstrated that the interaction between vTFs is essential for core complex formation and vTF transactivating activity.
As the late stage is an important step for viruses to complete their life cycles and produce infectious progeny virions, greater attention has been paid recently to the regulation of late gene expression. However, studies are mostly restricted to the functional level due to low accumulation of vTFs and difficulties in their mechanism of study. Recent work in KSHV demonstrated that ORF66, the homolog of pM49, was essential for viral late gene expression via interaction between ORF34, the homolog of pM95 (31, 32). Those studies, together with our results, provide a shred of evidence that the regulatory mechanism of viral late genes by vTFs is probably very conserved between betaherpesviruses and gammaherpesviruses. Glaunsinger’s group found that ORF66 and ORF24 (the homolog of pM87) occupied the promoter of L gene K8.1, but their promoter occupancy was dependent on the entire vTF complex assembly, indicating that these vTFs may function in a form of complex and regulate the localization of other vTFs on the viral genome.
Our previous study shed light on vTF complex assembly through IP experiments. In brief, the vTF complex from MCMV consisted of two components. pM49, pM95, pM79 and pM91 interacted directly or indirectly to form the core within the vTF complex; pM87 was thought to function as a hub to bridge pM92 and the core through the pM87-pM95 and pM87-pM92 interaction. Based on the research from Glaunsinger’s group (31) and our references, we propose a hypothesis for the mechanism of the MCMV vTF transcription regulation system; we speculate that the members of the core complex, pM49, pM95, pM91, and pM79, may have the same role in viral late gene regulation. Although viral late gene expression regulatory mechanism remains largely unknown, it has been reported that the pM79 analog pUL79 serves as an elongation factor, but with no effects on RNA polymerase II (RNAPII) recruitment (28). Since pM49 and pM79 belong to the same vTF complex component, we speculate that pM49, along with pM91 and pM95, may enhance transcription elongation as well. Besides the discovery of the role for pUL79, ORF66 and ORF30 were reported to be required for ORF24 occupancy in the KSHV genome (31). By analogy with KSHV vTFs, it is possible that pM49, pM91, and other core complex members are also involved in the regulation of pM87 localization to the MCMV genome. To confirm these hypotheses, further analyses are required to determine the distribution of vTFs in viral genomes.
In conclusion, our results suggest that pM49 functions as a transactivation factor in viral late gene expression, dependent on its interaction with pM95. This mechanistic insight could expand the development of antiviral drugs or vaccines for CMV infection. However, further research is required to investigate the underlying pM49 regulatory mechanisms in late gene expression.
MATERIALS AND METHODS
BAC mutagenesis and recombinant viruses.
The MCMV BAC pSMgfp which carries a full-length genome of the MCMV Smith strain, was used to produce wild-type control virus in this study. The green fluorescent (GFP) gene was inserted at the C terminus of the IE2 locus within the pSMgfp, under transcriptional control of the MCMV alpha promoter/enhancer, as IE2 was reported to be dispensable for MCMV infection (17, 33–35). All recombinant MCMV BAC clones used in this study were derived from pSMgfp using a Red BAC recombineering protocol as described previously (36–38). Recombination was carried out in GS1783, an Escherichia coli strain that harbored MCMV BAC, by electroporation. To construct pSMdd49, an IntN-kanamycin cassette was PCR amplified from pEBNA-IntN-Kans and recombined into pSMgfp 792 nucleotides (nt) downstream of the pM49 coding sequence start codon. Transformants were selected by kanamycin resistance. Then, the selection cassette was removed by arabinose-induced recombination. To generate pSM49Flag, a FLAG-kanamycin cassette was PCR amplified from pEBNA-FLAG-Kans and recombined into pSMgfp upstream of the pM49 stop codon. The selection process was performed as described above. The primers used for BAC recombination are listed in Table 2.
TABLE 2.
Primers used in BAC recombination
| Primer namea | Sequence (5ʹ–3ʹ) |
|---|---|
| M49-FLAG-F | ACAACACCGGCGCTGCCGGTCCGATCAACACTATTATCACTGAACTAAACATTATCACTGAACTAAACGACTACAAAGACCATGAC |
| M49-FLAG-R | TTAGTGAGTTTTGTCAGTTCAGAATTCCTTGTCATCGTCAAAAGTAGAGAGAGCTAGCTAGCTCGCTAGTTAGTGAGTTTTGTCAGTTCA |
| M49-dd-F | GCAGGCGCCCGCGCTTCCTCATCCCCATCGGCAATCGGACCCTGCGCTACCAATCGGACCCTGCGCTACGAATTCACCCGCAGCGGCTAC |
| M49-dd-R | CTACCCGGGTCTGAGGTCCGTCACTCCTTCACGTACAGGCGCGGCCGAGTTCCCGGAACCGCCGCCACCGCTACCCGGGTCTGAGGTCCG |
F, forward; R, reverse.
pSMdd49 and pSM49flag were used to generate SMdd49 and SM49flag viruses. MEF10.1control or MEF10.1IntC-pM49-C (for reconstituted SMdd49) cells were electroporated with 5 μg of pSMdd49 or pSM49flag BAC DNA and plated on a 10-cm plate. Culture medium was changed at 24 h postinfection. Virus supernatant were harvested at 7 days postinfection (dpi), when 80% of infected cells were lysed. Virus titer were determined in duplicate by a 50% tissue culture infectious dose (TCID50) assay in MEF10.1control or MEF10.1IntC-pM49-C cells. To amplify virus, cells were infected with virus at an MOI of 0.005, and cell-free supernatant was collected from infected culture at 7 dpi.
Plasmids, antibodies, and chemicals.
pM49 pM95, pM91, pM79, and pM87 coding sequences were amplified from pSMgfp and subcloned into pRetro-EBNA modified to include N- or C-terminal tags (3×FLAG or 3×hemagglutinin [HA]) (39). Plasmid pEBNA-pM49 mutants were engineered from pEBNA-3×FLAG-pM49. The plasmid pEBNA-HA-gp41IntC-pM49-C-terminal-domain was generated by inserting the HA-gp41IntC-pM49-C-terminal fragment into pRetro-EBNA. pEPKan-S2, which carried a kanamycin selection cassette bracketed by two I-SceI sites, was used for BAC recombination. pEBNA-FLAG-KanS, pEBNA-IntN-Kans, generated from pEPKan-S2, was used for FLAG tag or IntN insertion of BAC recombination (37, 38).
Primary antibodies used in this study included the following: anti-MCMV E1 and anti-MCMV M44 (generous gifts from Stipan Jonjic, University of Rijeka, Croatia), anti-MCMV gB (a generous gift from Anthony Scalzo, University of Western Australia), anti-HA (catalog no. MMS-101P; Covance), anti-FLAG (mouse, catalog no. M20008; Abmart), anti-FLAG (rabbit, F7425, Sigma-Aldrich), anti-Myc (mouse, catalog no. M20003; Abmart), and anti-α-tubulin (66031-1-Ig; proteintech). The secondary antibodies used for immunoblotting were horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG and goat anti-rabbit IgG (Jackson Laboratory). The secondary antibodies used for immunofluorescence were Alexa Fluor 555-conjugated goat anti-mouse IgG and Alexa Fluor 647-conjugated goat anti-rabbit IgG (Invitrogen). Other chemicals used in this study included Benzonase (catalog no. 70664; EMD Millipore), phosphonoacetic acid (catalog no. 284270-10G; Sigma-Aldrich), l-(+)-arabinose (catalog no. 5328-37-0; Urchem), and protein inhibitor cocktail (PIC) (Roche).
Cells.
Mouse embryonic fibroblast 10.1 cells (MEF10.1) (40), Phoenix cells (39), and HEK293T cells were propagated in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum, 1 mM sodium pyruvate, nonessential amino acids, and penicillin-streptomycin. To generate cells expressing 3×FLAG-pM49, 3FLAG-pM49 mutants, empty vector (MEF10.1control), or HA- IntC-pM49-C (MEF10.1IntC-pM49-C), retrovirus stocks were made by transfecting the retroviral vectors pEBNA- 3×FLAG-pM49, pEBNA-3FLAG-pM49 mutants, or pEBNA-HA-IntC-pM49-C, respectively, into Phoenix cells. Then, MEF10.1 cells were transduced two times with these polyethylene glycol (PEG)-condensed retroviruses, as described previously (41).
Viral growth analysis.
MEF10.1control or MEF10.1IntC-pM49-C cells were seeded in 12-well dishes. After 24 h, the cells were incubated with SMgfp or SMdd49 virus for 2 h at an MOI of 0.001. The inoculum was collected as input, the cells were rinsed with fresh medium, and finally, 1 ml fresh medium was added to each well. Cell-free medium from infected cultures was collected in duplicate at various time points postinfection, and virus titers in the media were determined by TCID50 assay in MEF10.1IntC-pM49-C cells. The infected cells were observed with a Leica fluorescence microscope. Images were captured with a Leica DFC450 digital camera.
Protein analysis.
Protein interactions were analyzed by coimmunoprecipitation assay as previously described (42). HEK293T cells were transfected with the indicated plasmids and collected after 48 h. Collected cells were lysed in 1 ml lysis buffer (40 mM HEPES [pH 7.4], 1 mM EDTA, 300 mM NaCl, and 0.5% NP-40) supplemented with 250 units of Benzonase nuclease and PIC, incubated at 4°C for 1 h, and centrifuged at 13,200 × g at 4°C for 15 min. Supernatant (40 μl) was saved as the input control and boiled in sodium dodecyl sulfate (SDS)-containing sample buffer. Then, the rest of the supernatant were incubated with FLAG M2 antibody-conjugated magnetic beads (Sigma-Aldrich) at 4°C for 2 h. Then, the beads were washed 4 times with 1 ml lysis buffer. The immunoprecipitants were eluted by 150 ng/μl FLAG peptide (Sigma-Aldrich). The input and elution were analyzed by immunoblotting with the indicated antibody.
Protein accumulation was analyzed by immunoblot assay as described previously. Briefly, cells were collected and lysed in SDS sample buffer. Proteins were resolved by SDS-polyacrylamide gel electrophoresis and transferred onto a polyvinylidene difluoride membrane, hybridized with primary antibodies, reacted with horseradish peroxidase-coupled secondary antibodies, and visualized by Clarity Western ECL substrate (Bio-Rad).
Intracellular localization of proteins of interest was analyzed by immunofluorescence assay as previously described (43). Cells were seeded onto coverslips and mock infected or infected with SM49flag at an MOI of 2. After 24 h, cells were washed, fixed and permeabilized, blocked, incubated with primary antibodies, and subsequently labeled with Alexa Fluor 647 (anti-rabbit) or Alexa Fluor 555 (anti-mouse)-conjugated secondary antibodies (Invitrogen). Cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; Beyotime) and mounted on slides with Prolong Gold antifade reagent (Invitrogen Molecular Probes). Confocal microscopic images were captured by an Olympus FV1200 confocal laser scanning microscope.
RNA and DNA analysis.
Intracellular relative mRNA levels were determined by reverse transcription-quantitative PCR (RT-qPCR) as previously described (29). Total RNA was extracted using TRIzol reagent (Invitrogen), cDNA was synthesized with a PrimeScript real-time (RT) reagent kit (TaKaRa) and quantified using SYBR Premix Ex Taq (TaKaRa) with primer pairs specific for viral genes or the mouse Gapdh gene. All reactions were performed in two biological and two technical replicates. The amounts of viral transcripts were normalized to the Gapdh gene. Intracellular DNA was measured by qPCR as previously described (17). MEF10.1control or MEF10.1IntC-pM49-C cells were infected with SMgfp virus or SMdd49 virus at an MOI of 2 and collected at the indicated time points postinfection. Viral and cellular DNA were quantified by qPCR using SYBR Premix Ex Taq (TaKaRa) with primer pairs specific for MCMV M123 (IE1) and the mouse actin gene, respectively. The amount of viral DNA was normalized by dividing IE1 equivalents over actin gene equivalents. The normalized amount of viral DNA in SMgfp-infected cells at 2 hpi was set at 1. The primers used for DNA and RNA analysis are listed in Table 3. Statistical analysis in this study was performed using Student’s t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
TABLE 3.
Primers used in quantitative PCR analysis
| Primera | Sequence (5′–3′) |
|---|---|
| MCMV m123(IE1)-F | CAGGGTGGATCATGAAGCCT |
| MCMV m123(IE1)-R | AGCGCATCGAAAGACAACG |
| MCMV M112/113 (E1)-F | GAATCCGAGGAGGAAGACGAT |
| MCMV M112/113 (E1)-R | GGTGAACGTTTGCTCGATCTC |
| MCMV M55 (gB)-F | GCGATGTCCGAGTGTGTCAAG |
| MCMV M55 (gB)-R | CGACCAGCGGTCTCGAATAAC |
| MCMV M99 (pp28)-F | GTCTGACGACGAAGACCAGG |
| MCMV M99 (pp28)-R | CTGTATCGGAGTGACCACGG |
| MCMV M32 (pp150)-F | GAGACGATGAGGATTGCGGT |
| MCMV M32 (pp150)-R | GGCCTCGTTGTCCACCATTA |
| Mouse Actin-F | GCTGTATTCCCCTCCATCGTG |
| Mouse Actin-R | CACGGTTGGCCTTAGGGTTCA |
| Mouse Gapdh-F | TGGAGAAACCTGCCAAGTATGA |
| Mouse Gapdh-R | CTGTTGAAGTCGCAGGAGACAA |
F, forward; R, reverse.
ACKNOWLEDGMENTS
We thank all of the members of the Herpesvirus and Molecular Virology Research Unit for helpful discussions and invaluable advice; Dong Yu (GSK Vaccines, USA), Martin Messerle (Hanover Medical School, Hanover, Germany), Wolfram Brune (Heinrich Pette Institute, Leibniz Institute for Experimental Virology, Hamburg, Germany), and Ulrich Koszinowski (Max von Pettenkofer Institute, Ludwig Maximilian University, Munich, Germany) for the MCMV BAC clone pSMgfp; Anthony Scalzo (University of Western Australia) for the gB antibody, Stipan Jonjic (University of Rijeka, Croatia) for the E1 antibodies; Gregory Smith (Northwestern University Feinberg School of Medicine) for E. coli strain GS1783; and members of the Ke Lan lab (Wuhan University, China) for technical support with BAC recombination.
This work was supported by the National Natural Science Foundation of China (grants 81371826 and 81572002 to Z.Q. and grants 31300148 and 31570169 to B.X.), the Ministry of Science and Technology of China (grant 2016YFA0502101), and the Chinese Academy of Sciences “100 Talents” program to Z.Q. The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Hahn G, Jores R, Mocarski ES. 1998. Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc Natl Acad Sci U S A 95:3937–3942. doi: 10.1073/pnas.95.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kondo K, Xu J, Mocarski ES. 1996. Human cytomegalovirus latent gene expression in granulocyte-macrophage progenitors in culture and in seropositive individuals. Proc Natl Acad Sci U S A 93:11137–11142. doi: 10.1073/pnas.93.20.11137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steininger C. 2007. Clinical relevance of cytomegalovirus infection in patients with disorders of the immune system. Clin Microbiol Infect 13:953–963. doi: 10.1111/j.1469-0691.2007.01781.x. [DOI] [PubMed] [Google Scholar]
- 4.Deayton JR, Prof Sabin CA, Johnson MA, Emery VC, Wilson P, Griffiths PD. 2004. Importance of cytomegalovirus viraemia in risk of disease progression and death in HIV-infected patients receiving highly active antiretroviral therapy. Lancet 363:2116–2121. doi: 10.1016/S0140-6736(04)16500-8. [DOI] [PubMed] [Google Scholar]
- 5.Krmpotic A, Bubic I, Polic B, Lucin P, Jonjic S. 2003. Pathogenesis of murine cytomegalovirus infection. Microbes Infect 5:1263–1277. doi: 10.1016/j.micinf.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 6.Rawlinson WD, Farrell HE, Barrell BG. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol 70:8833–8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fish KN, Britt W, Nelson JA. 1996. A novel mechanism for persistence of human cytomegalovirus in macrophages. J Virol 70:1855–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Honess RW, Roizman B. 1975. Regulation of herpesvirus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc Natl Acad Sci U S A 72:1276–1280. doi: 10.1073/pnas.72.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kozak M, Roizman B. 1974. Regulation of herpesvirus macromolecular synthesis: nuclear retention of nontranslated viral RNA sequences. Proc Natl Acad Sci U S A 71:4322–4326. doi: 10.1073/pnas.71.11.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isomura H, Stinski MF. 2003. The human cytomegalovirus major immediate-early enhancer determines the efficiency of immediate-early gene transcription and viral replication in permissive cells at low multiplicity of infection. J Virol 77:3602–3614. doi: 10.1128/jvi.77.6.3602-3614.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meier JL, Stinski MF. 1997. Effect of a modulator deletion on transcription of the human cytomegalovirus major immediate-early genes in infected undifferentiated and differentiated cells. J Virol 71:1246–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arumugaswami V, Wu TT, Martinez-Guzman D, Jia Q, Deng H, Reyes N, Sun R. 2006. ORF18 is a transfactor that is essential for late gene transcription of a gammaherpesvirus. J Virol 80:9730–9740. doi: 10.1128/JVI.00246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu TT, Park T, Kim H, Tran T, Tong L, Martinez-Guzman D, Reyes N, Deng H, Sun R. 2009. ORF30 and ORF34 are essential for expression of late genes in murine gammaherpesvirus 68. J Virol 83:2265–2273. doi: 10.1128/JVI.01785-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jia Q, Wu TT, Liao HI, Chernishof V, Sun R. 2004. Murine gammaherpesvirus 68 open reading frame 31 is required for viral replication. J Virol 78:6610–6620. doi: 10.1128/JVI.78.12.6610-6620.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis ZH, Hesser CR, Park J, Glaunsinger BA. 2016. Interaction between ORF24 and ORF34 in the Kaposi’s sarcoma-associated herpesvirus late gene transcription factor complex is essential for viral late gene expression. J Virol 90:599–604. doi: 10.1128/JVI.02157-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aubry V, Mure F, Mariame B, Deschamps T, Wyrwicz LS, Manet E, Gruffat H. 2014. Epstein-Barr virus late gene transcription depends on the assembly of a virus-specific preinitiation complex. J Virol 88:12825–12838. doi: 10.1128/JVI.02139-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chapa TJ, Johnson LS, Affolter C, Valentine MC, Fehr AR, Yokoyama WM, Yu D. 2013. Murine cytomegalovirus protein pM79 is a key regulator for viral late transcription. J Virol 87:9135–9147. doi: 10.1128/JVI.00688-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chapa TJ, Perng YC, French AR, Yu D. 2014. Murine cytomegalovirus protein pM92 is a conserved regulator of viral late gene expression. J Virol 88:131–142. doi: 10.1128/JVI.02684-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapa TJ, Du Y, Sun R, Yu D, French AR. 2017. Proteomic and phylogenetic coevolution analyses of pM79 and pM92 identify interactions with RNA polymerase II and delineate the murine cytomegalovirus late transcription complex. J Gen Virol 98:242–250. doi: 10.1099/jgv.0.000676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan D, Han T, Tang S, Xu W, Bao Q, Sun Y, Xuan B, Qian Z, Pan D, Han T, Tang S, Xu W, Bao Q, Sun Y, Xuan B, Qian Z. 2018. Murine cytomegalovirus protein pM91 interacts with pM79 and is critical for viral late gene expression. J Virol 92:e00675-18. doi: 10.1128/JVI.00675-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Omoto S, Mocarski ES. 2013. Cytomegalovirus UL91 is essential for transcription of viral true late (gamma2) genes. J Virol 87:8651–8664. doi: 10.1128/JVI.01052-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Omoto S, Mocarski ES. 2014. Transcription of true late (γ2) cytomegalovirus genes requires UL92 function that is conserved among beta- and gammaherpesviruses. J Virol 88:120–130. doi: 10.1128/JVI.02983-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gruffat H, Marchione R, Manet E. 2016. Herpesvirus late gene expression: a viral-specific pre-initiation complex is key. Front Microbiol 7:869. doi: 10.3389/fmicb.2016.00869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, Walsh A, Lam TT, Delecluse HJ, El-Guindy A. 2019. A single phosphoacceptor residue in BGLF3 is essential for transcription of Epstein-Barr virus late genes. PLoS Pathog 15:e1007980. doi: 10.1371/journal.ppat.1007980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Djavadian R, Chiu YF, Johannsen E. 2016. An epstein-barr virus-encoded protein complex requires an origin of lytic replication in cis to mediate late gene transcription. PLoS Pathog 12:e1005718. doi: 10.1371/journal.ppat.1005718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Serio TR, Cahill N, Prout ME, Miller G. 1998. A functionally distinct TATA box required for late progression through the Epstein-Barr virus life cycle. J Virol 72:8338–8343. doi: 10.1128/JVI.72.10.8338-8343.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gruffat H, Kadjouf F, Mariame B, Manet E. 2012. The Epstein-Barr virus BcRF1 gene product is a TBP-like protein with an essential role in late gene expression. J Virol 86:6023–6032. doi: 10.1128/JVI.00159-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perng YC, Campbell JA, Lenschow DJ, Yu D. 2014. Human cytomegalovirus pUL79 is an elongation factor of RNA polymerase II for viral gene transcription. PLoS Pathog 10:e1004350. doi: 10.1371/journal.ppat.1004350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan D, Xuan B, Sun Y, Huang S, Xie M, Bai Y, Xu W, Qian Z. 2016. An intein-mediated modulation of protein stability system and its application to study human cytomegalovirus essential gene function. Sci Rep 6:26167. doi: 10.1038/srep26167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qian Z, Xuan B, Hong TT, Yu D. 2008. The full-length protein encoded by human cytomegalovirus gene UL117 is required for the proper maturation of viral replication compartments. J Virol 82:3452–3465. doi: 10.1128/JVI.01964-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Didychuk AL, Castaneda AF, Kushnir LO, Huang CJ, Glaunsinger BA. 2019. Conserved CxnC motifs in Kaposi’s sarcoma-associated herpesvirus ORF66 are required for viral late gene expression and are essential for its interaction with ORF34. J Virol doi: 10.1128/JVI.01299-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watanabe T, Nishimura M, Izumi T, Kuriyama K, Iwaisako Y, Hosokawa K, Takaori-Kondo A, Fujimuro M. 2019. Kaposi’s sarcoma-associated herpesvirus ORF66 is essential for late gene expression and virus production via interaction with ORF34. J Virol doi: 10.1128/JVI.01300-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Busche A, Angulo A, Kay-Jackson P, Ghazal P, Messerle M. 2008. Phenotypes of major immediate-early gene mutants of mouse cytomegalovirus. Med Microbiol Immunol 197:233–240. doi: 10.1007/s00430-008-0076-3. [DOI] [PubMed] [Google Scholar]
- 34.Cardin RD, Abenes GB, Stoddart CA, Mocarski ES. 1995. Murine cytomegalovirus IE2, an activator of gene expression, is dispensable for growth and latency in mice. Virology 209:236–241. doi: 10.1006/viro.1995.1249. [DOI] [PubMed] [Google Scholar]
- 35.Manning WC, Mocarski ES. 1988. Insertional mutagenesis of the murine cytomegalovirus genome: one prominent alpha gene (ie2) is dispensable for growth. Virology 167:477–484. [PubMed] [Google Scholar]
- 36.Wagner M, Jonjic S, Koszinowski UH, Messerle M. 1999. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J Virol 73:7056–7060. doi: 10.1128/JVI.73.8.7056-7060.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tischer BK, Smith GA, Osterrieder N. 2010. En passant mutagenesis: a two step markerless Red recombination system. Methods Mol Biol 634:421–430. doi: 10.1007/978-1-60761-652-8_30. [DOI] [PubMed] [Google Scholar]
- 38.Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step Red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- 39.Kinsella TM, Nolan GP. 1996. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum Gene Ther 7:1405–1413. doi: 10.1089/hum.1996.7.12-1405. [DOI] [PubMed] [Google Scholar]
- 40.Harvey DM, Levine AJ. 1991. p53 alteration is a common event in the spontaneous immortalization of primary BALB/c murine embryo fibroblasts. Genes Dev 5:2375–2385. [DOI] [PubMed] [Google Scholar]
- 41.Qian Z, Xuan B, Chapa TJ, Gualberto N, Yu D. 2012. Murine cytomegalovirus targets transcription factor ATF4 to exploit the unfolded-protein response. J Virol 86:6712–6723. doi: 10.1128/JVI.00200-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qian Z, Xuan B, Gualberto N, Yu D. 2011. The human cytomegalovirus protein pUL38 suppresses endoplasmic reticulum stress-mediated cell death independently of its ability to induce mTORC1 activation. J Virol 85:9103–9113. doi: 10.1128/JVI.00572-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie M, Xuan B, Shan J, Pan D, Sun Y, Shan Z, Zhang J, Yu D, Li B, Qian Z. 2015. Human cytomegalovirus exploits interferon-induced transmembrane proteins to facilitate morphogenesis of the virion assembly compartment. J Virol 89:3049–3061. doi: 10.1128/JVI.03416-14. [DOI] [PMC free article] [PubMed] [Google Scholar]