Highlights
-
•
Pathogens evade cytosolic DNA sensing using divergent and overlapping strategies to target cGAS, 2′3′-cGAMP, and STING.
-
•
Studies of pathogen mechanisms of cGAS–STING evasion reveal new insights into cellular regulation of immune activation.
-
•
Newly discovered regulatory features of cGAS–STING signaling suggest yet undiscovered strategies which pathogens may employ for immune evasion.
Abstract
The cyclic GMP–AMP synthase (cGAS)– Stimulator of Interferon Genes (STING) pathway of cytosolic DNA sensing allows mammalian cells to detect and respond to infection with diverse pathogens. Pathogens in turn encode numerous factors that inhibit nearly all steps of cGAS–STING signal transduction. From masking of cytosolic DNA ligands, to post-translational modification of cGAS and STING, and degradation of the nucleotide second messenger 2′3′-cGAMP, pathogens have evolved convergent mechanisms to evade cGAS–STING sensing. Here we examine pathogen inhibitors of innate immunity in the context of newly discovered regulatory features controlling cellular cGAS–STING activation. Comparative analysis of these strategies provides insight into mechanisms of action and suggests aspects of cGAS–STING regulation and immune evasion that remain to be discovered.
Current Opinion in Immunology 2020, 66:27–34
This review comes from a themed issue on Host pathogens
Edited by Ashley L St. John and Thomas E Morrison
For a complete overview see the Issue and the Editorial
Available online 15th April 2020
https://doi.org/10.1016/j.coi.2020.04.002
0952-7915/© 2020 The Author(s). Published by Elsevier Ltd. This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Introduction
The cGAS–STING pathway detects cytosolic DNA and triggers a robust immune response that restricts replication of diverse pathogens. The enzyme cyclic GMP–AMP synthase (cGAS) directly senses cytosolic DNA and catalyzes synthesis of the nucleotide second messenger 2′3′-cGAMP [1, 2, 3, 4, 5]. 2′3′-cGAMP diffuses throughout the cell and binds to Stimulator of Interferon Genes (STING), a signaling adapter which oligomerizes and traffics from the endoplasmic reticulum (ER) to the ER-Golgi intermediate compartment (ERGIC), where it drives downstream innate immune responses through the type I interferon and NF-κB pathways to inhibit pathogen infection [6, 7, 8, 9, 10].
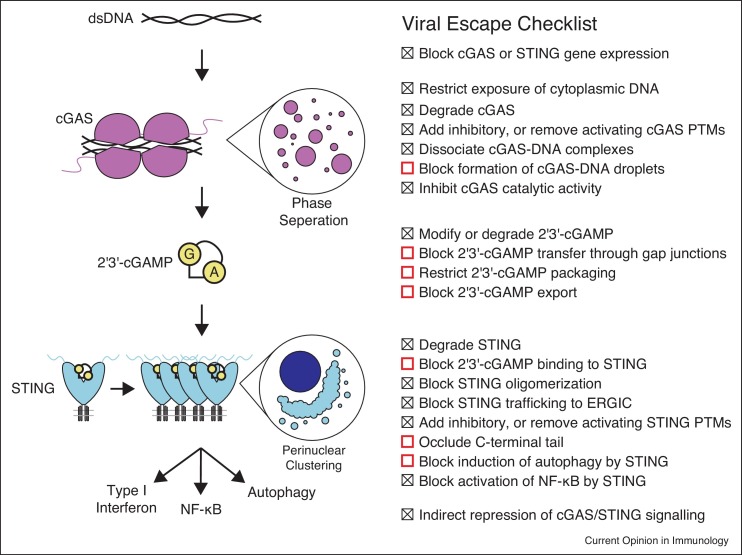
Activation of type I interferon and inflammatory signaling by cGAS–STING during infection places diverse bacterial and viral pathogens under selective pressure to disable this pathway. While the genomes of DNA viruses and bacteria may serve to directly activate cGAS, damage to the mitochondria during infection with RNA viruses can also trigger cGAS activation through exposure of mitochondrial DNA [11]. Therefore, pathogens from divergent groups encode proteins targeting cGAS, 2′3′-cGAMP, and STING to disrupt signaling. Despite the great diversity of these pathogens and the effectors they employ to restrict cGAS–STING activation, shared strategies can be identified which allow escape from the innate immune response (Figure 1 ).
Figure 1.
Summary of the cGAS–STING pathway and potential pathogen evasion strategies. Recognition of cytosolic double-stranded DNA triggers assembly of cGAS into DNA ladder structures, and liquid droplets. Oligomerization into a minimal 2:2 cGAS:DNA complex is required for activation to produce the second messenger 2′3′-cGAMP from ATP and GTP. 2′3′-cGAMP is subsequently recognized by STING which oligomerizes to form a signalosome complex and traffics to the ERGIC and perinuclear regions. This leads to recruitment of the downstream kinase TBK1 to promote innate immune signaling. Points in the pathway which are known (checked box) or potential (red open box) pathogen targets for restriction of cGAS–STING signaling are summarized on the right in the viral escape checklist.
Within the past two years, major advances have expanded our understanding of the mechanism of cGAS–STING signal transduction, providing new insight into how different viral and bacterial factors restrict signaling. In particular, formation of higher-order phase-separated droplets has emerged as a checkpoint for regulating cGAS activation [12•], new structures of STING in complex with TBK1 have provided a mechanism for assembly of the STING signalosome and activation of downstream signaling [13•,14•], and extracellular 2′3′-cGAMP signaling has emerged as an entirely new facet of cGAS–STING biology [15,16]. While other recent reviews provide in-depth analysis of these new discoveries [17,18], here we focus on conserved strategies utilized by different pathogens for evasion of cGAS–STING immunity. Study of these strategies has provided important insight into cellular regulation of cGAS–STING signaling, and likewise these pathogen immune escape factors will continue to serve as useful tools to gain mechanistic insight into pathway function and regulation. In turn, our deepening understanding of the cGAS–STING pathway creates new questions regarding the mechanisms of different groups of viral and bacterial factors which inhibit signaling and indicates new strategies which may be exploited by pathogens for immune evasion.
Pathogens target cGAS to evade cytosolic DNA sensing
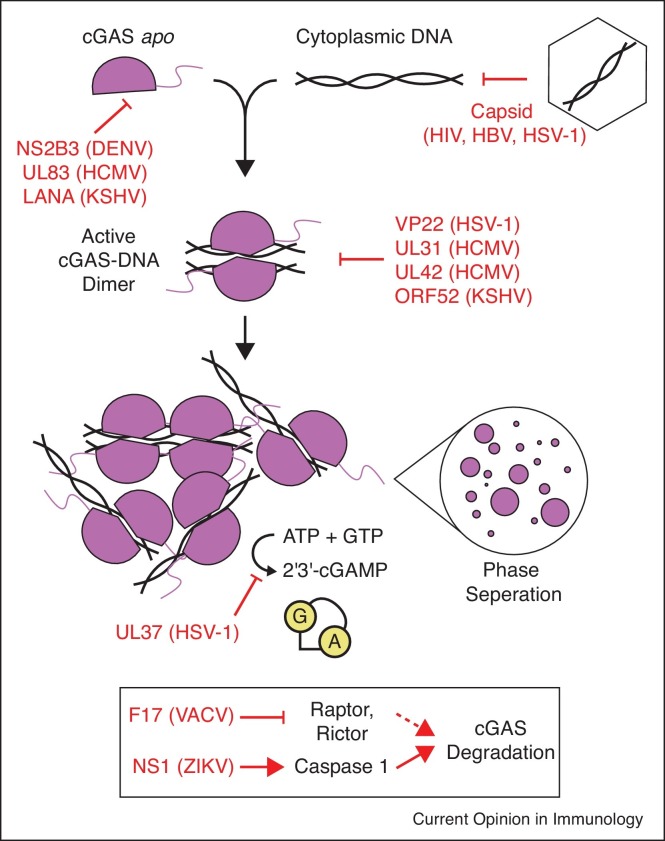
In order to productively infect a target cell, viruses and intracellular bacteria utilize diverse techniques to prevent activation of cGAS before they initiate replication (Figure 2 ). For example, viruses with a DNA genome or genome intermediate (e.g. herpesviruses, and retroviruses), must prevent exposure of DNA to the cytosol, or risk activation of cGAS. These viruses shield DNA from sensing within the viral capsid until it reaches the nucleus, and mutations altering capsid stability regulate cytosolic DNA sensing [19,20].
Figure 2.
Pathogen factors block cGAS activation. Upon DNA binding, cGAS undergoes a conformational change activating 2′3′-cGAMP synthesis. Assembly into a minimal 2:2 complex is required for catalytic activity, but further assembly into long DNA ladder structures and larger oligomers bridged by interactions with the unstructured N-terminus results in formation of phase-separated liquid droplets. ATP and GTP are converted in a two-step process to 2′3′-cGAMP within the cGAS active site, which is released into the cytosol and diffuses throughout the cell. Red text and arrows indicate steps in this process at which different viral and bacterial factors prevent or interfere with signaling. The box at the bottom indicates indirect viral strategies which trigger cGAS degradation.
Another strategy to prevent or restrict cGAS activation is to target cGAS for degradation, reducing levels of this sensor, and impairing 2′3′-cGAMP synthesis. This is accomplished in different ways by both DNA and RNA viruses. For example, the dengue virus (DENV) protease complex NS2B3 cleaves cGAS to prevent its activation by mitochondrial DNA during infection [11]. The related flavivirus Zika virus (ZIKV) utilizes an indirect strategy: its NS1 protein stabilizes caspase-1 leading to cGAS cleavage [21••]. Another indirect strategy is utilized by DNA viruses in the Poxviridae family, where the vaccinia virus (VACV) F17 protein dysregulates mTOR, leading to enhanced cGAS degradation and impaired cytosolic DNA sensing late in infection [22••].
In the event that viral DNA is exposed in the cytosol before replication and gene expression, the nuclear-replicating herpesviruses carry immune antagonists directly within the viral particle to disable DNA sensing after infection. Herpes simplex virus 1 (HSV-1) VP22 and human cytomegalovirus (HCMV) UL83 are both tegument proteins which bind cGAS and block downstream signaling [23,24]. Another member of the Herpesviridae, Kaposi's sarcoma-associated herpesvirus (KSHV), also encodes a tegument protein that inhibits cGAS, ORF52 (also named KicGAS) [25]. ORF52 blocks cGAS activation through a mechanism which requires both cGAS and DNA binding by ORF52. While these proteins from diverse herpesviruses seemingly vary in their requirement for DNA binding activity, they share striking similarities in that each is carried within the virion to bind cGAS and block immune activation after infection. HCMV expresses two additional proteins which interfere with cGAS-DNA binding and formation of higher-order complexes, UL31 and UL42 [26••, 27]. Likewise, cytoplasmic isoforms of the KSHV LANA protein have been shown to bind cGAS, and block downstream signaling [28]. Given new data demonstrating that cGAS phase separates with DNA [12], it is tempting to speculate that some or all of these proteins interfere individually or collaboratively with multivalent cGAS-DNA binding, thus restricting phase separation and altering the balance of innate immune sensing in favor of viral replication.
Pathogens also target cGAS through post-translational modification, resulting in impaired 2′3′-cGAMP synthesis. A second HSV-1 tegument protein, UL37, was recently demonstrated to inhibit cGAS by deamidating a critical residue in the cGAS activation loop [29••]. The activation loop is repositioned in a switch-like fashion after DNA binding, enabling 2′3′-cGAMP synthesis [2,30, 31, 32]. Deamidation of a single asparagine residue in this loop by UL37 results in significantly impaired 2′3′-cGAMP production, perhaps by blocking this switch-like conformational change.
Many open questions remain in the field regarding how cGAS senses pathogen infection. The nature of the cGAS ligand during infection with different pathogens remains unclear — whether it is always the viral or bacterial genome, or to what extent oxidative stress and exposure of mitochondrial DNA or cellular genomic DNA may play a role in cGAS activation during infection remains a topic of debate. Further, a striking number of viral cGAS antagonists serve as virion structural components, including several herpesvirus tegument proteins, and VACV F17. Given that virion stability and efficiency of viral DNA packaging may influence cytosolic DNA sensing during infection, further work to establish the exact mechanisms by which these proteins antagonize cGAS–STING signaling will provide important insight to help unite our understanding of their structural and immune antagonist roles.
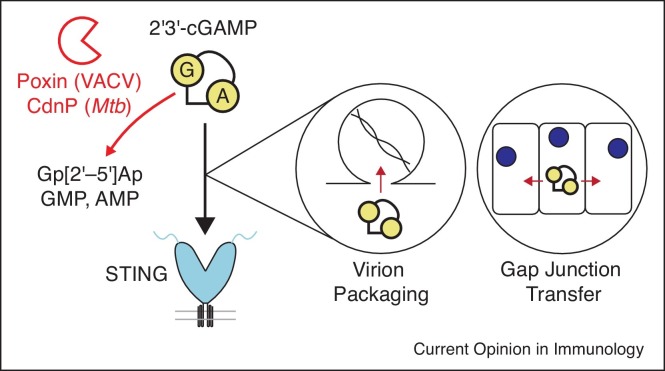
Pathogens degrade 2′3′-cGAMP to block STING activation
The nucleotide second messenger 2′3′-cGAMP is highly stable in the mammalian cytosol [33••]. This RNA signal can be disseminated through gap junctions to activate STING signaling in adjacent uninfected bystander cells, bypassing pathogen factors targeting STING in the infected cell [34]. Similarly, 2′3′-cGAMP can be packaged within nascent virions, driving a rapid immune response upon infection of a new target cell [35,36]. These aspects of 2′3′-cGAMP biology make it a critical target for elimination by viral and bacterial pathogens (Figure 3 ).
Figure 3.
Viral and bacterial enzymes degrade 2′3′-cGAMP. The second messenger 2′3′-cGAMP is highly stable in the mammalian cytosol. 2′3′-cGAMP can be packaged within budding virions to activate STING in newly infected cells, or spread cell-to-cell through gap junctions to activate bystander immunity in neighboring uninfected cells. Viral and bacterial enzymes degrade 2′3′-cGAMP in order to prevent binding to STING, and activation of downstream immune signaling.
Cyclic dinucleotide phosphodiesterase (CdnP) proteins of Mycobacterium and Streptococcus species enzymatically cleave cyclic dinucleotide molecules. Initially, these proteins were reported to cleave bacterial cyclic dinucleotides like cyclic di-AMP to avoid host immune recognition, as these endogenous bacterial signaling molecules can also be sensed by STING as pathogen-associated molecular patterns [10,37,38]. However, the Mycobacterium tuberculosis CdnP enzyme exhibits activity toward host 2′3′-cGAMP as well as bacterial cyclic di-AMP, indicating that it may serve a dual function in infection to prevent host recognition of bacterial cyclic di-AMP as well as degradation of host 2′3′-cGAMP [39].
Similarly, VACV and other related poxviruses encode a nuclease called poxvirus immune nuclease (poxin) which degrades 2′3′-cGAMP in order to prevent activation of the cGAS–STING pathway [33••]. Poxin is highly specific for host 2′3′-cGAMP, and deletion of poxin from the viral genome resulted in attenuation of VACV in a mouse model of infection. Functional poxin enzymes are also found in the genomes of insect viruses in the family Baculoviridae, and moths and butterflies, which serve as hosts to these viruses [33••]. The emerging role for cGAS–STING signaling in insects is likely to enable discovery of additional insect pathogen inhibitors of pathway activation [40, 41, 42]. In mammals, no cytosolic enzymes have been discovered which degrade 2′3′-cGAMP, but instead the extracellular enzyme ectonucleotide pyrophosphatase/phosphodiesterase family member 1 (ENPP1) has been shown to be the major source of 2′3′-cGAMP-degrading activity in mammalian tissue and plasma [43]. Interestingly, avian poxviruses lack a homolog of poxin, but do encode a predicted homolog of ENPP1, indicating these viruses may have obtained alternative means of degrading 2′3′-cGAMP during infection [44]. Together, these results indicate that 2′3′-cGAMP degradation is a more widespread mechanism for control of innate immune signaling than previously understood.
Given that 2′3′-cGAMP can be passed between cells through gap junctions, and infiltrate nascent virions [34, 35, 36], future study is required to understand if pathogens possess specific mechanisms for blocking these unique aspects of 2′3′-cGAMP biology. Recent studies showed that 2′3′-cGAMP can be exported from cells by an unknown transporter and imported into immune cells through the reduced folate transporter SLC19A1 to activate an immune response [15,16]. This indicates that 2′3′-cGAMP import and export may also be important steps at which viruses and bacteria interfere with immune signaling during infection.
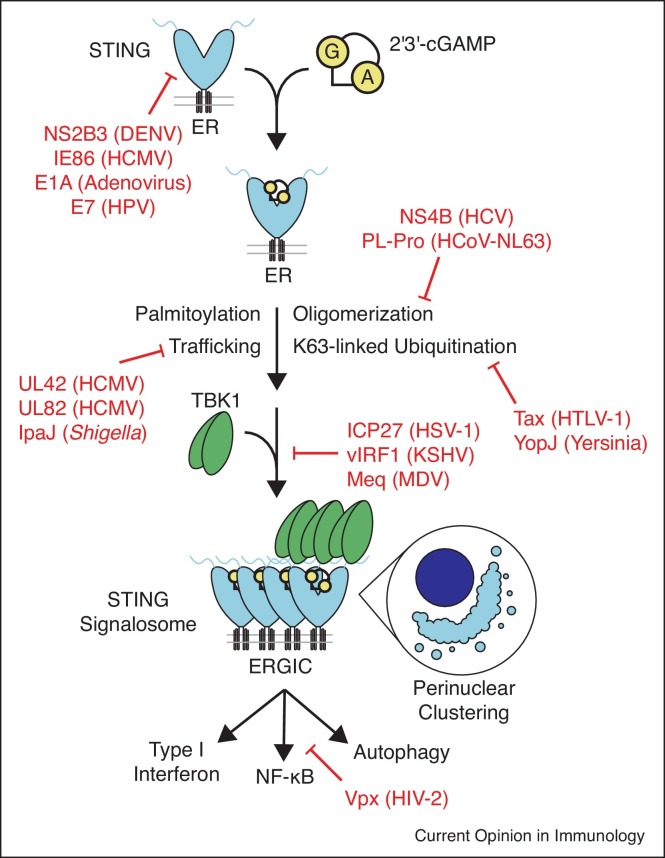
Pathogens block STING signalosome assembly
Several viruses employ strategies to degrade STING to prevent its activation by 2′3′-cGAMP (Figure 4 ). Similar to degradation of cGAS, DENV and other related flaviviruses proteolytically cleave STING using the viral protease complex NS2B3, blocking induction of signaling [45, 46••]. In addition, the HCMV IE86 protein promotes STING degradation in the proteasome, restricting activation of downstream signaling [47].
Figure 4.
Pathogen strategies for restriction of STING signaling. STING binding to 2′3′-cGAMP triggers a conformational change which drives STING oligomerization and assembly into a signalosome complex through palmitoylation, oligomerization, ER to ERGIC trafficking, and ubiquitination. The downstream kinase TBK1 is recruited into the STING signalosome, driving trans-phosphorylation and activation of this kinase to promote downstream IRF3 binding and activation of type I interferon signaling. STING also stimulates NF-κB and autophagy signaling to restrict pathogen replication. Red text and arrows show steps at which pathogens intervene to prevent activation of downstream signaling by STING.
Several processes, including oligomerization [13•, 48], palmitoylation [49,50], K63-linked ubiquitination [51,52], and ER to ERGIC trafficking [53,54] are reported to be important for assembly of the STING signalosome and activation of downstream signaling. It remains unclear how these different processes may regulate one another, but interestingly, a number of pathogen factors interfere with one or more steps. Two RNA virus factors, Hepatitis C virus NS4B, and the coronavirus HCoV-NL63 papain-like protease (PL-pro) both interfere with STING oligomerization [55,56]. Interestingly, it is the deubiquitinase activity rather than the protease activity of PL-pro that is required for antagonism of STING oligomer formation [56]. Several other pathogen factors have been discovered which appear to interfere specifically with STING ubiquitination, including the human T-lymphotrophic virus 1 Tax protein [57], and Yersinia YopJ protein, which also functions as a deubiquitinase [58]. YopJ is additionally reported to block STING ER to ERGIC trafficking, perhaps implying a connection between these processes [58]. Several viral proteins directly target STING trafficking to the ERGIC, which requires the iRhom2/TRAPβ complex [54]. The HCMV tegument protein UL82 disrupts the complex between STING, iRhom2 and TRAPβ [59]. Similarly, HCMV UL42, along with its role in antagonizing cGAS oligomerization, is reported to stimulate degradation of TRAPβ in order to block STING trafficking [27]. Last, the bacterium Shigella flexneri encodes IpaJ which inhibits STING trafficking by an indirect mechanism, targeting ARF GTPases to block traffic out of the ER [60].
After assembly of STING oligomers in the ERGIC, downstream signaling factors associate in order to drive antiviral and inflammatory responses. The mechanism is best understood for the kinase TBK1 and transcription factor IRF3 which associate with the STING C-terminal tail and become activated by trans-phosphorylation driven by assembly of multiple molecules on adjacent STING monomers [13•,14•,61]. The KSHV vIRF1 protein blocks this process, preventing association of TBK1 [62]. Mareck's disease virus is an avian oncogenic herpesvirus, and its oncoprotein Meq functions in a similar way, blocking recruitment of TBK1 to STING in chicken cells [63••]. Interestingly, the oncoproteins E1A and E7 from adenovirus and human papillomavirus also bind and inhibit STING, however the mechanistic consequences of binding have not yet been elucidated [64]. HSV-1 ICP27 functions differently, associating with the active STING/TBK1 complex, and preventing IRF3 recruitment and phosphorylation to block the downstream type I interferon response [65].
Certain pathogens have adapted to benefit from STING activation during infection. The intracellular bacterium Listeria monocytogenes secretes cyclic di-AMP directly into the cytosol of an infected cell, which binds and drives activation of STING [66]. Activation of STING in this context appears to prevent the induction of protective immunity to Listeria. This indicates that Listeria, and potentially other pathogens, may strategically manipulate STING for their own benefit, embracing and dysregulating signaling, rather than evading it.
Viral factors targeting the activation of STING provide a platform to gain a greater and more mechanistic understanding of this process. The details of how STING becomes activated after binding to 2′3′-cGAMP in cells remain incomplete — it is unclear how oligomerization might regulate STING trafficking, and likewise at what step post-translation modifications are applied to STING and exactly how they regulate signaling. Many factors produced by pathogens interfere with these processes, and future systematic study of those acting at different points may provide increased insight into the cell biological processes underlying STING activation. Further, recent STING structures in complex with TBK1 now provide a framework for structural study of factors like KSHV vIRF1 and HSV-1 ICP27 which act on the STING/TBK1 signalosome to prevent downstream immune activation. The details of how STING recruits other downstream factors aside from TBK1 and IRF3 are still unclear, but recent work shows that the HIV-2 Vpx protein can selectively inhibit STING-mediated NF-κB activation [67••], providing an opportunity to better understand this process.
Open questions and future prospects
In the short time since the discovery of cGAS–STING signaling, viral and bacterial factors targeting nearly every step in this pathway have been identified. However, for most of these factors, the specific molecular details cGAS and STING inhibition have not been determined. How do pathogen factors recognize and interact with cGAS and STING on a molecular level, and how has pathogen evasion shaped the structure and regulation of this innate immune pathway? Could viral or bacterial cGAS–STING pathway inhibitors make useful therapeutics for treating autoimmune diseases? cGAS was recently identified as part of a large family of bacterial cGAS/DncV-like nucleotidyltransferase (CD-NTase) enzymes, encompassing thousands of homologs which drive immunity to phage through the action of numerous distinct downstream effectors including STING-like proteins. [68, 69••, 70]. Could bacteriophages share strategies, or perhaps even conserved mechanisms of immune evasion with mammalian viruses? Phage are known to encode anti-CRISPR nucleases to degrade a cyclic oligoadenylate second-messenger for evasion of Type III CRISPR systems [71] raising the question of whether these viruses might also encode enzymes functionally similar to poxin for evasion of CD-NTase signaling. Intervention by pathogens has often provided important new insight into basic biological processes, and future studies of cGAS–STING signaling must continue to harness the power of viruses and bacteria to generate answers to these questions.
Authors’ contribution
Review was written by JBE and PJK, figures were prepared by JBE.
Conflict of interest
None declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgments
The authors acknowledge funding through the Richard and Susan Smith Family Foundation and the Parker Institute for Cancer Immunotherapy, and apologize to those in the field whose work was not cited in this short review due to space constraints.
References
- 1.Sun L., Wu J., Du F., Chen X., Chen Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao P., Ascano M., Wu Y., Barchet W., Gaffney B.L., Zillinger T., Serganov A.A., Liu Y., Jones R.A., Hartmann G. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013;153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diner E.J., Burdette D.L., Wilson S.C., Monroe K.M., Kellenberger C.A., Hyodo M., Hayakawa Y., Hammond M.C., Vance R.E. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 2013;3:1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang X., Shi H., Wu J., Zhang X., Sun L., Chen C., Chen Z.J. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ablasser A., Goldeck M., Cavlar T., Deimling T., Witte G., Röhl I., Hopfner K.-P., Ludwig J., Hornung V. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishikawa H., Ma Z., Barber G.N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun W., Li Y., Chen L., Chen H., You F., Zhou X., Zhou Y., Zhai Z., Chen D., Jiang Z. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci U S A. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin L., Waterman P.M., Jonscher K.R., Short C.M., Reisdorph N.A., Cambier J.C. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol Cell Biol. 2008;28:5014–5026. doi: 10.1128/MCB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong B., Yang Y., Li S., Wang Y.Y., Li Y., Diao F., Lei C., He X., Zhang L., Tien P. The adaptor protein MITA links virus-sensing receptors to IRF3 Transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 10.Burdette D.L., Monroe K.M., Sotelo-Troha K., Iwig J.S., Eckert B., Hyodo M., Hayakawa Y., Vance R.E. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aguirre S., Luthra P., Sanchez-Aparicio M.T., Maestre A.M., Patel J., Lamothe F., Fredericks A.C., Tripathi S., Zhu T., Pintado-Silva J. Dengue virus NS2B protein targets cGAS for degradation and prevents mitochondrial DNA sensing during infection. Nat Microbiol. 2017;2:1–11. doi: 10.1038/nmicrobiol.2017.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12•.Du M., Chen Z.J. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science. 2018;361:704–709. doi: 10.1126/science.aat1022. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study, the authors show that cGAS forms liquid droplets when exposed to DNA. These results define liquid phase separation as a new step required for cGAS–STING activation.
- 13•.Zhao B., Du F., Xu P., Shu C., Sankaran B., Bell S.L., Liu M., Lei Y., Gao X., Fu X. A conserved PLPLRT/SD motif of STING mediates the recruitment and activation of TBK1. Nature. 2019;569:718–722. doi: 10.1038/s41586-019-1228-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using X-ray crystallography and detailed biochemical analysis, the authors identify the motif in STING responsible for recruitment of TBK1 and explain a mechanism of TBK1 recruitment.
- 14•.Zhang C., Shang G., Gui X., Zhang X., Bai X.C., Chen Z.J. Structural basis of STING binding with and phosphorylation by TBK1. Nature. 2019;567:394–398. doi: 10.1038/s41586-019-1000-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using cryo-electron microscopy, the authors determine structures of STING in complex with TBK1 and describe the mechanism of how STING activation and oligomerization leads to TBK1 activation.
- 15.Luteijn R.D., Zaver S.A., Gowen B.G., Wyman S.K., Garelis N.E., Onia L., McWhirter S.M., Katibah G.E., Corn J.E., Woodward J.J. SLC19A1 transports immunoreactive cyclic dinucleotides. Nature. 2019;573:434–438. doi: 10.1038/s41586-019-1553-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ritchie C., Cordova A.F., Hess G.T., Bassik M.C., Li L. SLC19A1 is an importer of the immunotransmitter cGAMP. Mol Cell. 2019;75 doi: 10.1016/j.molcel.2019.05.006. 372–381.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ablasser A., Hur S. Regulation of cGAS- and RLR-mediated immunity to nucleic acids. Nat Immunol. 2020;21:17–29. doi: 10.1038/s41590-019-0556-1. [DOI] [PubMed] [Google Scholar]
- 18.Hu M., Shu H. Innate immune response to cytoplasmic DNA: mechanisms and diseases. Annu Rev Immunol. 2020;38:79–98. doi: 10.1146/annurev-immunol-070119-115052. [DOI] [PubMed] [Google Scholar]
- 19.Lahaye X., Satoh T., Gentili M., Cerboni S., Conrad C., Hurbain I., ElMarjou A., Lacabaratz C., Lelièvre J.D., Manel N. The capsids of HIV-1 and HIV-2 determine immune detection of the viral cDNA by the innate sensor cGAS in dendritic cells. Immunity. 2013;39:1132–1142. doi: 10.1016/j.immuni.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 20.Sun C., Schattgen S.A., Pisitkun P., Jorgensen J.P., Hilterbrand A.T., Wang L.J., West J.A., Hansen K., Horan K.A., Jakobsen M.R. Evasion of innate cytosolic DNA sensing by a gammaherpesvirus facilitates establishment of latent infection. J Immunol. 2015;194:1819–1831. doi: 10.4049/jimmunol.1402495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21••.Zheng Y., Liu Q., Wu Y., Ma L., Zhang Z., Liu T., Jin S., She Y., Li Y., Cui J. Zika virus elicits inflammation to evade antiviral response by cleaving cGAS via NS 1-caspase-1 axis. EMBO J. 2018;37:1–18. doi: 10.15252/embj.201899347. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this paper, the authors uncover that Zika virus induces an inflammatory response using its NS1 protein in order to evade sensing by cGAS, through caspase-1 mediated cGAS degradation. This observation shows an additional mechanism by which RNA viruses manipulate the host cell during infection to evade sensing of endogenous DNA by cGAS.
- 22••.Meade N., Furey C., Li H., Verma R., Chai Q., Rollins M.G., DiGiuseppe S., Naghavi M.H., Walsh D. Poxviruses evade cytosolic sensing through disruption of an mTORC1-mTORC2 regulatory circuit. Cell. 2018;174 doi: 10.1016/j.cell.2018.06.053. 1143–1157.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this paper, the authors demonstrate that vaccinia F17 alters mTOR signaling during infection, resulting in degradation of cGAS and defects in cytosolic DNA sensing. These results define a role for F17 in immune evasion, beyond its role as a virion component.
- 23.Huang J., You H., Su C., Li Y., Chen S., Zheng C. Herpes simplex virus 1 tegument protein VP22 abrogates cGAS/STING-mediated antiviral innate immunity. J Virol. 2018;92:1–11. doi: 10.1128/JVI.00841-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biolatti M., Oste D., Pautasso S., Gugliesi F., Von Einem J., Krapp C., Jakobsen M.R., Borgogna C., Gariglio M., Andrea M., De Human cytomegalovirus tegument protein pp65 (pUL83) Dampens type I interferon production by inactivating the DNA sensor cGAS without affecting STING. J Virol. 2018;92:1–18. doi: 10.1128/JVI.01774-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu J.J., Li W., Shao Y., Avey D., Fu B., Gillen J., Hand T., Ma S., Liu X., Miley W. Inhibition of cGAS DNA sensing by a herpesvirus virion protein. Cell Host Microbe. 2015;18:333–344. doi: 10.1016/j.chom.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26••.Huang Z.F., Zou H.M., Liao B.W., Zhang H.Y., Yang Y., Fu Y.Z., Wang S.Y., Luo M.H., Wang Y.Y. Human cytomegalovirus protein UL31 inhibits DNA sensing of cGAS to mediate immune evasion. Cell Host Microbe. 2018;24 doi: 10.1016/j.chom.2018.05.007. 69–80.e4. [DOI] [PubMed] [Google Scholar]; In this publication, the authors screen for HCMV inhibitors of cGAS–STING signaling, and identify UL31 as a potent inhibitor of pathway activation in cells. Using biochemistry and cell biology, the authors show that UL31 binds cGAS and drives its dissociation from DNA to block production of 2′3′-cGAMP.
- 27.Fu Y.Z., Guo Y., Zou H.M., Su S., Wang S.Y., Yang Q., Luo M.H., Wang Y.Y. Human cytomegalovirus protein UL42 antagonizes cGAS/MITA-mediated innate antiviral response. PLoS Pathog. 2019;15:1–22. doi: 10.1371/journal.ppat.1007691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang G., Chan B., Samarina N., Abere B., Weidner-Glunde M., Buch A., Pich A., Brinkmann M.M., Schulz T.F. Cytoplasmic isoforms of Kaposi sarcoma herpesvirus LANA recruit and antagonize the innate immune DNA sensor cGAS. Proc Natl Acad Sci U S A. 2016;113:E1034–E1043. doi: 10.1073/pnas.1516812113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29••.Zhang J., Zhao J., Xu S., Li J., He S., Zeng Y., Xie L., Xie N., Liu T., Lee K. Species-specific deamidation of cGAS by herpes simplex virus UL37 protein facilitates viral replication. Cell Host Microbe. 2018;24 doi: 10.1016/j.chom.2018.07.004. 234–248.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this publication, the authors use cell-based assays and in vitro biochemistry to show that the herpes simplex virus 1 tegument protein UL37 deamidates cGAS in order to inhibit its activation during infection.
- 30.Zhang X., Wu J., Du F., Xu H., Sun L., Chen Z., Brautigam C.A., Zhang X., Chen Z.J. The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop. Cell Rep. 2014;6:421–430. doi: 10.1016/j.celrep.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Civril F., Deimling T., de Oliveira Mann C.C., Ablasser A., Moldt M., Witte G., Hornung V., Hopfner K.-P. Structural mechanism of cytosolic DNA sensing by cGAS. Nature. 2013;498:332–337. doi: 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X., Shu C., Yi G., Chaton C.T., Shelton C.L., Diao J., Zuo X., Kao C.C., Herr A.B., Li P. Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity. 2013;39:1019–1031. doi: 10.1016/j.immuni.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33••.Eaglesham J.B., Pan Y., Kupper T.S., Kranzusch P.J. Viral and metazoan poxins are cGAMP-specific nucleases that restrict cGAS-STING signalling. Nature. 2019;566:259–263. doi: 10.1038/s41586-019-0928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper details the discovery of poxvirus immune nucleases (poxins) as 2′3′-cGAMP specific nucleases. Using a combination of molecular virology, biochemistry, and structural biology, the authors discover a family of viral and metazoan 2′3′-cGAMP nucleases and determine the mechanism of 2′3′-cGAMP degradation.
- 34.Ablasser A., Schmid-Burgk J.L., Hemmerling I., Horvath G.L., Schmidt T., Latz E., Hornung V. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature. 2013;503:530–534. doi: 10.1038/nature12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gentili M., Kowal J., Tkach M., Satoh T., Lahaye X., Conrad C., Boyron M., Lombard B., Durand S., Kroemer G. Transmission of innate immune signaling by packaging of cGAMP in viral particles. Science. 2015;349:1232–1236. doi: 10.1126/science.aab3628. [DOI] [PubMed] [Google Scholar]
- 36.Bridgeman A., Maelfait J., Davenne T., Partridge T., Peng Y., Mayer A., Dong T., Kaever V., Borrow P., Rehwinkel J. Viruses transfer the antiviral second messenger cGAMP between cells. Science. 2015;349:1228–1232. doi: 10.1126/science.aab3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andrade W.A., Firon A., Schmidt T., Hornung V., Fitzgerald K.A., Kurt-Jones E.A., Trieu-Cuot P., Golenbock D.T., Kaminski P.A. Group B Streptococcus degrades Cyclic-di-AMP to modulate STING-dependent type I interferon production. Cell Host Microbe. 2016;20:49–59. doi: 10.1016/j.chom.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woodward J.J., Lavarone A.T., Portnoy D.A. C-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dey R.J., Dey B., Zheng Y., Cheung L.S., Zhou J., Sayre D., Kumar P., Guo H., Lamichhane G., Sintim H.O. Inhibition of innate immune cytosolic surveillance by an M. tuberculosis phosphodiesterase. Nat Chem Biol. 2017;13:210–217. doi: 10.1038/nchembio.2254. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y., Gordesky-Gold B., Leney-Greene M., Weinbren N.L., Tudor M., Cherry S. Inflammation-induced, STING-dependent autophagy restricts zika virus infection in the drosophila brain. Cell Host Microbe. 2018;24 doi: 10.1016/j.chom.2018.05.022. 57–68.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin M., Hiroyasu A., Guzman R.M., Roberts S.A., Goodman A.G. Analysis of drosophila STING reveals an evolutionarily conserved antimicrobial function. Cell Rep. 2018;23 doi: 10.1016/j.celrep.2018.05.029. 3537–3550.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goto A., Okado K., Martins N., Cai H., Barbier V., Lamiable O., Troxler L., Santiago E., Kuhn L., Paik D. The kinase IKKβ regulates a STING- and NF-κB-dependent antiviral response pathway in drosophila. Immunity. 2018;49 doi: 10.1016/j.immuni.2018.07.013. 225–234.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li L., Yin Q., Kuss P., Maliga Z., Millán J.L., Wu H., Mitchison T.J. Hydrolysis of 2’3’-cGAMP by ENPP1 and design of nonhydrolyzable analogs. Nat Chem Biol. 2014;10:1043–1048. doi: 10.1038/nchembio.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laidlaw S.M., Anwar M.A., Thomas W., Green P., Shaw K., Skinner M.A. Fowlpox virus encodes nonessential homologs of cellular alpha-SNAP, PC-1, and an orphan human homolog of a secreted nematode protein. J Virol. 1998;72:6742–6751. doi: 10.1128/jvi.72.8.6742-6751.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aguirre S., Maestre A.M., Pagni S., Patel J.R., Savage T., Gutman D., Maringer K., Bernal-Rubio D., Shabman R.S., Simon V. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog. 2012;8:e1002934. doi: 10.1371/journal.ppat.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46••.Ding Q., Gaska J.M., Douam F., Wei L., Kim D., Balev M., Heller B., Ploss A. Species-specific disruption of STING-dependent antiviral cellular defenses by the Zika virus NS2B3 protease. Proc Natl Acad Sci U S A. 2018;115:E6310–E6318. doi: 10.1073/pnas.1803406115. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper analyzes STING cleavage by the NS2B3 protease complex of diverse flaviviruses. Using cell-based cleavage assays, the authors demonstrate that multiple flaviviruses specifically cleave human STING at a conserved site, and that STING cleavage activity is important for viral growth in cells.
- 47.Kim J.E., Kim Y.E., Stinski M.F., Ahn J.H., Song Y.J. Human cytomegalovirus IE2 86 kDa protein induces STING degradation and inhibits cGAMP-mediated IFN-β induction. Front Microbiol. 2017;8:1–14. doi: 10.3389/fmicb.2017.01854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shang G., Zhang C., Chen Z.J., Bai X., chen, Zhang X. Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP–AMP. Nature. 2019;567:389–393. doi: 10.1038/s41586-019-0998-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mukai K., Konno H., Akiba T., Uemura T., Waguri S., Kobayashi T., Barber G.N., Arai H., Taguchi T. Activation of STING requires palmitoylation at the Golgi. Nat Commun. 2016;7:11932. doi: 10.1038/ncomms11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haag S.M., Gulen M.F., Reymond L., Gibelin A., Abrami L., Decout A., Heymann M., Van Der Goot F.G., Turcatti G., Behrendt R. Targeting STING with covalent small-molecule inhibitors. Nature. 2018;559:269–273. doi: 10.1038/s41586-018-0287-8. [DOI] [PubMed] [Google Scholar]
- 51.Tsuchida T., Zou J., Saitoh T., Kumar H., Abe T., Matsuura Y., Kawai T., Akira S. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010;33:765–776. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 52.Zhang J., Hu M.M., Wang Y.Y., Shu H.B. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J Biol Chem. 2012;287:28646–28655. doi: 10.1074/jbc.M112.362608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ishikawa H., Barber G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luo W.W., Li S., Li C., Lian H., Yang Q., Zhong B., Shu H.B. iRhom2 is essential for innate immunity to DNA viruses by mediating trafficking and stability of the adaptor STING. Nat Immunol. 2016;17:1057–1066. doi: 10.1038/ni.3510. [DOI] [PubMed] [Google Scholar]
- 55.Yi G., Wen Y., Shu C., Han Q., Konan K.V., Li P., Kao C.C. Hepatitis C virus NS4B can suppress STING accumulation to evade innate immune responses. J Virol. 2016;90:254–265. doi: 10.1128/JVI.01720-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun L., Xing Y., Chen X., Zheng Y., Yang Y., Nichols D.B., Clementz M.A., Banach B.S., Li K., Baker S.C. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PLoS One. 2012;7:e30802. doi: 10.1371/journal.pone.0030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang J., Yang S., Liu L., Wang H., Yang B. HTLV-1 Tax impairs K63-linked ubiquitination of STING to evade host innate immunity. Virus Res. 2017;232:13–21. doi: 10.1016/j.virusres.2017.01.016. [DOI] [PubMed] [Google Scholar]
- 58.Cao Y., Guan K., He X., Wei C., Zheng Z., Zhang Y., Ma S., Zhong H., Shi W. Yersinia YopJ negatively regulates IRF3-mediated antibacterial response through disruption of STING-mediated cytosolic DNA signaling. Biochim Biophys Acta Mol Cell Res. 2016;1863:3148–3159. doi: 10.1016/j.bbamcr.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 59.Fu Y.Z., Su S., Gao Y.Q., Wang P.P., Huang Z.F., Hu M.M., Luo W.W., Li S., Luo M.H., Wang Y.Y. Human cytomegalovirus tegument protein UL82 inhibits STING-mediated signaling to evade antiviral immunity. Cell Host Microbe. 2017;21:231–243. doi: 10.1016/j.chom.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 60.Dobbs N., Burnaevskiy N., Chen D., Gonugunta V.K., Alto N.M., Yan N. STING activation by translocation from the ER is associated with infection and autoinflammatory disease. Cell Host Microbe. 2015;18:157–168. doi: 10.1016/j.chom.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao B., Shu C., Gao X., Sankaran B., Du F., Shelton C.L., Herr A.B., Ji J.Y., Li P. Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins. Proc Natl Acad Sci U S A. 2016;113:E3403–E3412. doi: 10.1073/pnas.1603269113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma Z., Jacobs S.R., West J.A., Stopford C., Zhang Z., Davis Z., Barber G.N., Glaunsinger B.A., Dittmer D.P., Damania B. Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses. Proc Natl Acad Sci U S A. 2015;112:E4306–E4315. doi: 10.1073/pnas.1503831112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63••.Li K., Liu Y., Xu Z., Zhang Y., Luo D., Gao Y., Qian Y., Bao C., Liu C., Zhang Y. Avian oncogenic herpesvirus antagonizes the cGAS-STING DNA-sensing pathway to mediate immune evasion. PLoS Pathog. 2019;15:1–25. doi: 10.1371/journal.ppat.1007999. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using cell-based assays, the authors of this study discover that the Mareck's disease virus Meq protein blocks recruitment of TBK1 to STING, inhibiting downstream signaling.
- 64.Lau A., Gray E.E., Brunette R.L., Stetson D.B. DNA tumor virus oncogenes antagonize the cGAS-STING DNA-sensing pathway. Science. 2015;350:568–571. doi: 10.1126/science.aab3291. [DOI] [PubMed] [Google Scholar]
- 65.Christensen M.H., Jensen S.B., Miettinen J.J., Luecke S., Prabakaran T., Reinert L.S., Mettenleiter T., Chen Z.J., Knipe D.M., Sandri-Goldin R.M. HSV-1 ICP 27 targets the TBK 1-activated STING signalsome to inhibit virus-induced type I IFN expression. EMBO J. 2016;35:1385–1399. doi: 10.15252/embj.201593458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Archer K.A., Durack J., Portnoy D.A. STING-dependent type I IFN production inhibits cell-mediated immunity to Listeria monocytogenes. PLoS Pathog. 2014;10:1–14. doi: 10.1371/journal.ppat.1003861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67••.Su J., Rui Y., Lou M., Yin L., Xiong H., Zhou Z., Shen S., Chen T., Zhang Z., Zhao N. HIV-2/SIV Vpx targets a novel functional domain of STING to selectively inhibit cGAS–STING-mediated NF-κB signalling. Nat Microbiol. 2019;4:2552–2564. doi: 10.1038/s41564-019-0585-4. [DOI] [PubMed] [Google Scholar]; In this paper, the authors identify Vpx as an inhibitor of STING-mediated NF-κB signaling. Their detailed biochemical and genetic approach demonstrates how a viral protein can serve as an important tool for understanding normal regulation and function of cellular cGAS–STING signaling.
- 68.Whiteley A.T., Eaglesham J.B., de Oliveira Mann C.C., Morehouse B.R., Lowey B., Nieminen E.A., Danilchanka O., King D.S., Lee A.S.Y., Mekalanos J.J. Bacterial cGAS-like enzymes synthesize diverse nucleotide signals. Nature. 2019;567:194–199. doi: 10.1038/s41586-019-0953-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69••.Cohen D., Melamed S., Millman A., Shulman G., Oppenheimer-Shaanan Y., Kacen A., Doron S., Amitai G., Sorek R. Cyclic GMP–AMP signalling protects bacteria against viral infection. Nature. 2019;574:691–695. doi: 10.1038/s41586-019-1605-5. [DOI] [PubMed] [Google Scholar]; This publication demonstrates that cGAS-like pathways in bacteria function to restrict bacteriophage infection. This work shows that prokaryotic and eukaryotic viruses must both evade sensing by cGAS-like systems, extending the field of viral cGAS evasion into the prokaryotic domain.
- 70.Ye Q., Lau R.K., Mathews I.T., Birkholz E.A., Watrous J.D., Azimi C.S., Jain M., Corbett K.D. HORMA domain proteins and a Pch2-like ATPase regulate bacterial cGAS-like enzymes to mediate bacteriophage immunity. Mol Cell. 2020;77:1–14. doi: 10.1016/j.molcel.2019.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Athukoralage J.S., McMahon S., Zhang C., Gruschow S., Graham S., Krupovic M., Whitaker R.J., Gloster T., White M.F. An anti-CRISPR viral ring nuclease subverts type III CRISPR immunity. Nature. 2020;577:572–575. doi: 10.1038/s41586-019-1909-5. [DOI] [PMC free article] [PubMed] [Google Scholar]