There is a great need to develop broadly reactive or universal vaccines against influenza viruses. Advanced, next-generation hemagglutinin (HA) head-based vaccines that elicit protective antibodies against H1N1 influenza viruses have been developed. This study focused on understanding the specific amino acids around the receptor binding site (RBS) that were important in elicitation of these broadly reactive antibodies. Specific glycan sites and amino acids located at the tip of the HA molecule enhanced the elicitation of these broadly reactive antibodies. A better understanding of the HA structures around the RBS will lead to more effective HA immunogens.
KEYWORDS: hemagglutination inhibition, influenza, H1N1, hemagglutinin inhibition assay
ABSTRACT
Vaccination is the most effective way to prevent influenza virus infections. However, the diversity of antigenically distinct isolates is a challenge for vaccine development. In order to overcome the antigenic variability and improve the protective efficacy of influenza vaccines, our research group has pioneered the development of computationally optimized broadly reactive antigens (COBRA) for hemagglutinin (HA). Two candidate COBRA HA vaccines, P1 and X6, elicited antibodies with differential patterns of hemagglutination inhibition (HAI) activity against a panel of H1N1 influenza viruses. In order to better understand how these HA antigens elicit broadly reactive immune responses, epitopes in the Cb, Sa, or Sb antigenic sites of seasonal-like and pandemic-like wild-type or COBRA HA antigens were exchanged with homologous regions in the COBRA HA proteins to determine which regions and residues were responsible for the elicited antibody profile. Mice were vaccinated with virus-like particles (VLPs) expressing one of the 12 modified HA antigens (designated V1 to V12), COBRA HA antigens, or wild-type HA antigens. The elicited antisera was assessed for hemagglutination inhibition activity against a panel of historical seasonal-like and pandemic-like H1N1 influenza viruses. Primarily, the pattern of glycosylation sites and residues in the Sa antigenic region, around the receptor binding site (RBS), served as signatures for the elicitation of broadly reactive antibodies by these HA immunogens. Mice were vaccinated with VLPs expressing HA antigens that lacked a glycosylation site at residue 144 and a deleted lysine at position 147 residue were more effective at protecting against morbidity and mortality following infection with pandemic-like and seasonal-like H1N1 influenza viruses.
IMPORTANCE There is a great need to develop broadly reactive or universal vaccines against influenza viruses. Advanced, next-generation hemagglutinin (HA) head-based vaccines that elicit protective antibodies against H1N1 influenza viruses have been developed. This study focused on understanding the specific amino acids around the receptor binding site (RBS) that were important in elicitation of these broadly reactive antibodies. Specific glycan sites and amino acids located at the tip of the HA molecule enhanced the elicitation of these broadly reactive antibodies. A better understanding of the HA structures around the RBS will lead to more effective HA immunogens.
INTRODUCTION
Influenza viruses have taken a heavy toll on public health through annual epidemics and occasional pandemics. Seasonal influenza outbreaks cause severe illness and deaths each year (1). Four times over the last century, a new influenza virus subtype has entered the human population and resulted in a pandemic. These new influenza virus strains arise from animal reservoirs and result in a human-transmissible virus (2). The influenza viruses can be classified into 3 types: A, B, and C. Type A viruses are divided into subtypes according to the combinations of hemagglutinin (HA) and neuraminidase on the surface of the virus. Of all the influenza A virus subtypes, only viruses of the H1, H2, and H3 HA subtypes are known to have adapted to circulate in humans.
The viral surface proteins, HA and neuraminidase (NA), often mutate, which enables the virus to escape host immune responses. In addition, posttranslational modifications to the HA protein, such as the addition of N-linked glycosaccharides (N-X-S/T, where X is any amino acid except proline), occur (3, 4). Glycosylation of the HA protein is critical for protein folding and stability (5), as well as for increasing the virulence and antigenicity of the virus (6). In addition, glycosylation can shield or redirect immune responses to other epitopes on the HA molecule (7–10). Virus resistance to neutralizing antibodies can occur following the addition of glycans on the HA protein. Therefore, the addition of glycans on the surface of HA from newly emerged pandemic viruses may cause the virus to evolve into seasonal-like humans strains.
While the effect of glycosylation on in vitro antigenicity and viral replication has been demonstrated, little is known about the role of glycosylation in in vivo immunogenicity. In this study, the effect of glycosylation on the elicitation of broadly reactive antibodies against H1N1 strains was explored. Our group has pioneered the development of computationally optimized broadly reactive antigens (COBRA) for HA as immunogens that elicit antibodies with hemagglutination inhibition (HAI) activity against both historical seasonal and current H1N1 influenza viruses isolated from humans and swine (11). Two candidate COBRA HA vaccines, P1 and X6, elicited antibodies with differential patterns of HAI activity against a panel of seasonal- and pandemic-like H1N1 viruses. X6 elicited antibodies that recognized primarily seasonal-like viruses. The COBRA P1 HA elicited antibodies that had HAI activity against both pandemic-like and some seasonal-like H1N1 viruses but notably not the vaccine strains from 2006 or 2007. In order to better understand the immunogenicity of these COBRA HA antigens, epitopes in the Cb and Sa sites were exchanged in the P1 and X6 COBRA HA proteins to determine which regions were responsible for the elicited antibody profile against a panel of H1N1 viruses. Overall, the glycosylation sites at residues 142 and 144, as well as the lysine residue at position 147, were responsible for the elicitation of broadly reactive antibodies by these two COBRA HA immunogens.
RESULTS
Design and characterization of modified COBRA P1 HA antigen.
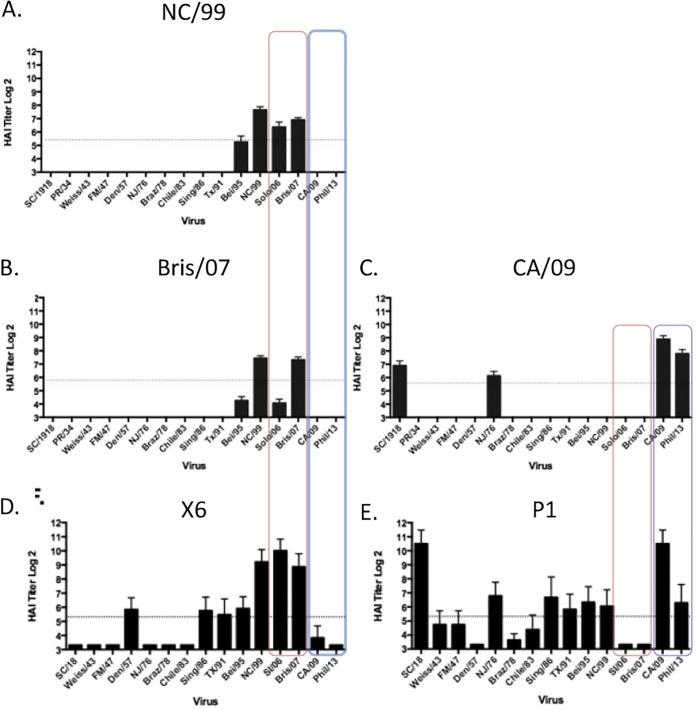
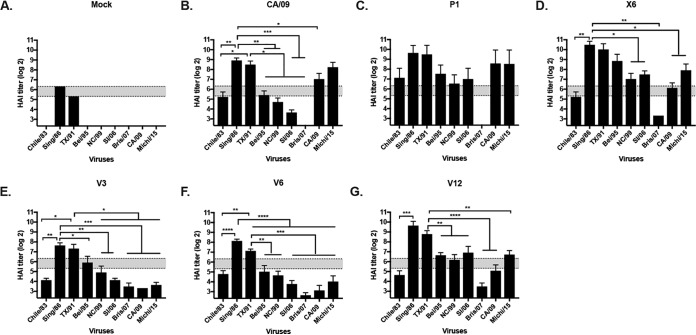
Previously, our group described the design and effectiveness of HA antigens that elicit broadly protective antibodies against H1N1 influenza viruses (11). These HA antigens are designed using a methodology called computationally optimized broadly reactive antigens (COBRA). Two of these HA antigens, P1 and X6, elicited antibodies with HAI activity against a broad number of H1N1 influenza viruses isolated over the past 30 years or more (Fig. 1). Vaccines based upon wild-type HA antigens from NC/99, Bris/07, or CA/09 virus elicited antibodies with a narrow range of HAI activity, recognizing 1 or 2 of the 15 viruses in the panel (Fig. 1). Specifically focusing on the differences in antibody breadth between P1 and X6, P1 elicits antibodies with HAI activity against pandemic H1N1 viruses (post-2009) but with no HAI activity against the 2006 or 2007 seasonal H1N1 viruses. In contrast, X6 elicits antibodies with HAI activity against the 2006 and 2007 seasonal H1N1 viruses but not the 2009 pandemic or postpandemic H1N1 strains (Fig. 1). The P1 HA was designed to cover the antigenic space from 1933 to 1957 and 2009 to 2011 and swine sequences from 1931 to 1998, and the X6 COBRA sequence covered antigenic space from 1999 to 2012. P1 and X6 COBRA HAs were used to determine potentially important epitopes that may lead to the difference in antibody breadth and virus recognition/neutralization.
FIG 1.
Hemagglutination inhibition (HAI) serum antibody titers induced by vaccination of mice with COBRA and wild-type HA VLP vaccines. HAI titers were determined for each group of immunologically naive mice (n = 11) vaccinated two times (weeks 0, 4, and 8) with either the X6 or P1 COBRA H1N3 VLP vaccine or H1N3 VLP vaccine expressing wild-type HA proteins from NC/99, Bris/07, or CA/09 against a panel of 15 H1N1 influenza viruses. Values are the log2 HAI titers of each individual animal from antisera collected on week 12. The dotted lines indicate the 1:40 HAI titer range. (A) NC/99 VLP; (B) Bris/07 VLP; (C) CA/09 VLP; (D) X6 VLP; (E) P1 VLP.
To better understand the epitopes involved in this HAI specificity, mutations were introduced in these two COBRA HA antigens to exchange amino acids or antigenic sites between COBRA HA sequences, as well as between wild-type H1N1 HA proteins. This technique is referred to as vaccines intelligently produced by epitope recombination (VIPER). VIPER technology incorporates specific changes into influenza virus HA sequences to induce increases in the elicited antibody breadth and effectiveness of these HA vaccines. Sequence analysis determined the potential epitopes of importance between COBRA P1, COBRA X6, CA/09 (pandemic H1N1), and Bris/07 HA antigens (Table 1). The Sa and Cb antigenic sites were identified as being similar in P1 and CA/09 but different in X6 and Bris/07 HA proteins. The Sa residues 142 to 148 in P1 and CA/09 (NHNTTKG) differ from the residues in X6 and Bris/07 (NHTVT-G). Cb residues 88 to 91 in P1 (LSAR) differ from the residues in X6 and Bris/07 (ISKE). Initially, H1N1 VIPER sequences utilized the COBRA P1 HA backbone sequence to exchange amino acids found in the COBRA X6 HA in the Sa and/or the Cb region (Table 1).
TABLE 1.
Amino acid residues in the Cb, Sa, and Sb antigenic sites in the vaccine HA antigensa
Portions of the Cb, Sa, and Sb antigenic residues are shown. Residues highlighted in blue are associated with the P1 HA vaccine, and residues highlighted in red are associated with the X6 HA antigen.
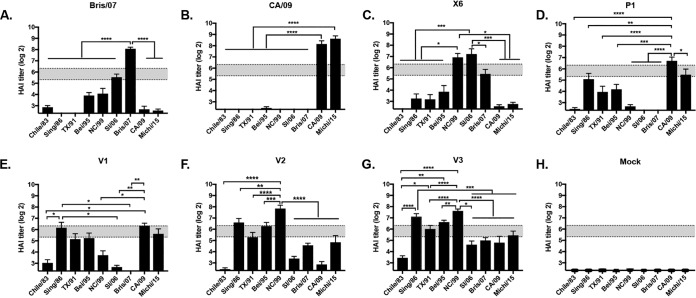
Three modified COBRA P1 HA antigens were produced and designated VIPER-1, -2, and -3. For VIPER-1 (V1), amino acids 142 to 148 (NHNTTKG) located in the Sa region were modified to match the residues in the X6 and Bris/07 HA (NHTVT-G) Sa region. This exchange introduced a deletion that is present in late seasonal H1N1 sequences at residue 147, where a Lys is present at this location in pandemic HA antigens (Table 1). For VIPER-2 (V2), the amino acids at residues 88 to 91 were changed from LSAR to ISKE in the Cb region of the COBRA P1 HA antigen (Table 1). The ISKE amino acids are located at this position in the NC/99, Bris/07, and COBRA X6 HA molecules and will transform the Cb antigenic site in the P1 HA from pandemic-like to seasonal-like. To generate VIPER-3 (V3), the amino acids located in both the Sa and Cb regions of the X6 COBRA HA antigen were introduced into the P1 HA sequence (Table 1).
Vaccinated mice challenged with H1N1 influenza viruses.
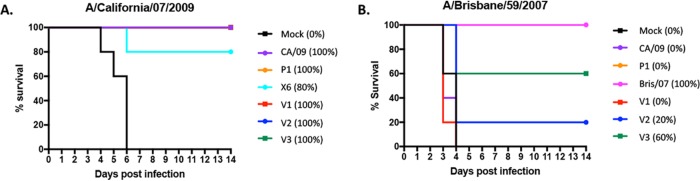
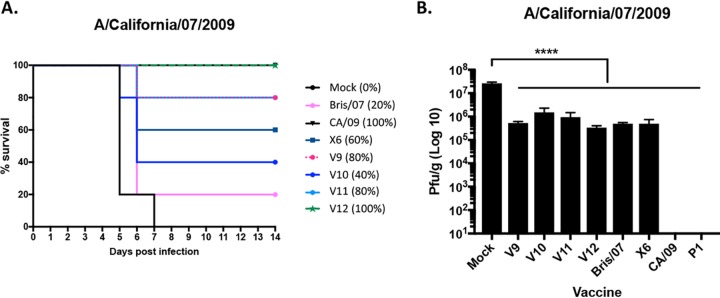
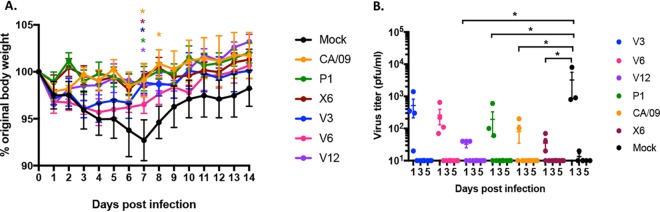
To determine the protective efficacy of each VIPER vaccine, BALB/c mice (n = 11) were vaccinated three times at 4-week intervals via intramuscular injection with purified virus-like particles (VLPs) expressing either VIPER, COBRA, or wild-type HA antigens (3 μg based upon HA content) plus an oil-in-water-based adjuvant, AF03 (provided by Sanofi Pasteur). Mice were then challenged with CA/09 influenza virus (1 × 106 PFU) on day 84 (Fig. 2A). Mice vaccinated with CA/09, P1, or VIPER V1 VLPs all survived challenge with little or no weight loss. Mock-vaccinated animals rapidly lost weight and reached experimental endpoints by day 6 postinfection. Mice vaccinated with the V2 or V3 VLP vaccine, as well as the COBRA X6 VLP vaccine, all survived challenge (Fig. 2A) but did lose ∼8 to 11% of body weight by day 6 postinfection and then slowly began to recover weight (Table 2). A different set of vaccinated mice were challenged with a Bris/07 PR8 (7:1) reassortant influenza virus (10 times the 50% lethal dose [LD50]). Mice vaccinated with the V2 VLP vaccine were not protected against Bris/07 challenge, and 60% of the V3 VLP-vaccinated mice survived, with the average weight loss of the survivors being ∼11% by day 6 postinfection (Table 2).
FIG 2.
Survival after H1N1 influenza virus challenges of mice. BALB/c mice (11 mice/group) were vaccinated on days 0, 28, and 56 with each vaccine plus the AF03 adjuvant and infected on day 84 with 1 × 106 PFU/ml of the H1N1 influenza virus. Kaplan-Meier survival curves for vaccinated mice challenged with A/California/07/2009 (CA/09) (A) or A/Brisbane/59/2007 (Bris/07) (B) influenza virus are shown. The percent survival per vaccine group is listed in parentheses.
TABLE 2.
Heat map of hemagglutination inhibition, weight loss, and survival of vaccinated micea
The average hemagglutination inhibition (HAI) titers per vaccine group are highlighted in blue for positive (POS) (HAI titer greater than 1:40), light blue for low positive (LOW) (HAI titer of 1:20), and red for negative (NEG) (HAI titer of less than 1:10). For weight loss, range from 0% as green and 20% weight loss as red. For survival, 100% as green and 0% as red. N.D., not done.
HA regions important for elicitation of antibodies with H1N1 HAI activity.
In order to determine the breadth of the vaccine-elicited antibodies, sera were collected from vaccinated mice at week 12, following the third vaccination. Mice vaccinated with Bris/07 VLPs had antisera with HAI activity against Bris/07, as well as SI/06 (Fig. 3A), and mice vaccinated with CA/09 VLPs had antisera with HAI activity against the CA/09 and Mich/15 pandemic-like viruses (Fig. 3B). In contrast, and as previously reported (11, 12), mice vaccinated with COBRA P1 VLPs had antisera against both H1N1pdm viruses and historical seasonal influenza viruses (Fig. 3D). Mice vaccinated with the VIPER V1 VLPs had antibodies with HAI activity against CA/09, Sing/86, TX/91, and Bei/95 (Fig. 3E), as well as SC/18 and NJ/76 (data not shown). These V1 VLP-elicited antibodies did not have HAI activity against SI/06 or Bris/07. This HAI pattern was similar to the HAI activity elicited by COBRA P1. Mice vaccinated with V2 or V3 VLPs had antibodies with HAI activity against almost all viruses in the panel to some degree (Fig. 3F and G). The HAI activity against CA/09 elicited by V2 VLPs was very low (Fig. 3F). V3 VLP vaccines were the best immunogen at eliciting antibodies with the broadest breadth of HAI activity, with the highest HAI titers against the panel (Fig. 3G).
FIG 3.
Hemagglutination inhibition (HAI) serum antibody titers induced by vaccination of mice with V1, V2, or V3 VLP vaccines. HAI titers were determined for each group of mice (n = 11) from blood collected at week 12 postvaccination. Mice were vaccinated with either COBRA, VIPER, or wild-type HA VLP vaccines and the collected sera were tested against a panel of 9 H1N1 influenza viruses. Values are the geometric mean titers plus standard errors of the means (SEM) (error bars). The dotted lines indicate the 1:40 to 1:80 HAI titer range. (A) Bris/07 VLP; (B) CA/09 VLP; (C) X6 VLP; (D) P1 VLP; (E) V1; (F) V2 VLP; (G) V3 VLP; (H) mock. All data are reported as absolute mean values ± SEM. HAI titers were compared using nonparametric one-way ANOVA. All statistical analysis was performed using GraphPad Prism 7 software (GraphPad, San Diego, CA, USA), and a P value of <0.05 was considered statistically significant (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
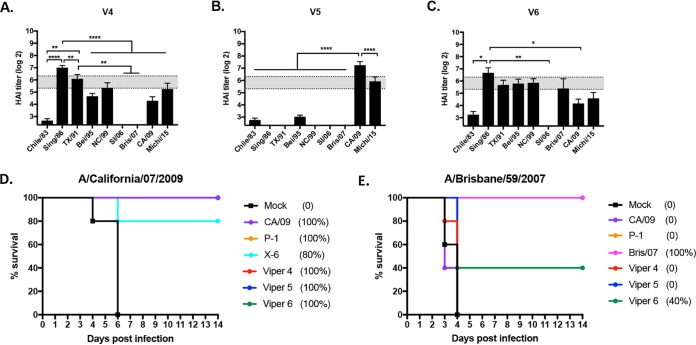
Introduction of the amino acids in the Sa site from seasonal-like influenza viruses into the V2 HA backbone increased the HAI titers to seasonal-like viruses and decreased the HAI activity in elicited sera to CA/09. In order to define in more detail the amino acids in the Sa region that are critical for the elicitation of antibodies with HAI activity against seasonal-like and pandemic-like H1N1 viruses, a second set of VIPER HA antigens was designed and designated VIPER V4 (Table 1). Utilizing the V2 HA amino acid sequence, the Sa site was changed to insert a Lys at residue 147. This results in a hybrid Sa site with residues 142 to 146 representing the amino acids, NHTVT, found in seasonal-like influenza virus HA antigens with the inserted Lys at residue 147 located in pandemic-like HA proteins (Table 1). Similar to the case for V2 and V3 HA antigens, this may shift the putative glycosylation site from position 144 to position 142, which may further alter the site structure and antigenicity of the V4 HA molecule. Mice vaccinated with V4 HA VLPs elicited antibodies with an HAI profile similar to that of antibodies elicited by V1 HA VLP vaccines. The antisera had HAI activity against Sing/86, TX/91, Bei/95, NC/99, and to a lesser extent CA/09 and Mich/15 but had no HAI activity against SI/06 or Bris/07 (Fig. 4A).
FIG 4.
Hemagglutination inhibition (HAI) serum antibody and survival after H1N1 influenza virus challenge. HAI titers were determined for each group of mice (n = 11) from blood collected at week 12 postvaccination. Mice were vaccinated with either V4, V5, or V6 HA VLP vaccines, and the collected sera were tested against a panel of 9 H1N1 influenza viruses. Values are the geometric mean titers plus standard errors of the means (SEM) (error bars). The dotted lines indicate the 1:40 to 1:80 HAI titer range. All data are reported as absolute mean values ± SEM. HAI titers were compared using nonparametric one-way ANOVA. All statistical analysis was performed using GraphPad Prism 7 software (GraphPad, San Diego, CA, USA), and a P value of <0.05 was considered statistically significant (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
To generate the V5 HA, the Thr at position 144 in the Sa region of V2 HA was converted into an Asn. This results in an Sa region that matches the seasonal-like Sa site, except for the Asn at position 144 that is found in pandemic-like HA sequences. Mice vaccinated with the V5 HA VLP had antibodies with HAI activity against CA/09 and Mich/15, both post-pandemic-like H1N1 viruses, with no detectable HAI titers against any of the seasonal-like H1N1 viruses (Fig. 4B). For V6 HA, the Asn at position 142 in V2 was converted to a Thr (Table 1). All seasonal-like and pandemic-like HA antigens have an Asn at position 142. Therefore, unlike the case for any of the wild-type, COBRA, or other VIPER HA antigens, there is no putative glycosylation site at position 142 or 144. Mice vaccinated with V6 HA VLPs had antibodies with HAI activity against most seasonal and pandemic H1N1 influenza viruses (Fig. 4C). The V6 HA antigen elicited antibodies with a pattern of HAI activity similar to that for V4. This HA had the Cb region from P1, no potential N-linked glycosylation sites in Sa, and the amino acid deletion at site 147.
Influenza virus challenge of vaccinated mice.
Interestingly, all mice vaccinated with the V4, V5, or V6 VLP vaccines, along with mice vaccinated with the CA/09 VLP vaccine, survived challenge with CA/09 (Fig. 4D). All mock-vaccinated mice died from infection by day 6 postchallenge. In contrast, only mice vaccinated with Bris/07 VLP or V6 VLP vaccines survived challenge with Bris/07. All the Bris/07 VLP-vaccinated mice survived, and 40% of the V6 VLP-vaccinated mice survived. All other mice vaccinated with the CA/09, V4, V5, or P1 VLP vaccine died or reached endpoints by day 3 to 4 postchallenge (Fig. 4E).
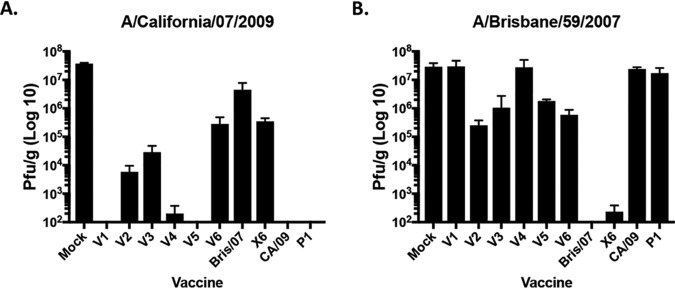
A set of mice were sacrificed on day 3 postchallenge, and the influenza virus titer in the lungs was assessed (Fig. 5). Mock-vaccinated mice had high lung titers (2 × 107 PFU/ml). Mice vaccinated with the CA/09, P1, or V1 VLP vaccine had no detectable virus in their lungs, and V2-, V3-, and V6 VLP-vaccinated mice had a 3- to 5-log drop in the average lung viral titer (average of 5 × 103 to 1 × 105 PFU/g) compared to that in mock-vaccinated mice (Fig. 5A). One mouse vaccinated with the X6 VLP vaccine had detectable virus, while the others had no detectable virus, and Bris/07 VLP-vaccinated mice had a 1-log drop in viral titer compared to that in mock-vaccinated mice (Fig. 5A). In contrast, mock-vaccinated mice challenged with Bris/07 had ∼2.2 × 107 PFU/g of virus in their lungs (Fig. 5B). Mice vaccinated with P1, V1, V4, or CA/09 VLPs had similar levels of virus in their lungs. However, mice vaccinated with V2, V3, V5, or V6 had a 1- to 2-log drop in lung viral titer compared to that in mock-vaccinated mice (Fig. 5B). One V3-vaccinated mouse and all of the Bris/07 and almost all of the X6 VLP-vaccinated mice had no detectable virus in the lungs (Fig. 5B).
FIG 5.
Viral lung titers in vaccinated mice. Vaccinated BALB/c mice challenged with A/California/07/2009 (A) or A/Brisbane/59/2007 (B) at week 13 postvaccination had lungs collected (3 mice/group/time point) on day 3 postinfection and percent survival assessed. Viral lung titers are listed as PFU per gram of lung tissue. The vaccines used for vaccination listed on the x axis.
Addition of glycosylation motifs to COBRA P1 HA enhances protective antibodies to seasonal H1 influenza virus strains.
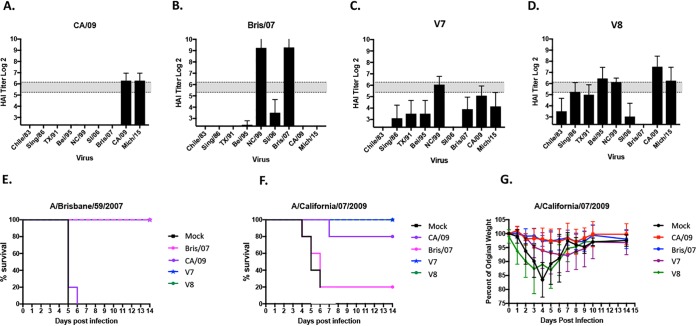
The CA/09 HA does not have a putative glycosylation motif at residue 144 but rather has an Asp (D) amino acid at this position. Seasonal influenza virus strains, such as Bris/07, also do not contain a glycosylation site at residue 144 (Thr), but the COBRA P1 HA has a putative glycosylation motif at this position (Table 1). The addition of the Asn (N) residue in P1 and V1 enhanced HAI activity against CA/09 but not Bris/07 (Fig. 5); however, the lack of a putative N-linked glycosylation site also enhanced HAI activity to NC/99 and Bris/07. Since CA/09 has a Asn at residue 146 and we hypothesized that there were N-linked glycosylations at all three Asn residues in the Sa site, we used the P1 COBRA HA sequence, inserted a third Asn into this region (residues 142, 144, and 146), and retained the Lys at residue 147 to determine the HAI activity against seasonal and pandemic-like H1N1 viruses (Fig. 6). Mice vaccinated with VLPs expressing a modified P1 COBRA HA with the addition of the Asn at position 146, as well as Asn and Glu at residues 179 and 180, respectively (V7), did not show a significantly altered phenotype (Fig. 6C), and all mice survived both Bris/07 and CA/09 challenges (Fig. 6E and F). Mice challenged with CA/09 had little or no weight loss (Fig. 6G), but mice challenged with Bris/07 lost between ∼7 and 11% of their body weight by day 7 postinfection and then recovered. Mice vaccinated with VLPs expressing the V8 HA, which has the same Sa sequence as the V7 HA except that the HA has the addition of only the Asn at 146 (Table 1), elicited protective antibodies similar to those with V7 but with a higher magnitude of HAI activity against the panel of H1N1 viruses (Fig. 6D), and again all the mice were protected against infection (Fig. 6E to G).
FIG 6.
Hemagglutination inhibition (HAI) serum antibody and challenge with H1N1 influenza viruses. HAI titers were determined for each group of mice (n = 11) from blood collected at week 12 postvaccination. (A to D) Mice were vaccinated with either CA/09 VLP (A), Bris/07 VLP (B), V7 VLP (C), or V8 VLP (D) vaccine, and the collected sera were tested against a panel of 9 H1N1 influenza viruses. Values are the geometric mean titers plus standard errors of the means (SEM) (error bars). The dotted lines indicate the 1:40 to 1:80 HAI titer range. (E and F) Kaplan-Meier survival curves for vaccinated mice challenged with A/Brisbane/59/2007 (E) or A/California/07/2009 (F). (G) Mice challenged with A/California/07/2009 were monitored daily for weight loss over a 14-day observation period.
Determining epitopes in Sa and Sb that contribute to the elicitation of antibodies with HAI activity against multiple H1N1 strains.
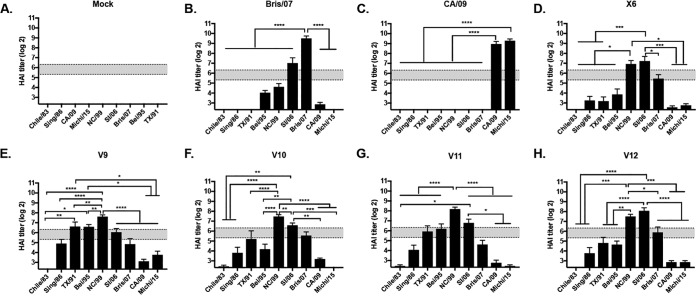
The region of Sa from amino acid 142 to 147 has been shown to be critical for eliciting antibodies with a seasonal-like or pandemic-like phenotype (Fig. 5 and 6). In order to determine if other regions of Sa (amino acids 170 to 181) or the Sb region (202 to 213) play a role in determining the breadth of elicited antibodies, mutations in the COBRA X6 HA were engineered to mimic homologous regions of CA/09 or COBRA P1 HA (Table 1). The corresponding Sb region from P1 was exchanged with X6 to generate V9 HA, and the corresponding Cb region was exchanged with X6 to generate V10 HA. For V11, both regions were exchanged. For V12, the Asn (N) at amino acid 177 in the Sa region was mutated to a Lys (K). Mice vaccinated with X6 VLP vaccine had antibodies with HAI activity against NC/99, SI/06, and Bris/07 (Fig. 7D), which was similar to the HAI activity from sera collected from mice vaccinated with VIPER V9, V10, V11, or V12 (Fig. 7E to H). None of these epitope exchanges significantly altered the pattern of elicited antibody recognition compared to that for antibodies elicited by COBRA X6 VLPs. Mice vaccinated with any of these VIPER vaccines had similar weight loss (peak of 15 to 20% of original weight) (Table 2); however, most mice, including all the V12-vaccinated mice, recovered and survived the CA/09 challenge (Fig. 8A). Viral lung titers were ∼2 logs lower at day 3 postchallenge in COBRA X6 or VIPER V9 to V12 VLP-vaccinated mice than in mock-vaccinated mice (Fig. 8B). Mice vaccinated with CA/09 VLPs had low to undetectable viral lung titers.
FIG 7.
Hemagglutination inhibition (HAI) serum antibody. HAI titers were determined for each group of mice (n = 11) from blood collected at week 12 postvaccination. Mice were vaccinated as follows: (A) mock; (B) Bris/07 VLP vaccine; (C) CA/09 VLP vaccine; (D) X6 VLP vaccine; (E) V9 VLP vaccine; (F) V10 VLP vaccine; (G) V11 VLP vaccine; (H) V12 VLP vaccine. The collected sera were tested against a panel of 9 H1N1 influenza viruses. The dotted lines indicate the 1:40 to 1:80 HAI titer range. All data are reported as absolute mean values ± SEM. HAI titers were compared using nonparametric one-way ANOVA. All statistical analysis was performed using GraphPad Prism 7 software (GraphPad, San Diego, CA, USA), and a P value of <0.05 was considered statistically significant (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
FIG 8.
Survival after H1N1 influenza virus challenges. BALB/c mice (11 mice/group) were vaccinated on days 0, 28, and 56 with each vaccine plus the AF03 adjuvant and infected on week 13 with 1 × 106 PFU of the A/California/07/2009 H1N1 influenza virus. (A) Kaplan-Meier survival curves for vaccinated mice challenged with each influenza virus. The percent survival per vaccine group is listed in parenthesis. (B) Viral lung titers are listed as PFU per gram of lung tissue on the y axis. Vaccines used for vaccination are listed on the x axis. All data are reported as absolute mean values ± SEM. HAI titers were compared using nonparametric one-way ANOVA. All statistical analysis was performed using GraphPad Prism 7 software (GraphPad, San Diego, CA, USA), and a P value of <0.05 was considered statistically significant (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
Elicitation of antibodies by VIPER HA antigens in ferrets preimmune to a historical H1N1 influenza virus.
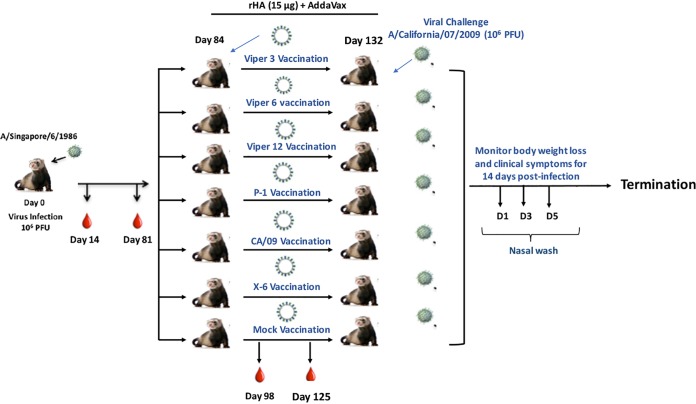
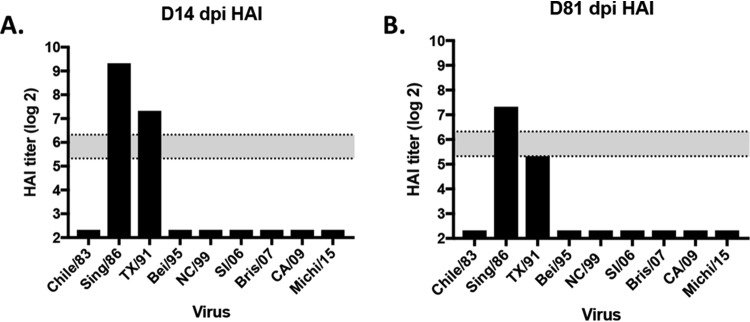
Using a previously developed preimmune ferret model, we infected ferrets with the Sing/86 H1N1 influenza virus (Fig. 9). As previously observed (12), all ferrets seroconverted and had high HAI titers to the Sing/86 and TX/91 viruses (Fig. 10), At day 84, ferrets were vaccinated with one of 6 recombinant HA (rHA) vaccines representing the wild-type CA/09, the COBRA P1 or X6, or the VIPER V3, V6, or V12 HA antigen (Fig. 11). Mock-vaccinated preimmune ferrets were used as controls. At day 98 (2 week postvaccination), ferrets vaccinated with P1, X6, or V12 had HAI titers against all 9 H1N1 viruses in the panel except Bris/07 (Fig. 11C, D, and G). Preimmune ferrets that were vaccinated with V3 or V6 had HAI titers elicited by the Sing/86 infection (Sing/86 and TX/91) and low HAI titers to the other H1N1 viruses in the panel (Fig. 11E and F). In contrast, ferrets vaccinated with CA/09 HA had high HAI titers to Sing/86 and TX/91, as well as CA/09 and Mich/15 (Fig. 11).
FIG 9.
Schematic of the infection schedule. Ferrets were infected intranasally (106 PFU) with A/Singapore/6/1986 H1N1 influenza virus. The ferrets were bled at days 14 and 81 postinfection. At day 84, all ferrets were mock vaccinated or vaccinated with one of 6 rHA vaccines plus AddaVax adjuvant. Blood was collected at day 96 (2 week postvaccination) and day 125 (6 weeks postvaccination). Ferrets were infected with the H1N1 influenza virus A/California/07/2009 (106 PFU) at day 132. The ferrets were observed for 2 weeks for clinical signs of infection. Nasal washes were collected at days 1, 3, and 5 postinfection, and the experiment was terminated at day 166.
FIG 10.
Hemagglutination inhibition (HAI) serum antibody titer induced by H1N1 viral infection. Ferrets were infected intranasally (106 PFU) with A/Singapore/6/1986 H1N1 influenza virus. At days 14 (A) and 81 (B), ferret sera were collected. HAI titers of pooled sera were tested against a panel of 9 H1N1 influenza viruses. Values are log2 HAI titers, and dotted lines indicate the 1:40 to 1:80 HAI titer range.
FIG 11.
Hemagglutination inhibition (HAI) serum antibody. HAI titers were determined for each group of preimmune ferrets (n = 4) from blood collected at day 125 postvaccination. Preimmune ferrets were vaccinated as follows: (A) mock; (B) CA/07 rHA vaccine; (C) P1 rHA vaccine; (D) X6 rHA vaccine; (E) V3 rHA vaccine; (F) V6 rHA vaccine; (G) V12 rHA vaccine. The collected sera were tested against a panel of 9 H1N1 influenza viruses. Values are the geometric mean titers plus standard errors of the means (SEM) (error bars). The dotted lines indicate the 1:40 to 1:80 HAI titer range. All data are reported as absolute mean values ± SEM. HAI titers were compared using nonparametric one-way ANOVA. All statistical analysis was performed using GraphPad Prism 7 software (GraphPad, San Diego, CA, USA), and a P value of <0.05 was considered statistically significant (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
Influenza virus challenge of vaccinated ferrets.
Preimmune ferrets vaccinated with the CA/09, P1, X6, and V12 HA vaccines survived challenge with CA/09 with little or no weight loss (Fig. 12A). Mock-vaccinated ferrets lost on average 8% of their body weight by day 6 postchallenge and then began to recover. In contrast, preimmune ferrets vaccinated with the V3 or V6 HA vaccine lost ∼5% of their body weight by day 3 postinfection and then began to recover (Fig. 12A). Virus was detectable in all ferret nasal washes at day 1 postinfection (Fig. 12B). However, all preimmune ferrets vaccinated with V12 or X6 had less than 40 PFU/ml at day 1 postinfection, which was statistically lower than that in preimmune ferrets vaccinated with the other vaccines or mock vaccinated (Fig. 12B).
FIG 12.
Challenge with A/California/07/2009 H1N1 influenza virus. Vaccinated preimmune ferrets (4 ferrets/group) were infected on day 132 with 1 × 106 PFU/ml of the A/California/07/2009 H1N1 influenza virus. (A) Percent original weight per vaccine group. (B) Viral nasal wash titers are listed as PFU on the y axis for each individual ferret collected at days 1, 3, and 5 postinfection. All data are reported as absolute mean values ± SEM. Weight losses among the different vaccinated groups were compared using nonparametric two-way ANOVA, and viral lung titers were compared using nonparametric one-way ANOVA. All statistical analysis was performed using GraphPad Prism 7 software (GraphPad, San Diego, CA, USA), and a P value of <0.05 was considered statistically significant (*, P < 0.05).
Viral neutralization titers.
In order to determine if vaccine-elicited antibodies would neutralize various H1N1 viruses, an in vitro focal reduction neutralization assay was performed (Fig. 13). Naive ferrets vaccinated with any of the wild-type, COBRA, or VIPER VLP vaccines had low to undetectable neutralizing antibody titers against all the H1N1 viruses tested. Ferrets preimmune to Sing/86 and then vaccinated with one of the wild-type, COBRA, or VIPER VLP vaccines had considerably higher neutralization titers than naive ferrets (data not shown). Sing/86-preimmune ferrets that were vaccinated with P1 VLPs had antisera with high (50% and 80%) neutralizing titers against all the H1N1 viruses except Bris/07. Only sera collected from ferrets preimmune to Sing/86 and vaccinated with CA/09 VLPs had a log2 titer equal to that for P1 against CA/09 virus (Fig. 13E). Sera from these preimmune, CA/09 VLP-vaccinated ferrets did not neutralize the other seasonal strains any better than those from ferrets vaccinated with the X6 COBRA or the VIPER HA antigens (Fig. 13A to D). Sing/86-preimmune ferrets that were vaccinated with the X6 VLP had a log2 serum dilution neutralization titer of 9.2 (50% inhibition) against CA/09, which was 2 to 3 logs lower than that for P1 or CA/09 VLP-vaccinated preimmune ferrets (Fig. 13E). Basically, all the elicited antisera had similar neutralization titers against Bris/07 (Fig. 13D), despite the wide range of observed HAI titers against the Bris/07 virus (Fig. 11).
FIG 13.
Focal reduction assay (FRA). Preimmune ferrets (n = 4) were vaccinated at days 0 and 84 with rHA vaccines. (A to E) At day 125, sera were collected and tested in an FRA against 5 H1N1 influenza viruses isolated between 1983 and 2009. Collected sera was assayed against Chile/83 (A), Sing/86 (B), NC/99 (C), Bris/07 (D), and CA/09 (E). For each virus, the virus concentration was standardized to 1.2 × 104 FFU/ml (corresponding to 600 FFU/50 μl which is the volume of virus added to each plate). A monolayer of MDCK SIAT cells (2.5 × 105 to 3 × 105 cells/ml) (100 μl/well in a 96-well plate) 1 day before assay is run. Cells were 95 to 100% confluent at the time of the assay. To determine the number of foci detected as percent infected cells normalized to 100% of the total. The dotted lines represent the 50% inhibition and the 80% inhibition by sera compared to virus-only control wells. (F to J) A heat map of the log2 serum dilution titer for the 50 and 80% inhibition per virus for each group of ferrets was also generated. Colors range from white (lowest level of inhibition) to dark blue (highest level of inhibition).
DISCUSSION
In this study, we explored how the residues and glycosylation sites around the receptor binding site impact the vaccine-induced immunogenicity by broadly reactive HA immunogens. The influenza virus A/Brisbane/59/2007 (Bris/07) was the last seasonal H1N1 strain recommended by the World Health Organization prior to the 2009 pandemic. Antibodies elicited by Bris/07 have HAI activity against Bris/07-like and a few other seasonal influenza viruses and no binding or HAI activity against the pandemic-like H1N1 strains, such as A/California/07/2009 (CA/09) (11). Similarly, CA/09-elicited antibodies have no HAI activity against Bris/07-like or other H1N1 seasonal strains (11). Our research group developed the next-generation COBRA algorithms as advanced H1 HA immunogens to address the elicitation of antibodies with HAI activity against strains across this seasonal-pandemic 2009 divide. The COBRA X6 HA elicits antibodies with HAI activity, which expands the number of seasonal H1N1 strains recognized compared to those recognized by antibodies elicited following Bris/07 virus vaccination or infection; however, these COBRA X6 HA-induced antibodies do not bind well or have HAI activity against Bris/07. The P1 COBRA HA elicits antibodies with HAI activity against pandemic-like H1N1 viruses, as well as some seasonal H1N1 strains, but not Bris/07. Therefore, we examined the amino acids in the Sa, Sb, and Cb regions of the CA/09, Bris/07, COBRA P1, and COBRA X6 HA antigens to identify residues that differed between these four HA antigens as part of possible epitopes associated with antibody binding, neutralization, and protection against H1N1 influenza viruses.
An initial review identified 53 amino acids that differed at 17 locations in these regions, with 2 being associated with putative glycosylation sites (Fig. 14). Since the introduction of H1N1 influenza viruses into the human population in 1918, the HAs of H1N1 influenza viruses have acquired multiple N-linked glycosylation sites. The sequon N-Xaa-S/T (where Xaa is any amino acid except Pro) is associated with glycosylation of accessible Asn (N) residues (3, 4). Glycosylation of the H1 HA globular head is associated with five dominant sites in various combinations (Asn at amino acids 142, 144, 172, 177, and 179 of HA) (13). From 1942 to 1985, H1N1 influenza viruses maintained several glycosylation combinations on HA at residues 144, 172, and 177/179. In 1986, the glycan shifted from residue 144 to residue 142, with the primary glycosylated residues on HA located at residues 142 and 177. Introduction of the pandemic-like H1N1 viruses in 2009 brought back naked, or glycan-free, globular head HA antigens (3, 4, 14). These glycans on HA not only shield critical neutralizing epitopes but also play a role in viral replication, virulence, and transmissibility (6–10, 15, 16).
FIG 14.
Schematic of the COBRA P1 H1N1 HA trimerized proteins. Each COBRA HA structure presented was generated using the 3D-JIGSAW algorithm, and renderings were performed using MacPyMol. The Sa site is shown in cyan, the Sb site in pink, and the Cb site in dark blue. The glycans at residues 142 and 144 are highlighted in green, and the K147 residue is highlighted in purple.
The COBRA X6 HA has Sa and Cb regions identical to those of Bris/07 and therefore has the same glycan pattern, with the putative glycan site at residue 142. The COBRA P1 HA has a glycosylation sequon in the Sa antigenic site at residue 144, but this differs from the case for the CA/09 HA, which has no putative glycosylation sites (Table 1). To determine how the Cb and Sa antigenic sites contributed to the immunogenicity of these COBRA HA antigens, a series of mutations were introduced to identify the specific amino acids or glycosylation sites that are critical for broadly reactive antibody responses to H1N1 viruses. Exchanging the Cb site in the COBRA P1 HA (LLSARS) with the Cb site located in the COBRA X6 HA (LISKES) to generate the V1 HA did not alter the elicited antibody profile of HAI activity against the panel of H1N1 influenza viruses, which was statistically similar to P1 HAI antibody profile (Fig. 3E). Addition of the Sa site from the COBRA X6 HA alone (V2 HA) or in combination with the Cb site (V3 HA) lowered the HAI titers against pandemic-like strains but raised the titers to seasonal-like strains, including Bris/07. Therefore, it appeared that the Cb antigenic site was not substantially involved the COBRA P1 or COBRA X6 HA-induced HAI activity; however, the Sa region appeared to be responsible for inducing these phenotypes.
Introduction of the COBRA X6 Sa region not only deleted a glycosylation site in the COBRA P1 HA but also deleted the Lys (K) at residue 147. The 1918-like H1N1 viruses had a Lys located at position 147. However, later viruses had an arginine (R) or isoleucine (I) at this position, or alternatively, the amino acid was deleted altogether. The amino acid residue at 147 was deleted from most seasonal-like H1N1 viruses isolated from 1995 to 2008. However, with the introduction of the pandemic-like swine H1N1 influenza viruses in 2009, the Lys at position 147 in HA returned to circulating human H1N1 influenza virus strains. The presence of this K147 residue in the CA/09 HA actually stabilizes the interaction between HA and sialic acid (13, 17, 18).
To determine if the K147 residue played a role in the immune responses elicited by the COBRA HA antigens, a second series of mutations in the COBRA P1 HA antigen was introduced. We examined how the HA residue at position 147 and the N-linked glycosylations cooperate to elicit broadly reactive antibodies with HAI activity against H1N1 influenza viruses. Adding the K147 residue to the V2 HA generated an HA antigen with a Cb region from COBRA P1 and the Sa region from the COBRA X6 HA with the addition of K147 (named V4 HA). Returning the Lys residue at position 147 with the N-linked glycan at residue 144 in HA elicited antibodies with HAI activities with similar profiles as for those antibodies elicited by P1 or V1 HA antigens (Fig. 4A). These antibodies had HAI activity against both seasonal and pandemic H1N1 influenza viruses, except for SI/06 and Bris/07. Therefore, in all four cases where the Lys was present at position 147 (CA/09, P1, V1, and V4 HA), the HA antigen elicited a COBRA P1 HAI antibody profile, regardless of whether the glycan was located at residue 142 or 144.
In HA antigens that lacked the K147 residue, the locations of the glycans were more important for the elicitation of antibodies with a breadth of HAI activity against the panel of H1N1 influenza viruses. The V5 HA lacks the K147 residue and the glycan located at residue 144. The V5 HA elicited antibodies with an HAI profile identical to those antibodies elicited by CA/09 HA (Fig. 4B). The V5 HA has a glycan at residue 144 but no K147 residue. In CA/09, the addition of the K147 residue compensates for the lack of glycans. However, the addition of one glycan at position 144, plus the K147 (P1 COBRA HA), expands the CA/09 HAI titers to include recognition of seasonal-like H1N1 viruses. Lastly, HA antigens with no glycan at residue 142 or 144 and no K147 (V6 HA) elicit antibodies with HAI activity with a profile similar to that for X6. Both the V6 HA and the CA/09 HA lack glycans at residues 142 and 144 and differ only at the K147 site, again emphasizing that the K147 found in CA/09 is a signature for the elicitation of the pandemic-like antibody phenotype. The V6 HA differs from Bris/07 HA only by the glycosylation site at position 142, but the HAI antibody profile is quite different. The glycan at position 142 without the K147 residue appears to be a signature for the elicitation of antibodies with HAI activity specific for recent seasonal H1N1 influenza viruses, such as SI/06 or Bris/07.
Overall, there were general signatures associated with each of these sequence combinations. The Cb mutations did not appear to have a significant impact on the elicitation of antibodies with specific HAI activity; therefore, we will focus on the specific amino acids in the Sa region. We were able to classify the HA antigens into five phenotypes: CA/09-like, P1-like, V2/V3-like, X6-like, and Bris/07-like. HA antigens elicited antibodies with HAI activity against pandemic-like viruses (i.e., CA/09) if the HA had either the 144 glycan or the K147 residue present. The CA/09 HA has no glycan at position 142 or 144 but does contain the K147 residue, and the V5 HA contains the 144 glycan but no K147 residue. The P1, V1, and V4 HA antigens, which have the K147 residue, have an expanded HAI antibody profile beyond the pandemic-like viruses to include seasonal H1N1 influenza viruses from 1986 to 2005. This HAI activity appeared to be independent of the presence of a glycan at residue 144, since the V4 HA antigen has a Thr at this position. All five of these HA antigens (CA/09, P1, V1, V4, and V5) protected mice against CA/09 infection, with little weight loss or signs of morbidity observed, but did not protect mice against Bris/07 challenge.
HA antigens, such as V2, V3, or V6, which do not have either the 144 glycan or the K147 residue elicits antibodies with HAI activity with a broad seasonal-like phenotype that recognizes H1N1 viruses from 1986 to 2005, with low HAI titers against viruses from seasonal viruses from 2006 to 2008 and pandemic viruses from 2009 to 2015. Mice vaccinated with these HA antigens were protected against CA/09 challenge, but they did lose some weight. When these vaccinated mice were challenged with the Bris/07 virus, they had mild morbidity, with ∼50% of the mice surviving challenge.
Even though we were able to document that the Sa region, and specifically the glycans at positions 142 and 144 and K147 residue, were as important in order to elicit broadly protective antibodies, we acknowledge that other regions or antigenic sites play a role in the HA structure and contribute to the elicitation of broadly reactive antibodies. For example, the targeted Sa and Cb sequences in the Bris/07, X6, and V3 HA antigens are identical, but the X6 and V3 HA antigens can provide protection against CA/09 challenge, whereas the Bris/07 HA vaccine cannot protect mice against CA/09 challenge (Fig. 5). Therefore, other regions of HA that were not included in this analysis may contribute to protection. A second region of Sa is located at amino acids 170 to 181. Seven of the 12 amino acids differ between COBRA P1 and COBRA X6 HA and may play a role in the overall HA structure, along with other regions of HA that contribute to the immunogenicity of these immunogens.
The HA immunogen structure is only one component for the effectiveness of the elicited antibodies. The antigenic HA target plays a significant role in the overall protective capacity of the elicited antibodies. These COBRA or VIPER HA-based vaccines did not effectively elicit antibodies against the A/Chile/1/1983 (Chile/83) strain in our H1N1 panel. The Chile/83 virus has a glycosylation site at residue 144 on its HA, along with the K147 residue (3). H1N1 swine isolates with an N-linked glycosylation site at residue 144 render these influenza viruses resistant to antibody-meditated neutralization, whereas viruses with an N142 residue instead of an N144 residue are sensitive to neutralization (19). Therefore, even if the glycan is missing on the HA immunogens and the putative neutralizing epitope is exposed to elicit high-titer antibodies to this specific epitope, if the epitope is shielded on the target HA antigen, the effectiveness of the vaccine to protect against influenza infection is decreased. This is a critical understanding in order to develop universal vaccines based upon the globular HA head. The development of HA immunogens that can elicit high-titer antibodies to multiple HA epitopes on antigenically distinct viruses will advance the development of new influenza vaccine designs. A mixture of more than one broadly reactive HA immunogen may be needed to elicit protective antibodies against all influenza virus strains within a subtype. Overall, the elicitation of broadly reactive HA antigens can be improved by understanding how the residues around the receptor binding site are structured and exposed to the immune system.
MATERIALS AND METHODS
Cells and viruses.
The human embryonic kidney (HEK) 293T cells and Madin-Darby canine kidney (MDCK) cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin solution (10,000 U/ml). All influenza A H1N1 viruses used in this study were grown in eggs and purchased from Virapur (NY): A/Chile/1/1983, A/Singapore/6/1986, A/Texas/36/1991, A/Beijing/262/1995, A/New Caledonia/20/1999, A/Solomon Islands/3/2006, A/Brisbane/59/2007, A/California/07/2009, and A/Michigan/45/2015.
VIPER.
Using site-directed mutagenesis, specific amino acids in the P1 COBRA HA antigen (11, 20) were mutated to corresponding sites in wild-type H1 influenza virus HA proteins according to the manufacturer’s protocol (Genewiz, South Plainfield, NJ, USA). For VIPER 1, the amino acids at residues 88 to 91 were changed from LSAR to ISKE in the Cb region of the COBRA P1 HA antigen (Table 1). The ISKE amino acids are located at this position in the NC/99, Bris/07, and COBRA X6 HA molecules. This will transform this antigenic site from pandemic-like to seasonal-like. For VIPER 2, the amino acids at residues 144 to 147 were changed from NTTK to TVT- in the Sa region of the COBRA P1 HA antigen (Table 1). The ISKE amino acids are located at this position in the NC/99, Bris/07, and COBRA X6 HA molecules. There are no corresponding amino acids in these seasonal HA proteins at position 147 (Table 1). For VIPER 3, both the Cb and Sa site mutants were introduced into the P1 COBRA HA backbone sequence simultaneously.
For VIPER 4, the sequence is identical to that for VIPER 2 except that a Lys is included at position 147. For VIPER 5, the sequence is identical to that for VIPER 1 except there is no amino acid located at position 147. For VIPER 6, the sequence is the same as that for VIPER 5 except the Asn at position 142 has been mutated to a Thr. For VIPER 7, the amino acids at residues 144 to 147 were changed from NTTK to NTNK in the Sa region of the COBRA P1 HA antigen (Table 1). In addition, the amino acids at positions 179 and 180 were changed from SK to NQ to match the amino acids located in A/Michigan/45/2015. For VIPER 8, the amino acid at residue 146 was changed from T to N in the Sa region of the COBRA P1 HA antigen (Table 1). Each HA gene was fully sequenced, and the amino acid integrity was verified.
VIPER 9 to 12 HA vaccines were designed based on the COBRA X6 HA sequence (11, 20). For VIPER 9, the Cb antigenic site from COBRA X6 was exchanged for the Cb site from COBRA P1 (residues 87 to 92). For VIPER 10, the Sb antigenic site from COBRA X6 was exchanged for the Sa site from COBRA P1 (residues 202 to 213), and for VIPER 11, both the Cb and Sb antigenic sites from COBRA X6 were exchanged for the same sites in COBRA P1. Lastly, for VIPER 12, the putative N-linked glycosylation in the Sa antigenic site located at residue 177 was mutated from the amino acid sequence NLS to KLS (Table 1). The HA genes were codon optimized for expression in mammalian cells and then biochemically synthesized by Genewiz (South Plainfield, NJ, USA).
Vaccine preparation.
DNA (3 μg) expressing each COBRA or wild-type HA gene cassette plus the influenza virus neuraminidase (A/mallard/Alberta/24/01, H7N3), the HIV p55 Gag sequences, and one of the various H1N1 wild-type-, COBRA HA-, or VIPER HA-expressing plasmids was used to transiently transfect HEK 293T cells (1 × 106) as previously described (21). Following 72 h of incubation at 37C, supernatants from transiently transfected cells were collected, centrifuged to remove cellular debris, and filtered through a 0.22-μm-pore membrane. Virus-like particles (VLPs) were purified by ultracentrifugation on a 20% glycerol cushion at 27,000 × g for 4 h at 4°C. VLPs were resuspended in phosphate-buffered saline (PBS). The total protein concentration was assessed by conventional bicinchoninic acid assay (BCA). The HA concentration was quantified by Western blotting.
For Western blotting, cell lysates were then electrophoresed on a 10% sodium dodecyl sulfate-polyacrylamide gel and transferred to a polyvinylidene difluoride (PVDF) membrane. The blot was probed with pooled mouse antisera elicited by A/Brisbane/57/2007 and A/California/07/2009 viral infections. HA-antibody complexes were then detected using goat anti-mouse IgG labeled with horseradish peroxidase (HRP) (Southern Biotech, Birmingham, AL, USA). HRP activity was detected using a chemiluminescent substrate (Pierce Biotechnology, Rockford, IL, USA) and exposed to X-ray film (Thermo Fisher, Pittsburgh, PA, USA).
To determine hemagglutination activity, each VLP preparation added in equal volumes with turkey red blood cells (RBCs) to a V-bottom 96-well plate and incubated with serially diluted volumes of VLPs at room temperature for 30 min. The highest dilution of VLPs with full agglutination of RBCs was considered the endpoint HA titer.
Quantification of HA in purified VLPs.
Purified VLPs were electrophoresed on a 10% sodium dodecyl sulfate-polyacrylamide gel and transferred to a PVDF membrane. Recombinant HA (HA1 H1N1 A/California/07/2009) at different concentrations was used as a standard. The blot was probed with monoclonal antibody 15B7 (Immune Technology Corporation, New York, NY, USA). Goat anti-mouse IgG labeled with HRP (Southern Biotech, Birmingham, AL, USA) was used to probe for HA-antibody complexes. HRP activity was detected using the Clarity Western ECL substrate (Bio-Rad, Hercules, CA, USA), and the digital images were captured with the Chemi-Doc imaging system (Bio-Rad, Hercules, CA, USA). Linear regression standard curve analysis was done using the known concentrations of recombinant standard HA. The HA content in each VLP was calculated by interpolation from the data of the standard curve. Experiments were performed in duplicate, and multiple exposure times were analyzed for all iterations.
Determination of HA content.
A high-affinity, 96-well flat bottom enzyme-linked immunosorbent assay (ELISA) plate was coated with 5 to 10 μg of total protein of VLPs and serial dilutions of a recombinant H3 antigen (3006_H3_Vc; Protein Sciences, Meriden, CT) in ELISA carbonate buffer (50 mM carbonate buffer, pH 9.5), and the plate was incubated overnight at 4°C on a rocker. The next morning, plates were washed in PBS with 0.05% Tween 20 (PBST), and then nonspecific epitopes were blocked with 1% bovine serum albumin (BSA) in PBST solution for 1 h at room temperature (RT). Buffer was removed, and then stalk-specific group 1 antibody (22) was added to the plate and incubated for 1 h at 37°C. Plates were washed and then probed with goat anti-human IgG horseradish peroxidase-conjugated secondary antibody (2040-05; Southern Biotech, Birmingham, AL) for 1 h at 37°C. Plates were washed, and then freshly prepared o-phenylenediamine dihydrochloride (OPD) (Sigma-Aldrich, St. Louis, MO, USA) substrate in citrate buffer (Sigma-Aldrich, St. Louis, MO, USA) was added to the wells, followed by 1 N H2SO4 stopping reagent. Plates were read at 492-nm absorbance using a microplate reader (Powerwave XS; BioTek, Winooski, VT), and background was subtracted from negative wells. Linear regression standard curve analysis was performed using the known concentrations of recombinant standard antigen to estimate the HA content in VLP lots.
Mouse vaccination and infection.
BALB/c mice (Mus musculus, females, 6 to 8 weeks old) were purchased from Envigo (Indianapolis, IN, USA), housed in microisolator units, and allowed free access to food and water; they were cared for under USDA guidelines for laboratory animals. All procedures were reviewed and approved by the University of Georgia Institutional Animal Care and Use Committee (IACUC) (no. 2016-02-011-Y3-A7). Mice were randomly divided (n = 11/group), vaccinated intramuscularly with either 1 μg of VIPER 1 to 12, COBRA P1, X6, Brisbane/59/2007, or California/07/2009 VLPs or PBS formulated with AF03 (oil-in-water emulsion) for a final volume of 50 μl, and then boosted with the same amount of VLPs or PBS at weeks 4 and 8 postvaccination. The final concentration after mixing 1:1 with VLPs was 2.5% squalene. Blood collected at weeks 6, 10, and 12 postvaccination was processed, and the sera were harvested and frozen at −20°C.
At week 13 postvaccination, all mice were challenged with 1 × 106 PFU (10× LD50) of the wild-type A/California/07/2009 (H1N1) or with 8.75 × 106 PFU A/Brisbane/59/2007xPR8 (6:2) in a volume of 50 μl. Mice were monitored daily for 14 days for weight loss, disease signs, and deaths. Three mice from each group were euthanized on days 2 and 3 postinfection, respectively, and lungs were harvested and snap-frozen on dry ice and then stored at −80°C for viral titration in the future. Mice were humanly euthanized when they reached the humane endpoint by losing 20% of their original body weight or accumulated a disease score of up to 3 (lethargy = 1, hunched posture = 1, rough fur = 1, weight loss of 15% to 20% of original body weight = 1, weight loss of >20% of original body weight = 3). The experimental endpoint was defined as >20% weight loss. All procedures were performed in accordance with Guide for the Care and Use of Laboratory Animals (23), the Animal Welfare Act (24), and Biosafety in Microbiological and Biomedical Laboratories (25).
HAI assay.
The hemagglutination inhibition (HAI) assay was used to assess functional antibodies to the HA that are able to inhibit agglutination of turkey erythrocytes. The protocols were adapted from the WHO laboratory influenza surveillance manual (26). A panel of nine H1N1 viruses was used for the HAI assay, including A/Chile/1/1983, A/Singapore/6/1986, A/Texas/36/1991, A/Beijing/262/1995, A/New Caledonia/20/1999, A/Brisbane/59/2007, A/Solomon Islands/3/2006, A/California/07/2009, and A/Michigan/45/2015. The HAI assay was performed as previously described (11). Briefly, to inactivate nonspecific inhibitors, sera were treated with receptor-destroying enzyme (RDE) (Denka Seiken, Co., Japan) prior to being tested. Three parts of RDE were added to one part of serum and incubated overnight at 37°C. RDE was inactivated by incubation at 56°C for 30 min. RDE-treated sera were diluted in a series of 2-fold serial dilutions in V-bottom microtiter plates. An equal volume of each H1N1 virus, adjusted to approximately 8 hemagglutination units (HAU)/50 μl, was added to each well. The plates were covered and incubated at room temperature for 30 min, and then 0.8% of turkey erythrocytes (Lampire Biologicals, Pipersville, PA, USA) in PBS was added. Red blood cells (RBCs) were stored at 4°C and used within 72 h of preparation. The plates were mixed by agitation and covered. RBCs were incubated at room temperature for 30 min. The HAI titer was determined as the reciprocal dilution of the last well that contained nonagglutinated RBCs. Positive and negative serum controls were included for each plate. All mice with an HAI of ≤1:10 for preexisting antibodies to currently circulating human influenza viruses prior to vaccination were considered negative. Seroprotection was defined as an HAI titer of >1:40 and seroconversion as a 4-fold increase in titer compared to baseline, as per the WHO and European Committee for Medicinal Products guidelines to evaluate influenza vaccines (27); however, we often examined a more stringent threshold of >1:80. Mice were naive and seronegative at the time of vaccination, and thus seroconversion and seroprotection rates are interchangeable in this study.
FRA.
The focus reduction assay (FRA) used in this study was initially developed by the World Health Organization collaborating center in London, UK (28, 29) and modified by U.S. Centers for Disease Control and Prevention (CDC) (Thomas Rowe, personal communication). One day prior to the assay, MDCK-SIAT1 cells were cells plated at 2.5 to 3 × 105 cells/ml (100 μl/well in 96-well plate). Cells need to be 95 to 100% confluent at the time of the assay in 96-well plates overnight to form a confluent monolayer in Dulbecco’s modified Eagle’s medium (DMEM) containing 5% heat-inactivated fetal bovine serum and antibiotics. The following day, the cell monolayers were rinsed with 0.01 M PBS, pH 7.2 (Gibco), followed by the addition of 2-fold serially diluted RDE-treated serum at 50 μl per well, starting with a 1:20 dilution, in virus growth medium containing 1 μg/ml tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin and VGM-T (DMEM containing 0.1% BSA, penicillin-streptomycin, and 1 μg/ml TPCK-treated trypsin [Sigma, St. Louis, MO, USA]). Afterwards, 50 μl virus for all FRAs shown here, standardized to 1.2 × 104 focus-forming units (FFU)/ml (corresponds to 600 FFU/50 μl), in VGM-T was added to each plate, or VGM-T was added to cell control wells. The virus stocks were standardized by previous titration in the FRA. Following 2 h of incubation at 37°C with 5% CO2, the cells in each well were overlaid with 100 μl of equal volumes of 1.2% Avicel RC/CL (28) (type RC581 NF; FMC Health and Nutrition, Philadelphia, PA, USA) in 2× modified Eagle’s medium (MEM) containing 1 μg/ml TPCK-treated trypsin, 0.1% BSA, and antibiotics. Plates were incubated for 18 to 22 h at 37°C and 5% CO2. The overlays were then removed from each well, and the monolayer was washed once with PBS to remove any residual Avicel. The plates were fixed with ice-cold 4% formalin in PBS for 30 min at 4°C, followed by a PBS wash and permeabilization using 0.5% Triton X-100 in PBS-glycine at room temperature for 20 min. Plates were washed three times with wash buffer (PBS, 0.1% Tween 20) and incubated for 1 h with a monoclonal antibody against influenza A virus nucleoprotein (30) (International Reagent Resource [IRR]) in ELISA buffer (PBS, 10% horse serum, 0.1% Tween 80). After washing three time with wash buffer, the cells were incubated with goat anti-mouse peroxidase-labeled IgG (SeraCare, Inc., Milford, MA, USA) in ELISA buffer for 1 h at RT. Plates were washed three times with wash buffer, and infectious foci (spots) were visualized using TrueBlue substrate (SeraCare, Inc., Milford, MA, USA) containing 0.03% H2O2 incubated at room temperature for 10 to 15 min. The reaction was stopped by washing five times with distilled water. Plates were dried and foci enumerated using a CTL BioSpot Analyser with ImmunoCapture 6.4.87 software (CTL, Shaker Heights, OH). The FRA titer was reported as the reciprocal of the highest dilution of serum corresponding to 50% focus reduction compared to the virus control (VC) minus the cell control (CC).
In order for a plate to pass quality control, both the average of the octuplet VC wells and the average of the octuplet CC wells must pass. The virus controls initially were between 150 to 650 foci (spots), and the cell controls must be fewer than 21 foci. The virus control wells were subsequently expanded to between 200 and 1,600 spots. Additionally, the reference (vaccine strain) virus was run in triplicate plates in each individual assay; at least two out of three plates must pass VC and CC criteria, and homologous ferret antisera must have the same titer. Each assay plate (one virus per plate) contained a panel of reference antisera as well as a human vaccine serum control to assess overall assay consistency.
Viral lung titers.
Lungs were homogenized in DMEM (1 ml), and the supernatant was collected by spinning the homogenized samples at 2,000 rpm for 5 min. Low-passage MDCK cells were plated at a confluence of 1 × 106 in each well of a six-well plate (Greiner Bio-One, Monroe, NC, USA). Following 24 h of incubation, MDCK cells were infected with different dilutions of samples in 100 μl of DMEM supplemented with penicillin-streptomycin. After 1 h of incubation at room temperature, the medium was removed, cells were washed twice with fresh DMEM, and 2 ml of MEM plus 0.8% agarose (Cambrex, East Rutherford, NJ, USA) was added. Cells were incubated for 72 h at 37°C with 5% CO2. Agarose was removed, and the cells were then fixed with 10% buffered formalin and stained with 1% crystal violet (Fisher Science Education, Waltham, MA, USA) for 15 min. The crystal violet was removed by rinsing thoroughly in distilled water. The numbers of plaques were counted, and the virus titer, presented as PFU/lung, in lung tissue was calculated.
Determination of viral nasal wash titers.
MDCK cells were seeded at 5 × 105 in each well of a six-well plate. Samples were diluted (final dilution factors of 100 to 10−6), overlaid onto the cells in 100 μl of DMEM supplemented with penicillin-streptomycin, and incubated for 1 h with intermittent shaking every 15 min. Samples were removed, cells were washed twice, the medium was replaced with 2 ml of L15 medium plus 0.8% agarose (Cambrex, East Rutherford, NJ, USA), and cells were incubated for 72 h at 37°C with 5% CO2. Agarose was removed and discarded. The cells were fixed with 10% buffered formalin and then stained with 1% crystal violet for 15 min. The plates were then thoroughly washed in distilled water to remove excess crystal violet before being air dried, and then the numbers of plaques were counted and the numbers of PFU per milliliter calculated.
Statistical analysis.
All data are reported as absolute mean values ± standard errors of the means (SEM). Weight losses among the different vaccinated groups were compared using nonparametric two-way analysis of variance (ANOVA), and viral lung titers and HAI titers were compared using nonparametric one-way ANOVA. All statistical analysis was performed using GraphPad Prism 7 software (GraphPad, San Diego, CA, USA), and a P value of <0.05 was considered statistically significant (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
ACKNOWLEDGMENTS
We thank Amanda Skarlupka for helpful discussions and reviews. We also thank James D. Allen and Dawn Taylor-Mulniex for technical assistance. The H1N1 influenza A viruses were obtained through the Influenza Reagent Resource, Influenza Division, WHO Collaborating Center for Surveillance, Epidemiology and Control of Influenza, Centers for Disease Control and Prevention, Atlanta, GA, USA.
This work, including the efforts of T.M.R., was funded by University of Georgia (UGA) (MRA-001) and the Vaccine and Gene Therapy Institute of Florida (VGTIFL-001). In addition, T.M.R. is supported by the Georgia Research Alliance as an Eminent Scholar.
REFERENCES
- 1.WHO. 31 January 2018. Influenza (seasonal). http://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal). Accessed 7 September 2018.
- 2.Palese P. 2004. Influenza: old, and new threats. Nat Med 10:S82–S87. doi: 10.1038/nm1141. [DOI] [PubMed] [Google Scholar]
- 3.Kim JI, Park MS. 2012. N-linked glycosylation in the hemagglutinin of influenza A viruses. Yonsei Med J 53:886–893. doi: 10.3349/ymj.2012.53.5.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei CJ, Boyington JC, Dai K, Houser KV, Pearce MB, Kong WP, Yang ZY, Tumpey TM, Nabel GJ. 2010. Cross-neutralization of 1918 and 2009 influenza viruses: role of glycans in viral evolution and vaccine design. Sci Transl Med 2:24ra21. doi: 10.1126/scitranslmed.3000799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hebert DN, Zhang JX, Chen W, Foellmer B, Helenius A. 1997. The number and location of glycans on influenza hemagglutinin determine folding and association with calnexin and calreticulin. J Cell Biol 139:613–623. doi: 10.1083/jcb.139.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, Zhu J, Li Y, Bradley KC, Cao J, Chen H, Jin M, Zhou H. 2013. Glycosylation on hemagglutinin affects the virulence and pathogenicity of pandemic H1N1/2009 influenza A virus in mice. PLoS One 8:e61397. doi: 10.1371/journal.pone.0061397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zost SJ, Parkhouse K, Gumina ME, Kim K, Diaz Perez S, Wilson PC, Treanor JJ, Sant AJ, Cobey S, Hensley SE. 2017. Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. Proc Natl Acad Sci U S A 114:12578–12583. doi: 10.1073/pnas.1712377114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pentiah K, Lees WD, Moss DS, Shepherd AJ. 2015. N-linked glycans on influenza A H3N2 hemagglutinin constrain binding of host antibodies, but shielding is limited. Glycobiology 25:124–132. doi: 10.1093/glycob/cwu097. [DOI] [PubMed] [Google Scholar]
- 9.Tate MD, Job ER, Deng YM, Gunalan V, Maurer-Stroh S, Reading PC. 2014. Playing hide and seek: how glycosylation of the influenza virus hemagglutinin can modulate the immune response to infection. Viruses 6:1294–1316. doi: 10.3390/v6031294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laursen NS, Wilson IA. 2013. Broadly neutralizing antibodies against influenza viruses. Antiviral Res 98:476–483. doi: 10.1016/j.antiviral.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carter DM, Darby CA, Lefoley BC, Crevar CJ, Alefantis T, Oomen R, Anderson SF, Strugnell T, Cortes-Garcia G, Vogel TU, Parrington M, Kleanthous H, Ross TM. 2016. Design and characterization of a computationally optimized broadly reactive hemagglutinin vaccine for H1N1 influenza viruses. J Virol 90:4720–4734. doi: 10.1128/JVI.03152-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carter DM, Darby CA, Johnson SK, Carlock MA, Kirchenbaum GA, Allen JD, Vogel TU, Delagrave S, DiNapoli J, Kleanthous H, Ross TM. 2017. Elicitation of protective antibodies against a broad panel of H1N1 viruses in ferrets preimmune to historical H1N1 influenza viruses. J Virol 91:e01283-17. doi: 10.1128/JVI.01283-17. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Das SR, Puigbo P, Hensley SE, Hurt DE, Bennink JR, Yewdell JW. 2010. Glycosylation focuses sequence variation in the influenza A virus H1 hemagglutinin globular domain. PLoS Pathog 6:e1001211. doi: 10.1371/journal.ppat.1001211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong M, Lee PS, Hoffman RM, Zhu X, Krause JC, Laursen NS, Yoon SI, Song L, Tussey L, Crowe JE Jr, Ward AB, Wilson IA. 2013. Antibody recognition of the pandemic H1N1 influenza virus hemagglutinin receptor binding site. J Virol 87:12471–12480. doi: 10.1128/JVI.01388-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun X, Jayaraman A, Maniprasad P, Raman R, Houser KV, Pappas C, Zeng H, Sasisekharan R, Katz JM, Tumpey TM. 2013. N-linked glycosylation of the hemagglutinin protein influences virulence and antigenicity of the 1918 pandemic and seasonal H1N1 influenza A viruses. J Virol 87:8756–8766. doi: 10.1128/JVI.00593-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Medina RA, Stertz S, Manicassamy B, Zimmermann P, Sun X, Albrecht RA, Uusi-Kerttula H, Zagordi O, Belshe RB, Frey SE, Tumpey TM, Garcia-Sastre A. 2013. Glycosylations in the globular head of the hemagglutinin protein modulate the virulence and antigenic properties of the H1N1 influenza viruses. Sci Transl Med 5:187ra170. doi: 10.1126/scitranslmed.3005996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu R, McBride R, Nycholat CM, Paulson JC, Wilson IA. 2012. Structural characterization of the hemagglutinin receptor specificity from the 2009 H1N1 influenza pandemic. J Virol 86:982–990. doi: 10.1128/JVI.06322-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuzaki Y, Sugawara K, Nakauchi M, Takahashi Y, Onodera T, Tsunetsugu-Yokota Y, Matsumura T, Ato M, Kobayashi K, Shimotai Y, Mizuta K, Hongo S, Tashiro M, Nobusawa E. 2014. Epitope mapping of the hemagglutinin molecule of A/(H1N1)pdm09 influenza virus by using monoclonal antibody escape mutants. J Virol 88:12364–12373. doi: 10.1128/JVI.01381-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hause BM, Stine DL, Sheng Z, Wang Z, Chakravarty S, Simonson RR, Li F. 2012. Migration of the swine influenza virus delta-cluster hemagglutinin N-linked glycosylation site from N142 to N144 results in loss of antibody cross-reactivity. Clin Vaccine Immunol 19:1457–1464. doi: 10.1128/CVI.00096-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ross TM, Crevar CJ, Carter DM. January 2017. US patent 9,555,095.
- 21.Green TD, Montefiori DC, Ross TM. 2003. Enhancement of antibodies to the human immunodeficiency virus type 1 envelope by using the molecular adjuvant C3d. J Virol 77:2046–2055. doi: 10.1128/jvi.77.3.2046-2055.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan GS, Lee PS, Hoffman RM, Mazel-Sanchez B, Krammer F, Leon PE, Ward AB, Wilson IA, Palese P. 2014. Characterization of a broadly neutralizing monoclonal antibody that targets the fusion domain of group 2 influenza A virus hemagglutinin. J Virol 88:13580–13592. doi: 10.1128/JVI.02289-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 24.U.S. Congress. 1966. Animal Welfare Act of 1966 intended to regulate the transport, sale and handling of dogs, cats, guinea pigs, nonhuman primates, hamsters and rabbits intended to use for research or other purposes. Public law 89-544 United States code, title 7, sections 2131–2156. U.S. Congress, Washington, DC. [Google Scholar]
- 25.U.S. Department of Health and Human Services. 2009. Biosafety in microbiological and biomedical laboratories, 5th ed HHS publication no. (CDC) 21-1112 U.S. Department of Health and Human Services, Washington, DC. [Google Scholar]
- 26.WHO Global Influenza Surveillance Network. 2011. Manual for the laboratory diagnosis and virological surveillance of influenza. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 27.European Medicines Agency. 2014. Guideline on influenza vaccines: non-clinical and clinical module (draft). European Medicines Agency, London, UK. [Google Scholar]
- 28.Matrosovich M, Matrosovich T, Garten W, Klenk HD. 2006. New low-viscosity overlay medium for viral plaque assays. Virol J 3:63. doi: 10.1186/1743-422X-3-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sullivan K, Kloess J, Qian C, Bell D, Hay A, Lin YP, Gu Y. 2012. High throughput virus plaque quantitation using a flatbed scanner. J Virol Methods 179:81–89. doi: 10.1016/j.jviromet.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 30.Walls HH, Harmon MW, Slagle JJ, Stocksdale C, Kendal AP. 1986. Characterization and evaluation of monoclonal antibodies developed for typing influenza A and influenza B viruses. J Clin Microbiol 23:240–245. [DOI] [PMC free article] [PubMed] [Google Scholar]