Abstract
Decades of research on the Aryl hydrocarbon Receptor (AhR) has unveiled its involvement in the toxicity of halogenated and polycyclic aromatic hydrocarbons, and a myriad of normal physiological processes. The molecular dissection of AhR biology has centered on a canonical signaling pathway in an effort to mechanistically reconcile the diverse pathophysiological effects of exposure to environmental pollutants. As a consequence, we now know that canonical signaling can explain many but not all of the AhR-mediated effects. Here we describe recent findings that point to non-canonical signaling pathways, and focus on a novel AhR interaction with the Krüppel-like Factor 6 protein responsible for previously un-recognized epigenetic changes in the chromatin affecting gene expression.
1. Introduction
Since its discovery in the 1980s, the aryl hydrocarbon receptor (AhR) has been a major focus in toxicology due to the fact that it mediates the effects of the halogenated aromatic hydrocarbon environmental pollutants [1]. AhR studies have mainly focused on understanding molecular basis of toxicity induced by the prototypical AhR ligand 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). As a result, the AhR has been implicated in regulatory processes affecting the immune system, liver homeostasis, cardiac development, wound healing, cell proliferation and apoptosis, tumor promotion and metabolic diseases [2–7]. Studies into the underlying molecular mechanisms have centered on a now well-defined canonical signaling pathway. This commentary seeks to compare and contrast canonical signaling with a recently identified non-canonical pathway that promises to enrich our understanding of AhR biology in response to toxic insults as well as normal physiological processes.
2. The aryl hydrocarbon receptor
The murine AhR was first purified in 1991 and subsequently cloned in 1992 [8,9]. This work revealed that the AhR is a member of the basic helix-loop-helix (bHLH) PAS family of transcription factors defined by the Period (PER), AhR nuclear translocator (Arnt), and single-minded (SIM) proteins [10]. AhR expression is essentially ubiquitous in mammals consistent with a broad-spectrum homeostatic role, however expression levels varying widely across tissues with the liver, thymus, lung, kidney, spleen, and placenta exhibiting greatest expression [11]. Additionally, AhR expression is developmentally regulated [12], and more recent evidence indicates a role for the AhR in developmental process affecting hematopoiesis, immune system biology, neural differentiation, and liver architecture [13–17]. Structurally, the N-terminal half of the mammalian AhR is well-conserved, suggestive of functional importance in fundamental physiological activities. In contrast, the receptor’s C-terminal region exhibits species differences reflected as polymorphisms and variations in protein length which largely account for the size differences observed between species [18]. The functional impact of the polymorphisms remains largely undefined. A basic sequence and juxtaposed helix-loop-helix region in the (bHLH) domain located near the N-terminus is responsible for DNA binding and Arnt protein dimerization, respectively [16]. The PAS domain, specifically the PAS-B region overlaps with the AhR’s ligand binding domain, and confers protein–protein interactions with Hsp90 and the Retinoblastoma tumor suppressor protein [19,20]. The C-terminal half of the AhR protein encompasses a glutamine-rich transactivation domain, and harbors the binding sites for several cofactors including p300, SMRT, SRC1, and RIP140 [21–24]. It is noteworthy that the C-terminal region is also responsible for the interaction with the Krüppel-like factor 6 (KLF6) protein [25,26], the focus of the discussion on non-canonical signaling below.
3. Canonical AhR signaling
Canonical AhR signaling has received attention for decades, and is described in detail in several excellent reviews [5,27,28]. In order to provide context in this commentary, a few of the salient findings are presented here. Canonical signaling begins in the cytoplasm where unliganded AhR is bound by a chaperone complex. This chaperone complex includes heat shock protein 90 (Hsp90) [29], immunophilin-like protein XAP2 (also known as ARA9 or AIP) [30–32], and p23 [33]. Transformation of the AhR by ligand binding is thought to involve a conformational change that reveals the nuclear localization sequence to mediate nuclear translocation of the receptor–chaperone complex via β-importins [34]. The conformational change in the PAS-A domain following ligand binding also facilitates subsequent dissociation of the chaperonins and heterodimerization with Arnt protein through the HLH domain inside the nucleus [35]. The AhR–Arnt heterodimer binds DNA at xenobiotic response elements (XRE) defined by the 5’-GCGTG-3’ core consensus motif [36] (Fig. 1, canonical signaling pathway). It should be noted however, that while the core sequence is essential for AhR–Arnt complex binding, evidence exists implicating a role for nucleotides flanking the core motif in DNA binding and function [37,38]. Studies revealed 1) that the AhR’s DNA binding affinity was influenced by the nucleotide sequence flanking the core motif, 2) that DNA binding affinity and transcriptional responsiveness were not strictly correlated, and 3) that flanking residues could affect AhR-DNA binding in an agonist-specific manner. However, the veracity of the last observation is challenged by more recent findings [39].
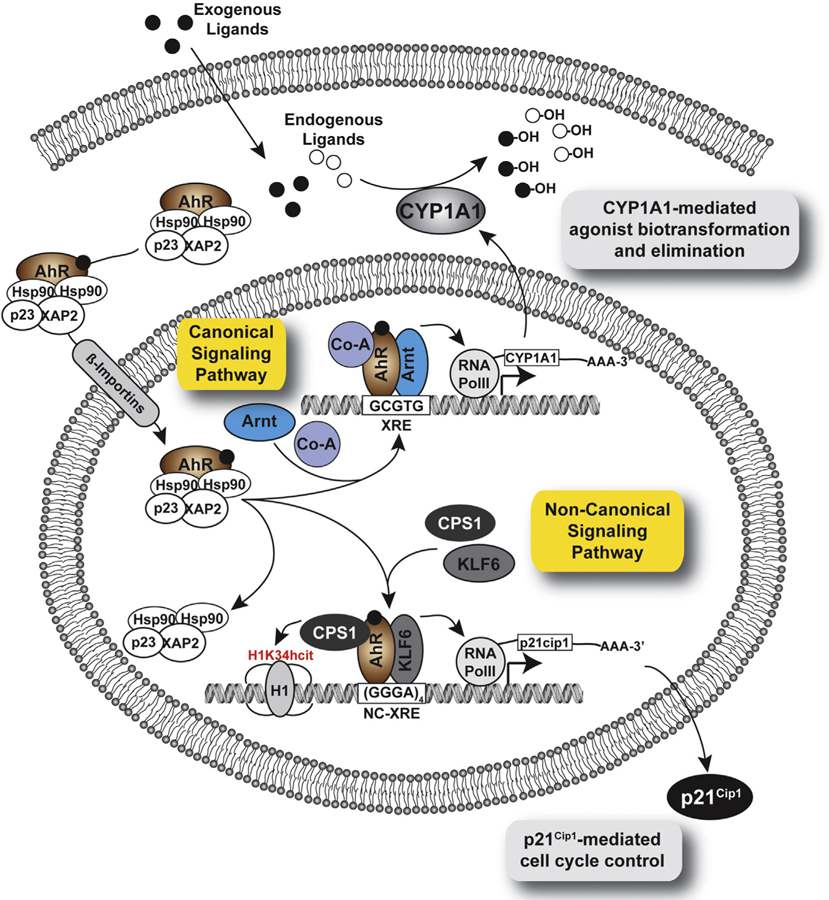
Fig. 1.
The diagram depicts the canonical and non-canonical signaling pathways. The unliganded cytosolic AhR in complex with the chaperones (Hsp90, XAP2, p23) is common to each pathway. Upon ligand binding—by either exogenous or endogenous agonists—the complex translocates into the nucleus, dissociates from the chaperones and forms a DNA-bound complex with the Arnt protein at XRE sites defined by the 5’ -GCGTG-3’ core recognition motif (canonical signaling), or the KLF6 protein at NC-XRE sites defined by the 5’-(GGGA)4-3’ tetranucleotide repeat motif (non-canonical signaling), respectively. The AhR–Arnt complex is known to recruit several co-activators (Co-A) in a context dependent manner, and induces CYP1A1 expression resulting in the biotransformation and depletion of AhR agonists. The AhR-KLF6 complex has been shown to recruit CPS1 as a cofactor responsible for histone H1 homocitrullination on lysine 34 (H1K34hcit). The non-canonical signaling pathway is active in cell cycle control through regulated expression of p21Cip1.
The AhR–Arnt–XRE interactions regulate the expression of a plethora of genes including those encoding both phase I and phase II xenobiotic metabolism enzymes (e.g., the Cyp1a1, Cyp1a2, Cyp1b1, and the Gst-ya genes) associated with adaptive or toxic responses to exogenous agonists [40–42]. While much is now know about the molecular events that lead to AhR activation and DNA binding, the mechanistic basis for AhR-mediated dioxin toxicity is not yet fully understood. Nevertheless, the widely held consensus is that most, if not all of the toxic manifestations caused by TCDD are AhR mediated [43], and several of the better characterized toxicities are clearly mediated by the canonical AhR–Arnt signaling complex [44].
4. Non-canonical signaling
To better understand TCDD toxicity, several groups sought to identify AhR target genes through DNA microarray studies using cell culture and whole animal models [45–48]. A general observation stemming from these studies is that despite computational analyses covering 3 Kb [47] or 6 Kb [48] of the sequence flanking the transcription start sites of responsive genes, many did not contain a readily identifiable XRE. One possible explanation is that expression of genes lacking a XRE reflects indirect AhR-mediated signaling, and indeed, such changes in expression may be attributed to latent secondary effects. However, it is formally possible that the AhR alters transcription directly through a site(s) distinct from the consensus XRE. For instance, the ligand-activated AhR–Arnt dimer can interact directly with unliganded estrogen receptor to promote formation of a transcriptionally active complex binding to estrogen response elements [49]. Correspondingly, the AhR can form a quaternary complex with p300, pRb and E2F to suppress S phase gene expression [50,51], or with an unknown protein(s) upstream of the CYP1A2 gene [52]. Evidence is also accumulating for direct AhR-DNA binding in conjunction with the RelB protein to a distinct response element located in the interleukin-8 gene regulatory region [53].
5. KLF6 is a novel AhR DNA-binding partner
The plasminogen activator inhibitor-1 (PAI-1) gene represents an example where direct TCDD responsiveness is attributed to a regulatory region devoid of a canonical XRE [54]. Huang and Elferink [55] recently characterized a novel non-consensus XRE (NC-XRE) consisting of a 5’-GGGA-3’ tetranucleotide motif within the PAI-1 promoter that supports direct DNA binding and function by the AhR independently of the Arnt protein. The lack of sequence homology between the XRE and NC-XRE (Fig. 2) presupposes recruitment of a unique AhR DNA binding complex. Our studies confirmed that AhR-DNA binding to the NC-XRE is absolutely dependent on Krüppel-like Factor 6 (KLF6), a novel AhR partner protein [25]. KLF6 (also known as Zf9 or CPBP) is a ubiquitously expressed Cys2-His2 transcription factor belonging to a growing family (17 currently) of Krüppel-like zinc finger transcription factors that regulate processes including cell proliferation, signal transduction, differentiation, and development [56,57]. KLFs are evolutionarily highly conserved across vertebrate species, sharing an 81 amino acid C-terminal zinc finger DNA-binding domain that can interact with “GC-box” or “CACCC-box” DNA motifs in responsive promoters [58,59]. Therefore, it was intriguing to discover that the 5’ basic region in KLF6 juxtaposing the first zinc finger conferred binding to the NC-XRE [25]. Likewise, targeted deletion of the AhR’s basic region necessary for XRE binding had no noticeable impact on DNA binding to the NC-XRE, suggesting that AhR binding to the NC-XRE is fundamentally different from binding to the XRE.
Fig. 2.
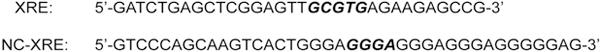
The CYP1A1 XRE and PAI-1 NC-XRE sequences are presented with the core nucleotides necessary for protein-DNA binding depicted in bold italic type.
Mutations in KLF6 and loss-of-heterozygosity have been associated with several human malignancies including prostate, colorectal and liver cancer suggesting that KLF6 is a tumor suppressor [60], functioning by regulating expression of the p21Cip1 cyclin-dependent kinase inhibitor [61]. Jackson et al. [62] recently showed that transient expression of the G1 phase cyclin-dependent kinase inhibitor p21Cip1 during liver regeneration is dependent on NC-XRE-mediated AhR-KLF6 regulatory control [62]. This study also demonstrated that sustained p21Cip1 induction by TCDD is responsible for the previously observed inhibition of normal liver regeneration [63]. Moreover, these data confirm that the AhR-KLF6 complex is responsible for both normal cell cycle control processes, and the deleterious effects of TCDD resulting in disrupted liver regeneration.
6. Carbamoyl phosphate synthase 1 (CPS1) is a novel AhR-KLF6 cofactor
Using the NC-XRE as a DNA affinity reagent to purify the TCDD-inducible AhR-KLF6 complex, mass spectrometry peptide sequencing identified CPS1 as a component of the complex [64]. CPS1 is an ≈160 kDa multidomain protein that catalyzes the irreversible reaction: [65]. While CPS1 is customarily regarded as a mitochondrial enzyme involved in the detoxification of ammonia through the urea cycle [66], the characterization of CPS1 as a component of the NC-XRE-bound AhR-KLF6 complex revealed a novel role for this protein. Specifically, carbamoyl phosphate, the product of CPS1 catalysis, was shown to carbamylate lysine-rich histones to produce homocitrulline through a non-enzymatic mechanism [67]. The proposed mechanism involves nucleophilic attack of lysine residues by the highly electrophilic acylphosphate group on carbamoyl phosphate. A similar process was previously reported for 1,3-bisphosphoglycerate forming a specific, functionally important post-translational modification on glyceral-dehyde-3-phosphate dehydrogenase [68]. Joshi et al. [64] recently demonstrated that a functionally important lysine residue in histone H1 (K34) is reversibly homocitrullinated (H1K34hcit) following AhR-KLF6 recruitment to NC-XRE sites in the genome. Formation of H1K34hcit is dependent on CPS1 expression. It is noteworthy that acetylation of a K34 on the histone H1.4 variant (H1.4K34Ac) is associated with increased H1 mobility and transcriptional activation [69]. In comparison to core histones that remain in place for several hours, H1 proteins are substantially more mobile with a mean residence time at any one binding site estimated to be ≈3 min [70]. It is tempting to speculate that the H1K34hcit post-translational modification may reduce the mean residence time, resulting in a more open chromatin conformation conducive to increased transcription. It is also possible that the H1K34hcit may represent a distinctive modification in the “histone code” read by regulatory proteins to promote transcription [71].
Peptidylarginine deiminase (PADI) enzymes convert arginine and methylarginine residues to citrulline via a hydrolytic process called citrullination or deimination [72]. Recruitment of PADI2 to the genome was shown to citrullinate arginine 26 on histone H3 culminating in local chromatin decondensation and transcriptional activation [73]. Hence the discovery that the PADI2 gene is a transcriptional target for the AhR-KLF6-CPS1 complex implies that CPS1 and PADI2 function in concert to modify histones H1 (H1K24hcit) and H3 (H3R26cit), respectively, to alter the epigenome in response to AhR activation [64].
7. Conclusions
The discovery of non-canonical AhR signaling pathways represents new opportunities to reconcile TCDD toxicity at a molecular level. The recent identification of the AhR-KLF6 complex dramatically increases the repertoire of AhR target genes, and ongoing studies will ascertain how they contribute to toxic and normal physiological processes. The complexity in understanding AhR biology is highlighted by findings showing that carcinogenic receptor agonists induce the canonical AhR signaling pathway to promote tumor formation, yet AhR-regulated expression of p21Cip1 through the non-canonical pathway is consistent with tumor suppression. Hence, future studies will need to reconcile the contribution of both signaling pathways in order to reach a comprehensive understanding. Since both the AhR and KLF6 are ubiquitously expressed, it is reasonable to assume that this complex is functional in many different tissues, however the role for CPS1 is envisioned to be somewhat restricted given that its expression is predominantly hepatic. Hence, involvement of homocitrullination in epigenetic programming may be largely specific to liver functions. Nevertheless, this does not preclude the AhR-KLF6 complex from recruiting other co-factors to regulate tissue-specific gene expression—be they hitherto known, or ones yet to be discovered. What is evident from the recent studies on non-canonical signaling is that the AhR still holds secrets that need to be revealed before a complete understanding of receptor biology is realized.
Acknowledgements
This work was supported by the National Institutes of Health Grants R01ES026874, R21ES024607 and P30ES006676 (to C. J. E.), K01DK102514 (to A. D. J.), and by the National Council for the Improvement of Higher Education (CAPES) (K.P.C).
Abbreviations
- AhR
aryl hydrocarbon receptor
- XRE
xenobiotic response element
- hcit
homocitrulline
- NC-XRE
non-consensus xenobiotic response element
- TCDD
2,3,7,8-tetrachlorodibenzo-p-dioxin
- KLF6
Krüppel-like factor 6
- CPS1
carbamoyl phosphate synthase 1
Footnotes
The authors declare that they have no conflicts of interest with the contents of this article.
References
- 1.Poland A, Knutson JC: 2,3,7,8-tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: examination of the mechanism of toxicity. Ann Rev Pharmacol Toxicol 1982, 22:517–554. [DOI] [PubMed] [Google Scholar]
- 2.Esser C, Rannug A: The aryl hydrocarbon receptor in barrier organ physiology, immunology, and toxicology. Pharmacol Rev 2015, 67:259–279. [DOI] [PubMed] [Google Scholar]
- 3.Bock KW, Kohle C: The mammalian aryl hydrocarbon (Ah) receptor: from mediator of dioxin toxicity toward physiological functions in skin and liver. Biol Chem 2009, 390: 1225–1235. [DOI] [PubMed] [Google Scholar]
- 4.Carreira VS, Fan Y, Kurita H, Wang Q, Ko CI, Naticchioni M, et al. : Disruption of Ah receptor signaling during mouse development leads to abnormal cardiac structure and function in the adult. PLoS One 2015, 10:e0142440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murray IA, Patterson AD, Perdew GH: Aryl hydrocarbon receptor ligands in cancer: friend and foe. Nat Rev Cancer 2014, 14:801–814.This review provides an excellent account of the receptor’s role in tumor biology
- 6.Ma C, Marlowe JL, Puga A: The aryl hydrocarbon receptor at the crossroads of multiple signaling pathways. Exs 2009, 99: 231–257. [DOI] [PubMed] [Google Scholar]
- 7.Jaeger C, Tischkau SA: Role of aryl hydrocarbon receptor in circadian clock disruption and metabolic dysfunction. Environ Health Insights 2016, 10:133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradfield CA, Glover E, Poland A: Purification and N-terminal amino acid sequence of the Ah receptor from the C57BL/6J mouse. Mol Pharmacol 1991, 39:13–19. [PubMed] [Google Scholar]
- 9.Burbach KM, Poland A, Bradfield CA: Cloning of the Ah-receptor cDNA reveals a distinctive ligand-activated transcription factor. Proc Natl Acad Sci U S A 1992, 89:8185–8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Labrecque MP, Prefontaine GG, Beischlag TV: The aryl hydrocarbon receptor nuclear translocator (ARNT) family of proteins: transcriptional modifiers with multi-functional protein interfaces. Curr Mol Med 2013, 13:1047–1065. [DOI] [PubMed] [Google Scholar]
- 11.Harper PA, Riddick DS, Okey AB: Regulating the regulator: factors that control levels and activity of the aryl hydrocarbon receptor. Biochem Pharmacol 2006, 72:267–279. [DOI] [PubMed] [Google Scholar]
- 12.Abbott BD, Birnbaum LS, Perdew GH: Developmental expression of two members of a new class of transcription factors: I. Expression of aryl hydrocarbon receptor in the C57BL/6N mouse embryo. Dev Dyn 1995, 204:133–143. [DOI] [PubMed] [Google Scholar]
- 13.Gasiewicz TA, Singh KP, Casado FL: The aryl hydrocarbon receptor has an important role in the regulation of hematopoiesis: implications for benzene-induced hematopoietic toxicity. Chem Biol Interact 2010, 184:246–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiss EA, Vonarbourg C, Kopfmann S, Hobeika E, Finke D, Esser C, et al. : Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science 2011, 334:1561–1565. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Innocentin S, Withers DR, Roberts NA, Gallagher AR, Grigorieva EF, et al. : Exogenous stimuli maintain intra-epithelial lymphocytes via aryl hydrocarbon receptor activation. Cell 2011, 147:629–640. [DOI] [PubMed] [Google Scholar]
- 16.Akahoshi E, Yoshimura S, Ishihara-Sugano M: Over-expression of AhR (aryl hydrocarbon receptor) induces neural differentiation of Neuro2a cells: neurotoxicology study. Environ Health 2006, 5:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walisser JA, Glover E, Pande K, Liss AL, Bradfield CA: Aryl hydrocarbon receptor-dependent liver development and hepatotoxicity are mediated by different cell types. Proc Natl Acad Sci U S A 2005, 102:17858–17863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hahn ME: Aryl hydrocarbon receptors: diversity and evolution. Chem Biol Interact 2002, 141:131–160. [DOI] [PubMed] [Google Scholar]
- 19.Antonsson C, Whitelaw ML, McGuire J, Gustafsson JA, Poellinger L: Distinct roles of the molecular chaperone hsp90 in modulating dioxin receptor function via the basic helix-loop-helix and PAS domains. Mol Cell Biol 1995, 15: 756–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elferink CJ, Ge NL, Levine A: Maximal aryl hydrocarbon receptor activity depends on an interaction with the retinoblastoma protein. Mol Pharmacol 2001, 59:664–673. [DOI] [PubMed] [Google Scholar]
- 21.Kumar MB, Tarpey RW, Perdew GH: Differential recruitment of coactivator RIP140 by Ah and estrogen receptors. Absence of a role for LXXLL motifs. J Biol Chem 1999, 274: 22155–22164. [DOI] [PubMed] [Google Scholar]
- 22.Tohkin M, Fukuhara M, Elizondo G, Tomita S, Gonzalez FJ: Aryl hydrocarbon receptor is required for p300-mediated induction of DNA synthesis by adenovirus E1A. Mol Pharmacol 2000, 58:845–851. [DOI] [PubMed] [Google Scholar]
- 23.Rushing SR, Denison MS: The silencing mediator of retinoic acid and thyroid hormone receptors can interact with the aryl hydrocarbon (Ah) receptor but fails to repress Ah receptor-dependent gene expression. Arch Biochem Biophys 2002, 403: 189–201. [DOI] [PubMed] [Google Scholar]
- 24.Kumar MB, Perdew GH: Nuclear receptor coactivator SRC-1 interacts with the Q-rich subdomain of the AhR and modulates its transactivation potential. Gene Expr 1999, 8:273–286. [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson SR, Joshi AD, Elferink CJ: The tumor suppressor Kruppel-like factor 6 is a novel aryl hydrocarbon receptor DNA binding partner. J Pharmacol Exp Ther 2013, 345: 419–429.The first article describing the AhR-KLF6 interaction and DNA binding to the NC-XRE
- 26.Jackson DP, Joshi AD, Elferink CJ: Ah receptor pathway in-tricacies; Signaling through diverse protein partners and DNA-motifs. Toxicol Res 2015, 4:1143–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beischlag TV, Luis Morales J, Hollingshead BD, Perdew GH: The aryl hydrocarbon receptor complex and the control of gene expression. Crit Rev Eukaryot Gene Expr 2008, 18:207–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McMillan BJ, Bradfield CA: The aryl hydrocarbon receptor sans xenobiotics: endogenous function in genetic model systems. Mol Pharmacol 2007, 72:487–498. The review provides a detailed description of endogenous AhR functions in both invertebrate and vertebrate systems.
- 29.Perdew GH: Association of the Ah receptor with the 90-kDa heat shock protein. J Biol Chem 1988, 263:13802–13805. [PubMed] [Google Scholar]
- 30.Meyer BK, Pray-Grant MG, Vanden Heuvel JP, Perdew GH: Hepatitis B virus X-associated protein 2 is a subunit of the unliganded aryl hydrocarbon receptor core complex and exhibits transcriptional enhancer activity. Mol Cell Biol 1998, 18:978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma Q, Whitlock JP Jr: A novel cytoplasmic protein that interacts with the Ah receptor, contains tetratricopeptide repeat motifs, and augments the transcriptional response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. J Biol Chem 1997, 272:8878–8884. [PubMed] [Google Scholar]
- 32.Carver LA, LaPres JJ, Jain S, Dunham EE, Bradfield CA: Characterization of the Ah receptor-associated protein, ARA9. J Biol Chem 1998, 273:33580–33587. [DOI] [PubMed] [Google Scholar]
- 33.Kazlauskas A, Poellinger L, Pongratz I: Evidence that the co-chaperone p23 regulates ligand responsiveness of the dioxin (Aryl hydrocarbon) receptor. J Biol Chem 1999, 274: 13519–13524. [DOI] [PubMed] [Google Scholar]
- 34.Petrulis JR, Kusnadi A, Ramadoss P, Hollingshead B, Perdew GH: The hsp90 Co-chaperone XAP2 alters importin beta recognition of the bipartite nuclear localization signal of the Ah receptor and represses transcriptional activity. J Biol Chem 2003, 278:2677–2685. [DOI] [PubMed] [Google Scholar]
- 35.Swanson HI: DNA binding and protein interactions of the AHR/ARNT heterodimer that facilitate gene activation. Chem Biol Interact 2002, 141:63–76. [DOI] [PubMed] [Google Scholar]
- 36.Dere E, Lo R, Celius T, Matthews J, Zacharewski TR: Integration of genome-wide computation DRE search, AhR ChIP-chip and gene expression analyses of TCDD-elicited responses in the mouse liver. BMC genomics 2011, 12:365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lusska A, Shen E, Whitlock JP Jr: Protein-DNA interactions at a dioxin-responsive enhancer. Analysis of six bona fide DNA-binding sites for the liganded Ah receptor. J Biol Chem 1993, 268:6575–6580. [PubMed] [Google Scholar]
- 38.Matikainen T, Perez GI, Jurisicova A, Pru JK, Schlezinger JJ, Ryu HY, et al. : Aromatic hydrocarbon receptor-driven Bax gene expression is required for premature ovarian failure caused by biohazardous environmental chemicals. Nat Genet 2001, 28:355–360. [DOI] [PubMed] [Google Scholar]
- 39.DeGroot DE, Hayashi A, Denison MS: Lack of ligand-selective binding of the aryl hydrocarbon receptor to putative DNA binding sites regulating expression of Bax and paraoxonase 1 genes. Arch Biochem Biophys 2014, 541:13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hankinson O: The aryl hydrocarbon receptor complex. Annu Rev Pharmacol Toxicol 1995, 35:307–340. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt JV, Bradfield CA: Ah receptor signaling pathways. Annu Rev Cell Dev Biol 1996, 12:55–89. [DOI] [PubMed] [Google Scholar]
- 42.Nebert DW, Gonzalez FJ: P450 genes: structure, evolution, and regulation. Annu Rev Biochem 1987, 56:945–993. [DOI] [PubMed] [Google Scholar]
- 43.Mimura J, Fujii-Kuriyama Y: Functional role of AhR in the expression of toxic effects by TCDD. Biochim Biophys Acta 2003, 1619:263–268. [DOI] [PubMed] [Google Scholar]
- 44.Nukaya M, Walisser JA, Moran SM, Kennedy GD, Bradfield CA: Aryl hydrocarbon receptor nuclear translocator in hepatocytes is required for aryl hydrocarbon receptor-mediated adaptive and toxic responses in liver. Toxicol Sci 2010, 118:554–563.This article provides conclusive evidence for involvement of the canonical signaling pathway in many of the hepatotoxic responses mediated by TCDD.
- 45.Puga A, Maier A, Medvedovic M: The transcriptional signature of dioxin in human hepatoma HepG2 cells. Biochem Pharmacol 2000, 60:1129–1142. [DOI] [PubMed] [Google Scholar]
- 46.Frueh FW, Hayashibara KC, Brown PO, Whitlock JP Jr: Use of cDNA microarrays to analyze dioxin-induced changes in human liver gene expression. Toxicol Lett 2001, 122:189–203. [DOI] [PubMed] [Google Scholar]
- 47.Boverhof DR, Burgoon LD, Tashiro C, Chittim B, Harkema JR, Jump DB, et al. : Temporal and dose-dependent hepatic gene expression patterns in mice provide new insights into TCDD-Mediated hepatotoxicity. Toxicol Sci 2005, 85:1048–1063. [DOI] [PubMed] [Google Scholar]
- 48.Tijet N, Boutros PC, Moffat ID, Okey AB, Tuomisto J, Pohjanvirta R: Aryl hydrocarbon receptor regulates distinct dioxin-dependent and dioxin-independent gene batteries. Mol Pharmacol 2006, 69:140–153. [DOI] [PubMed] [Google Scholar]
- 49.Ohtake F, Takeyama K, Matsumoto T, Kitagawa H, Yamamoto Y, Nohara K, et al. : Modulation of oestrogen receptor signalling by association with the activated dioxin receptor. Nature 2003, 423:545–550. [DOI] [PubMed] [Google Scholar]
- 50.Puga A, Barnes SJ, Dalton TP, Chang C, Knudsen ES, Maier MA: Aromatic hydrocarbon receptor interaction with the retinoblastoma protein potentiates repression of E2F-dependent transcription and cell cycle arrest. J Biol Chem 2000, 275: 2943–2950. [DOI] [PubMed] [Google Scholar]
- 51.Marlowe JL, Knudsen ES, Schwemberger S, Puga A: The aryl hydrocarbon receptor displaces p300 from E2F-dependent promoters and represses S phase-specific gene expression. J Biol Chem 2004, 279:29013–29022. [DOI] [PubMed] [Google Scholar]
- 52.Sogawa K, Numayama-Tsuruta K, Takahashi T, Matsushita N, Miura C, Nikawa J, et al. : A novel induction mechanism of the rat CYP1A2 gene mediated by Ah receptor-Arnt heterodimer. Biochem Biophys Res Commun 2004, 318:746–755. [DOI] [PubMed] [Google Scholar]
- 53.Vogel CF, Sciullo E, Matsumura F: Involvement of RelB in aryl hydrocarbon receptor-mediated induction of chemokines. Biochem Biophys Res Commun 2007, 363:722–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Son DS, Rozman KK: 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) induces plasminogen activator inhibitor-1 through an aryl hydrocarbon receptor-mediated pathway in mouse hepatoma cell lines. Arch Toxicol 2002, 76:404–413. [DOI] [PubMed] [Google Scholar]
- 55.Huang G, Elferink CJ: A novel nonconsensus xenobiotic response element capable of mediating aryl hydrocarbon receptor-dependent gene expression. Mol Pharmacol 2012, 81:338–347.This manuscript provided the first characterization of AhR activity through the non-consensus xenobiotic response element (NC-XRE), including the lack of a requiremnt for the Arnt protein.
- 56.Dang DT, Pevsner J, Yang VW: The biology of the mammalian Kruppel-like family of transcription factors. Int J Biochem Cell Biol 2000, 32:1103–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsumoto N, Kubo A, Liu H, Akita K, Laub F, Ramirez F, et al. : Developmental regulation of yolk sac hematopoiesis by Kruppel-like factor 6. Blood 2006, 107:1357–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Philipsen S, Suske G: A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res 1999, 27:2991–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bieker JJ: Kruppel-like factors: three fingers in many pies. J Biol Chem 2001, 276:34355–34358. [DOI] [PubMed] [Google Scholar]
- 60.Bureau C, Hanoun N, Torrisani J, Vinel JP, Buscail L, Cordelier P: Expression and function of Kruppel like-factors (KLF) in carcinogenesis. Curr Genomics 2009, 10:353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Narla G, Heath KE, Reeves HL, Li D, Giono LE, Kimmelman AC, et al. : KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science 2001, 294:2563–2566. [DOI] [PubMed] [Google Scholar]
- 62.Jackson DP, Li H, Mitchell KA, Joshi AD, Elferink CJ: Ah receptor-mediated suppression of liver regeneration through NC-XRE-driven p21Cip1 expression. Mol Pharmacol 2014, 85:533–541.This manuscript demonstrated that TCDD-induced sustained AhR activation through non-canonical signaling is responsible for the deleterious effects of TCDD during normal liver regeneration following injury.
- 63.Mitchell KA, Lockhart CA, Huang G, Elferink CJ: Sustained aryl hydrocarbon receptor activity attenuates liver regeneration. Mol Pharmacol 2006, 70:163–170. [DOI] [PubMed] [Google Scholar]
- 64.Joshi AD, Mustafa MG, Lichti CF, Elferink CJ: Homocitrullination is a novel histone H1 epigenetic mark dependent on aryl hydrocarbon receptor recruitment of carbamoyl phosphate synthase 1. J Biol Chem 2015, 290:27767–27778.This is the first report showing that the AhR-KLF6 complex recruits carbamoyl phosphate synthase 1 to NC-XRE sites in the chromatin resulting in homocitrullination of histone H1, which constitutes a novel epigenetic mark believed to alter chromatin achitecture.
- 65.Metzenberg RL, Marshall M, Cohen PP: Carbamyl phosphate synthetase: studies on the mechanism of action. J Biol Chem 1958, 233:1560–1564. [PubMed] [Google Scholar]
- 66.Martinez AI, Perez-Arellano I, Pekkala S, Barcelona B, Cervera J: Genetic, structural and biochemical basis of carbamoyl phosphate synthetase 1 deficiency. Mol Genet Metab 2010, 101:311–323. [DOI] [PubMed] [Google Scholar]
- 67.Ramponi G, Leaver JL, Grisolia S: Homocitrulline formation following carbamylation of histones with carbamyl phosphate. FEBS Lett 1971, 16:311–314. [DOI] [PubMed] [Google Scholar]
- 68.Moellering RE, Cravatt BF: Functional lysine modification by an intrinsically reactive primary glycolytic metabolite. Science 2013, 341:549–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kamieniarz K, Izzo A, Dundr M, Tropberger P, Ozretic L, Kirfel J, et al. : A dual role of linker histone H1.4 Lys 34 acetylation in transcriptional activation. Genes Dev 2012, 26:797–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brown DT, Izard T, Misteli T: Mapping the interaction surface of linker histone H1(0) with the nucleosome of native chromatin in vivo. Nat Struct Mol Biol 2006, 13:250–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seet BT, Dikic I, Zhou MM, Pawson T: Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol 2006, 7:473–483. [DOI] [PubMed] [Google Scholar]
- 72.Klose RJ, Zhang Y: Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol 2007, 8:307–318. [DOI] [PubMed] [Google Scholar]
- 73.Zhang X, Bolt M, Guertin MJ, Chen W, Zhang S, Cherrington BD, et al. : Peptidylarginine deiminase 2-catalyzed histone H3 arginine 26 citrullination facilitates estrogen receptor alpha target gene activation. Proc Natl Acad Sci U S A 2012, 109: 13331–13336. [DOI] [PMC free article] [PubMed] [Google Scholar]