Abstract
Successful cancer treatment continues to elude modern medicine and its arsenal of therapeutic strategies. Therapy resistance is driven by significant tumor heterogeneity, complex interactions between malignant, microenvironmental and immune cells and cross talk between signaling pathways. Advances in molecular characterization technologies such as next generation sequencing have helped unravel this network of interactions and identify druggable therapeutic targets. Tyrosine kinase inhibitors (TKI) are a class of drugs seeking to inhibit signaling pathways critical to sustaining proliferative signaling, resisting cell death, and the other hallmarks of cancer. While tumors may initially respond to TKI therapy, disease progression is near universal due to mechanisms of acquired resistance largely involving cellular signaling pathway reprogramming. With the ultimate goal of improved TKI therapeutic efficacy our group has developed intracellular paired agent imaging (iPAI) to quantify drug target interactions and oligonucleotide conjugated antibody (Ab-oligo) cyclic immunofluorescence (cycIF) imaging to characterize perturbed signaling pathways in response to therapy. iPAI uses spectrally distinct, fluorescently labeled targeted and untargeted drug derivatives, correcting for non-specific drug distribution and facilitating quantitative assessment of the drug binding before and after therapy. Ab-oligo cycIF exploits in situ hybridization of complementary oligonucleotides for biomarker labeling while oligonucleotide modifications facilitate signal removal for sequential rounds of fluorescent tagging and imaging. Ab-oligo CycIF is capable of generating extreme multi-parametric images for quantifying total and phosphorylated protein expression to quantify protein activation, expression, and spatial distribution. Together iPAI and Ab-oligo cycIF can be applied to interrogate drug uptake and target binding as well as changes to heterogenous cell populations within tumors that drive variable therapeutic responses in patients.
Keywords: cyclic immunostaining, fluorescence imaging, cancer heterogeneity, cell signaling, tyrosine kinase inhibitors, paired agent imaging
1. INTRODUCTION
The promise of molecularly targeted therapy for cancer patients has largely not been realized due to adaptive resistance following monotherapy, resulting in short term response but lacking in long term cures. Unraveling the complexities of adaptive resistance is challenging because it is a complex interplay of drug distribution and target engagement as well as multifaceted and dynamic interactions between the tumor and its microenvironment. Important to the understanding of adaptive resistance mechanisms is that our comprehension of cancer and successful treatment strategies has evolved from a collection of rapidly proliferating cells to a diverse population that evades death through oncogenic dysregulation of signaling pathways and complex interactions with the tumor microenvironment.1, 2 Large-scale cancer genome sequencing efforts have been used extensively to identify novel drug targets, but success has been tempered since cancers are known to be heterogenous both within a single tumor as well as between the primary tumor and metastatic spread. Additionally, DNA and RNA analyses of single patients are indirect measures of functional cell-signaling proteins – the actual target of the majority of molecular therapeutics.3 Proteomics has since emerged as a powerful tool in cancer therapy prediction and evaluation, where the significance of protein expression, function, protein-protein interactions, and spatial organization of key tumor biomarkers has been directly validated. Despite these advances, significant challenges still exist to translate our knowledge of cancer genomics and proteomics into personalized medicine, including: identification of druggable targets, interpreting genomic heterogeneity, and designing durable therapeutic regimens in the context of adaptive resistance.4–7 Further, it is now understood that although monotherapy may achieve diminishment of sensitive subpopulations, it may inadvertently result in the outgrowth of “resistant persister cells.”8 These resistant cell subpopulations eventually lead to disease progression with limited therapeutic options. Identification of resistant cell subpopulations and unraveling their mechanisms of resistance in response to therapy is critical to highly personalized treatments to address both genetic aberrations and resistance vulnerabilities, resulting in potent cancer cures. Therefore, proteomic quantification tools to assess plasticity at the cellular level in response to therapy are necessary to evaluate new treatment regimens that will effectively address adaptive resistance mechanisms in distinct inter- and intra-tumoral subpopulations.
Efficacious therapeutics capable of treating heterogenous tumors require robust drug target engagement in the complex setting of the diseased tissue, where outcomes are dictated by duration, completeness and cellular heterogeneity of drug target engagement.9, 10 Major contributing factors to therapeutic failure are insufficient drug target engagement, off-target activity and cell signaling pathway reprogramming as a mechanism of adaptive resistance. Drug target engagement and off-target activity are typically assessed by bulk sampling of plasma or tissue, yielding a heterogenous average, and thus lacking single cell resolution of drug distribution and target binding.11–13 An assortment of small molecule drugs and protein targets with labeling modifications have been generated to facilitate direct visualization of tissue distribution and target engagement.13–18 While useful for spatial assessment, labeling modifications can vastly alter drug distribution, fail to assess untargeted drug accumulation and lack single cell resolution.19–22 Recently, a novel method using the parent drug and a companion fluorescently labeled drug detected by fluorescence polarization microscopy was developed, but requires multiple doses or repeated imaging to achieve quantitative measures.10
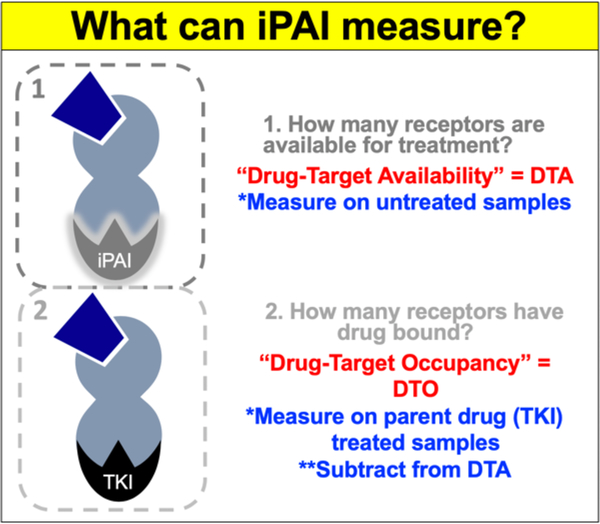
To facilitate quantification of cell-to-cell variation in tissues, we have adapted a technique from autoradiography termed Paired Agent Imaging (PAI). PAI was originally created for quantitative imaging in vivo, where signal from non-specific accumulation of protein-based, radiolabeled affinity reagents in malignant tissue was corrected for by co-administration of a control antibody labeled with an isotope of different energy.23 This technique has been reinvigorated by the optics community and used extensively by our group and others, where spectrally-distinct targeted and untargeted imaging agents are used to correct for non-specific uptake, providing quantitative in vivo or ex vivo assessment of receptor density.24–28 We have recently extended the PAI technique to small molecule therapeutics using spectrally-distinct, fluorescently-labeled targeted and untargeted drug derivatives, such as tyrosine kinase inhibitors (TKIs) to label intracellular targets for intracellular PAI (iPAI). By collecting images of the targeted and untargeted agents, we can calculate (1) the number of available drug targets in untreated samples (drug target availability, DTA), aiding in prediction of effective dose, and (2) the number of occupied drug targets in treated samples (drug targeted occupancy, DTO), quantifying intracellular drug target engagement on individual cells (Figure 1). Notably, although our technique relies on fluorescently-labeled drugs for quantification, all treatment is completed with the parent drug and is thus classified as a label-free method with quantitative assessment of the interaction of the parent drug with its native target.
Figure 1. Intracellular Paired Agent Imaging (iPAI).
iPAI can quantify (1) DTA by measuring untreated samples and (2) DTO by measuring treated samples and subtracting the value from untreated samples (DTA).
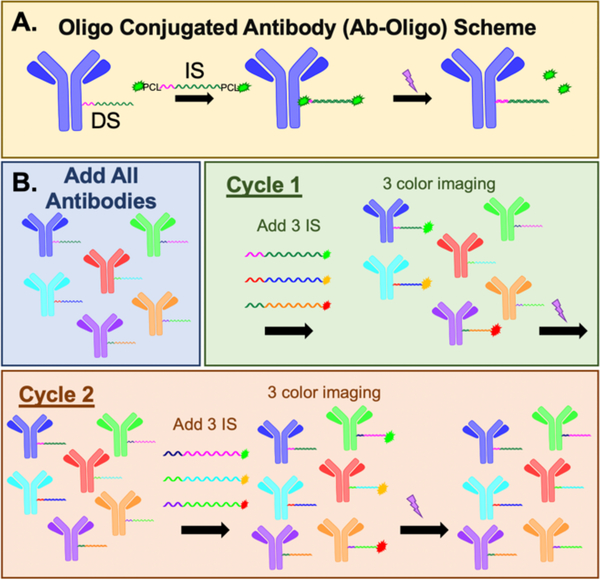
Additionally, there still remains a need for methods of measuring mechanisms of resistance via cell signaling pathway reprogramming. It is necessary to characterize cell signaling pathway reprogramming on the same cell-by-cell basis as drug target engagement is identified for an accurate measure of therapeutic response. Typically cell signaling is measured in cell lysates, which lack spatial context. Alternatively, highly multiplexed immunostaining technologies offer in situ measurement of cell signaling perturbations with single cell resolution. Highly-multiplexed immunostaining techniques have evolved as a popular means for quantitative spatial proteomics. Two main methods employ (1) conventional antibody staining techniques (i.e., immunofluorescence [IF] or immunohistochemistry [IHC]) in a cyclic fashion or (2) mass spectroscopy imaging using rare earth metal labeled antibodies.29–36 These advanced immunostaining strategies have seen widespread adoption due to their natural integration into conventional staining protocols. In order to generate high-dimensional images, cycles of staining, imaging and signal removal (e.g., antibody stripping34, 36, 37 or fluorophore bleaching29, 30, 38) are utilized to create spatial maps of the proteome. While effective, these cyclic protocols generally require weeks to complete antibody staining and the harsh signal removal conditions commonly damage antigenicity and tissue integrity as cycle number increases.39 Mass spectroscopy imaging (i.e., CyTOF,40 MIBI,31, 33 etc.) does not require cycling due to the detection of unique target mass-to-charge ratios, resulting in reduced imaging times to generate the same highly multiplexed dataset. However, mass spectroscopy imaging resolution is limited by laser spot size, resulting in poor detection of individual cells. Additionally, low abundance antigens, such as phosphoproteins, which will be required to interpret cell signaling pathways, can be challenging to detect by mass spectroscopy imaging due to detection sensitivity of the technique. To bridge the utility of cyclic immunostaining and mass spectroscopy imaging, hybrid techniques use unique antibody tags, such as DNA barcodes,41 that are assigned to antibodies of interest, analogous to the rare earth metal tagged antibodies. This permits one staining step with a “master mix” of antibodies, rather than multiple cycles of antibody staining and signal removal. To date, detection methods include cleaving DNA barcodes from antibodies for hybridization to complementary fluorescent barcodes for protein expression quantification (i.e. Nanostring41, 42) or hybridization to complementary oligonucleotide sequences labeled with fluorophore as developed and optimized by our team. We utilize oligonucleotide labeled antibody (Ab-oligo) cyclic immunofluorescence (cycIF) to generate highly multiplexed, high resolution images on a single sample. Ab-oligo cycIF uses photocleavable linkers located on the complementary imaging strands (IS), for signal removal to facilitate cyclic rounds of labeling (Figure 2).
Figure 2. Ab-oligo scheme.
A. Primary antibodies are conjugated to single stranded oligos (DS = docking stand) termed Ab-oligo. After Ab-oligo sample labeling, the complementary oligo (IS = imaging strand) conjugated to two fluorophores and two photocleavable linkers (PCL) is used to specifically label the antigen of interest in situ. After imaging, UV light is applied to cleave the PCLs and fluorophores washed from the sample. B. An illustrative example of a 3 color, 2 cycle Ab-oligo staining experiment is shown, where each Ab-oligo has a unique DS. The complementary IS are labeled with conventional fluorophores. The sample is stained with a mixed cocktail of the 6 Ab-oligos. In cycle 1, 3 Ab-oligos are labeled with their complementary IS with 3 different fluorophores. 3 color imaging is completed, then the signal is removed with UV light. This is followed by a second cycle with complementary IS with the same 3 fluorophores that hybridize to the other 3 Ab-oligos for imaging followed by UV light induced signal removal.
Fully realized personalized therapy will include curated drug selection based on inter- and intra-tumoral heterogeneity and how these dictate therapeutic response and efficacy. Our complementary iPAI and Ab-oligo cycIF imaging technologies facilitate quantification of drug target engagement, tumor biology and adaptive resistance mechanisms, resulting in the only methodology to simultaneously visualize and quantify the complex interactions that define effective cancer therapy on a single cell basis. Herein, we describe the combination of iPAI and Ab-oligo CycIF to generate a high dimensional image set characterizing with single cell resolution DTA, EGFR cell signaling and cell state.
2. METHODOLOGY
2.1. Mouse xenograft model
Athymic nude mice (Homozygous 490, Charles River Laboratories, Wilmington, MA) were subcutaneously implanted with A-431 human skin cancer cells, which were expanded in vitro in appropriate growth media (DMEM + 10% fetal bovine serum [FBS, VWR Scientific, Radnor, PA] + 1% penicillin/streptomycin). Cells were trypsinized, counted and resuspended in growth media to a concentration of 2 × 107 cells/ml. Mice were anesthetized using a dose of 100 mg/kg ketamine (Hospira Inc., Lake Forest, IL) and xylazine was administered at a dose of 10 mg/kg (AnaSed, Shenandoah, IA) by intraperitoneal injection. The mice were then implanted with A-431 cells into each rear flank at a final concentration of 1 × 106 cells/flank in 50% v/v Matrigel (Corning Inc., Corning, NY), resulting in two tumors/mouse. For implantation, the syringe was inserted and held in place for 5 seconds following completion of the injection to prevent any cell suspension leakage. The mice were monitored daily after implantation for tumor growth.
2.2. iPAI imaging agents
Two fluorescent agents, both derivatives of the Erlotinib which targets epidermal growth factor receptor (EGFR) were used in this study: 1) Erl-SiTMR was used as the cell membrane permeant iPAI targeted agent to be imaged in the Cy5 channel; 2) Erl-TMR was used as the cell membrane permeant iPAI untargeted agent to be imaged in the Cy3 channel. The agents were injected into the cell line derived xenograft (CDX) bearing mice simultaneously via tail vein injection at a concentration of 3.5 mg/kg for both the targeted and untargeted iPAI agents.
2.3. Ab-oligo conjugation
Monoclonal antibodies were purchased from AbCam (Cambridge, UK), Thermo Fisher Scientific (Waltham, MA), or Cell Signaling Technology (Danvers, MA) to the following targets: cytokeratin 8 (CK8), Ki-67, E-Cadherin (E-Cad), CoxIV, and phosphorylated EGFR (pEGFR). A unique dibenzocyclooctyne (DBCO) or amine-terminated single-stranded oligonucleotide (docking strand, DS, 28 mer in length) was obtained from Integrated DNA Technologies (IDT, Coralville, IA) to conjugate to each primary antibody. Prior to modification, the IgG antibodies were purified from storage buffers including excess azide reagent using 50 kDa Amicon filters (EMD Millipore, Burlington, MA). Antibody modification and oligonucleotide (oligo) conjugation were performed using the SiteClick™ Antibody Azido modification kit (Thermo Fisher Scientific) following the manufacturer’s instructions. Excess oligo was removed using a 100 kDa Amicon filter. After purification, the absorbance of the Ab-oligo conjugate was measured on a Spectramax M5 (Molecular Devices, San Jose, CA). The maximum absorbance of the antibody and conjugated DS were measured at 280 and 260 nm, respectively. Using the estimated extinction coefficient of 210,000 M−1cm−1 for all antibodies and the manufacturer reported extinction coefficient for each DS, the approximate Ab-oligo conjugation ratios were calculated.
2.4. iPAI fluorescence microscopy and signal removal
Frozen A431 xenograft tissue blocks were sectioned at 10 μm thickness and one section adhered to each slide. The slides were then imaged on a Zeiss Axio Scan.Z1 (Carl Zeiss AG, Oberkochen, Germany), equipped with a Colibri 7 (Carl Zeiss) light source and Orca Flash4 v.2 (Hamamatsu, Hamamatsu City, Japan). The targeted iPAI probe image, Erl-SiTMR was collected in the Cy5 channel and the untargeted probe, Erl-TMR, was collected in the Cy3 channel. After image acquisition, iPAI signal was removed from the tissue by washing 3x-5 minutes (min) with 1X PBS, pH 7.4. The tissue was reimaged to confirm complete removal of iPAI probe from the tissue prior to antibody staining.
2.5. Multiplexed Ab-oligo staining, imaging and signal removal
Following iPAI signal removal, the coverslip was removed and tissue incubated in 2% paraformaldehyde (PFA, Sigma-Aldrich) at room temperature (RT) for 15 min and then washed with 1X PBS, pH 7.4 (3 × 5 min). Tissue was then permeabilized using 1X PBS, pH 7.4 + 0.3% Triton X-100 for 15 min at RT and washed with pH 7.4 PBS (3 × 5 min). The slides were blocked at RT for 30 min in Ab-oligo blocking and dilution buffer which contained 2% bovine serum albumin (BSA, bioWORLD, Dublin, OH), 0.5 mg/mL sheared salmon sperm DNA (Thermo Fisher Scientific) and 0.5% dextran sulfate (Sigma-Aldrich, St. Louis, MO) in 1X PBS, pH 7.4. The 5 selected Ab-oligo conjugates were combined into a single cocktail solution and diluted in the Ab-oligo blocking and dilution buffer to a final protein concentration for each conjugate of 15 μg/mL. The tissue sections were covered with 40 μL of the diluted Ab-oligo conjugate cocktail and incubated at 4 °C overnight in a humidified chamber. The next day, the sections were washed with 2X saline-sodium citrate (SSC) buffer, pH 7 (VWR, Radnor, PA) for 3 × 5 min. The sections were fixed in 2% PFA for 15 min at RT, then washed again in 2X SSC buffer (3 × 5 min).
The tissues stained with the Ab-oligo conjugates were labeled with IS in rounds, where IS labeled with distinct fluorophores and complementary to specific Ab-oligos were stained at 350 nM in each staining round. Serial sections were used for staining with the Ab-oligo cocktail or IS only as a negative control. A DNA-selective dye that is fluorogenic in the Cy2 channel, Nuclear Green from AbCam (Cambridge, UK), was applied to the slides at a final concentration of 100 nM for 10 min at RT before washing with 2x SCC (2 × 5 min).
Antibody stained slides were imaged on a Zeiss AxioScan.Z1. Images were collected in all 4 channels (Cy2, Cy3, Cy5, Cy7). All stained slides were treated with UV light for 15 mins through the coverslip followed by washing 10 times with 0.1X SSC and remounted with Fluoromount-G. Finally, the slides were imaged with the same settings used prior to UV treatment to quantify any remaining signal. Subsequent rounds of IS addition, imaging and signal removal were repeated until all Ab-oligo conjugates were imaged.
2.6. Image processing and visualization
The iPAI model used in this study calculated the drug target availability (DTA) by first normalizing the fluorescence data between the untargeted probe in the Cy3 channel and targeted probe in the Cy5 channel. This was calculation was performed with a custom MATLAB script. The calculation for the DTA was then made with the following equation:
To visualize the Ab-oligo CycIF image data, the separate whole tissue images from each round of Ab-oligo imaging were registered in QiTissue (Pittsburgh, PA) using the DAPI signal in each round. The fluorescence signal intensity was then adjusted manually to display the marker specific staining pattern and false colored to create the visualization.
3. RESULTS AND DISCUSSIONS
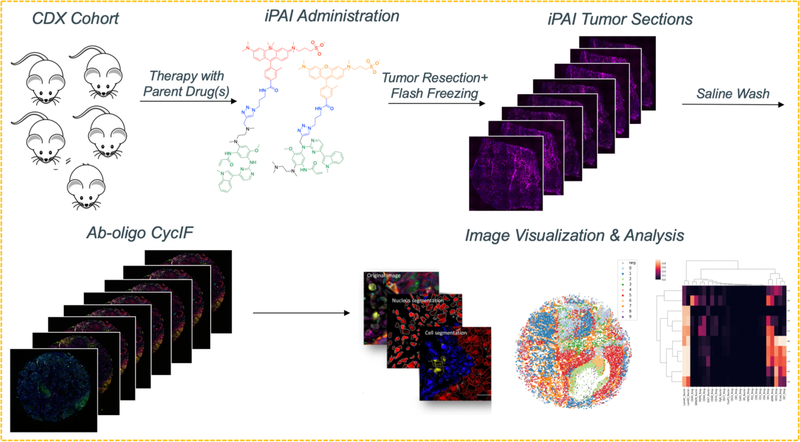
The application of iPAI and Ab-oligo cycIF on the same tissue demonstrates the ability for high dimensional analysis measuring drug target availability (DTA) as well as cell phenotype and signaling pathway perturbations with single cell resolution. The process of combining the two technologies into a single methodology is performed by imaging iPAI probes, removing the iPAI signal, staining and imaging an Ab-oligo CycIF panel followed by image registration to facilitate high dimensional image analysis.
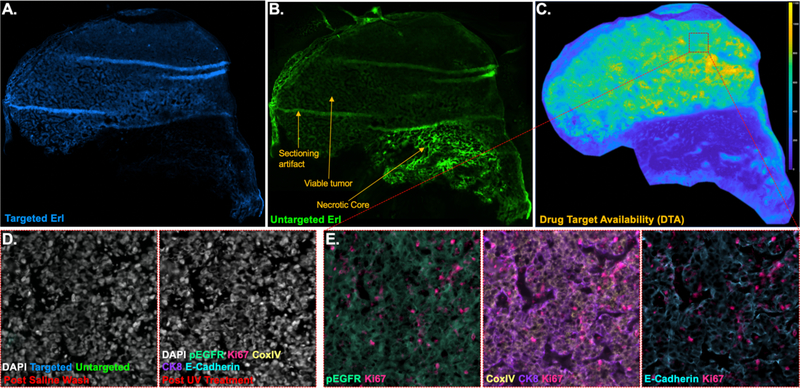
Here, an A431 xenograft was grown in athymic nude mice, injected with Erlotinib iPAI targeted and untargeted probes and resected. Whole tissue imaging of the targeted and untargeted iPAI probes (Figure 3A, B) revealed tissue characteristics such as viable tumor region, a necrotic core and cutting artifact produced during cryosectioning. These images and their false coloring were rendered in QiTissue where contrast levels were adjusted to show the staining pattern from each iPAI probes across the tissue. Interestingly, targeted probe uptake was minimal in the necrotic core and largely limited to the viable tumor regions. This is in contrast to the untargeted probe which was observed to have increased uptake in the necrotic core versus the viable tumor region. After performing the ratiometric calculation to generate the DTA image (Figure 3C) it was clear that the highest concentration of EGFR binding sites available were in the viable tumor region as expected. The ratiometric calculation also accounted for the non-specific uptake of both agents in the necrotic core and resulted in the a near zero DTA in the necrotic region. It is also worth noting that the ratiometric calculation was able to remove the cutting artifacts, which are not prominent in the DTA image. Further investigation will be necessary to determine if regions of tissue with sectioning artifact should be excluded from analysis even after sectioning artifact is no longer visible in the DTA image (Figure 3C).
Figure 3. iPAI & CycIF provide complementary data on a single tissue sample.
Erl iPAI agents were systemically administered to an EGFR (+) CDX mouse four hours prior to sacrifice, where resected tumor was flash frozen and cryosectioned. Images were collected and tiled on the Zeiss AxioScan, enabling visualization of the A. targeted and B. untargeted Erl iPAI fluorescence as well as calculation of C. DTA using QiTissue. Tumor sections contained both viable and necrotic regions, where targeted drug uptake and DTA were minimal in the necfrotic region. DTA calculations also negated the cryosectioning artifacts. D. Targeted and untargeted Erl iPAI fluorescence was readily removed using a saline wash, while tissue integrity was maintained (left), permitting E. Ab-oligo cycIF staining with varied biomarkers (i.e., pEGFR, Ki-67, CoxIV, CK8 and E-Cadherin), which could be readily visualized using QiTissue. Ab-oligo cycIF positive staining signal was removed between staining rounds using UV light treatment (part D, right).
Following imaging of the iPAI agents, the tissue was washed with 2x SSC and reimaged to show complete signal removal of the iPAI agents (Figure 3D, left). Subsequently, Ab-oligo cycIF imaging was performed where five proteins, pEGFR, Ki67, CoxIV, CK8, and E-Cadherin, were stained and imaged over the course of three rounds imaging and signal removal (Figure 3E). Signal removal was achieved with the described UV light exposure method to cleave the fluorophores off the IS (Figure 3D, right). The total number of rounds employed was due to the fact that some markers had complementary IS conjugated to the same fluorophore so they needed to be imaged in separate rounds. The fluorophores used for each marker was AF546 for pEGFR, Ki67, CoxIV and AF750 for CK8 and E-Cadherin. All rounds of whole tissue images were registered and contrast levels of the signal of each marker adjusted with QiTissue. Each marker was then also false colored for visualizations of each marker specific staining pattern (Figure 3D, E).
The Ki-67 positivity observed across the whole tissue is represented in the region of interest shown where there was a high percentage of Ki-67 expressing cells. Also observed was the pEGFR expression demonstrating the sensitivity of Ab-oligo cycIF to detect phosphorylated proteins of interest to monitor cell signaling pathways. In future studies where tissue will be treated, this capability will enable detection of cell signaling pathway changes in response to treatment and quantification of how that response varies across the xenograft. This potential combined with the DTA image generated from the iPAI images will facilitate robust analysis where DTA can be correlated with cell phenotypes. Further developments of this methodology will incorporate cohorts of tissue treated with parent drugs of iPAI agent derivatives. Thus, after establishing DTA in control (untreated) tissues drug target occupancy (DTO) can be calculated in treated tissues where adaptive resistance mechanisms, specifically involving cell signaling pathway reprogramming, can be measured with Ab-oligo CycIF (Figure 4).
Figure 4. Image quantification using QiTissue.
(1) All fluorescence images from iPAI and cycIF staining will be (2) registered via nuclear staining, permitting (3) segmentation at the single cell level and feature extraction. (4) Thresholding of each antibody stain will be used to determine the positive cell population, which will permit spatial analysis by (5) subpopulation clustering and (6) heatmapping.
4. CONCLUSION
While effective, targeted therapy with tyrosine kinase inhibitors inevitably leads to disease progression as tumors adapt to become resistant to therapy through significant tumor heterogeneity. Necessary to overcoming these resistance mechanisms are molecular characterization technologies capable of measuring if the drug is reaching its target and how the presence or absence of the drug on a cancer cell impacts its cell signaling pathways and viability to understand mechanisms of adaptive resistance. We have combined our iPAI and Ab-oligo cycIF imaging technologies to fill this gap in molecular characterization technologies. iPAI is demonstrated here to be able to quantify drug target receptors available for treatment and then on the same tissue Ab-oligo cycIF is able to monitor cell signaling pathway proteins. Further development of this methodology will be necessary to correlate drug target occupancy and local drug concentration with cell signaling pathway reprogramming and cell viability on a single cell basis.
ACKNOWLEDGEMENTS
This work was generously funded by the Prospect Creek Foundation, the Mark Foundation for Cancer Research & the NCI (1R44CA224994-01). The authors thank the Advanced Light Microscopy Core for their support in fluorescent image acquisition.
REFERENCES
- 1.Brockman RW Mechanisms of Resistance to Anticancer Agents. Adv Cancer Res 7, 129–234 (1963). [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D & Weinberg RA Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Lu Y et al. Using reverse-phase protein arrays as pharmacodynamic assays for functional proteomics, biomarker discovery, and drug development in cancer. Semin Oncol 43, 476–483 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collisson EA, Cho RJ & Gray JW What are we learning from the cancer genome? Nature Reviews Clinical Oncology 9, 621–630 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garraway Levi A. & Lander, Eric S. Lessons from the Cancer Genome. Cell 153, 17–37 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Konieczkowski DJ, Johannessen CM & Garraway LA A Convergence-Based Framework for Cancer Drug Resistance. Cancer Cell 33, 801–815 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yap TA, Omlin A & de Bono JS Development of Therapeutic Combinations Targeting Major Cancer Signaling Pathways. Journal of Clinical Oncology 31, 1592–1605 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Ramirez M et al. Diverse drug-resistance mechanisms can emerge from drug-tolerant cancer persister cells. Nat Commun 7, 10690 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bunnage ME, Chekler ELP & Jones LH Target validation using chemical probes. Nature Chemical Biology 9, 195–199 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Dubach JM et al. Quantitating drug-target engagement in single cells in vitro and in vivo. Nature Chemical Biology 13, 168–173 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arrowsmith J & Miller P Phase II and Phase III attrition rates 2011–2012. Nature Reviews Drug Discovery 12, 569–569 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Allison M Reinventing clinical trials. Nature Biotechnology 30, 41–49 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Rutkowska A et al. A Modular Probe Strategy for Drug Localization, Target Identification and Target Occupancy Measurement on Single Cell Level. ACS Chemical Biology 11, 2541–2550 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Cohen MS, Hadjivassiliou H & Taunton J A clickable inhibitor reveals context-dependent autoactivation of p90 RSK. Nature Chemical Biology 3, 156–160 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao M et al. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature 465, 96–100 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller MA et al. Tumour-associated macrophages act as a slow-release reservoir of nano-therapeutic Pt(IV) pro-drug. Nature Communications 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee H, Landgraf R & Wilson JN Synthesis and photophysical properties of a fluorescent cyanoquinoline probe for profiling ERBB2 kinase inhibitor response. Bioorg Med Chem 25, 6016–6023 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Carney B et al. Target engagement imaging of PARP inhibitors in small-cell lung cancer. Nat Commun 9, 176 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lomenick B et al. Target identification using drug affinity responsive target stability (DARTS). Proc Natl Acad Sci U S A 106, 21984–21989 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matthews PM, Rabiner EA, Passchier J & Gunn RN Positron emission tomography molecular imaging for drug development. Br J Clin Pharmacol 73, 175–186 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munteanu B et al. Label-free in situ monitoring of histone deacetylase drug target engagement by matrix-assisted laser desorption ionization-mass spectrometry biotyping and imaging. Anal Chem 86, 4642–4647 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Simon GM, Niphakis MJ & Cravatt BF Determining target engagement in living systems. Nat Chem Biol 9, 200–205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pressman D & E.D. & Blau, M. The use of paired labeling in the determination of tumor-localizing antibodies. Cancer research, 845–850 (1957). [PubMed] [Google Scholar]
- 24.Baeten J, Haller J, Shih H & Ntziachristos V In vivo investigation of breast cancer progression by use of an internal control. Neoplasia 11, 220–227 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barth CW, Schaefer JM, Rossi VM, Davis SC & Gibbs SL Optimizing fresh specimen staining for rapid identification of tumor biomarkers during surgery. Theranostics 7, 4722–4734 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis SC, Gibbs SL, Gunn JR & Pogue BW Topical dual-stain difference imaging for rapid intraoperative tumor identification in fresh specimens. Opt Lett 38, 5184–5187 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tichauer KM et al. Dual-tracer background subtraction approach for fluorescent molecular tomography. J Biomed Opt 18, 16003 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tichauer KM et al. Tumor endothelial marker imaging in melanomas using dual-tracer fluorescence molecular imaging. Mol Imaging Biol 16, 372–382 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerdes MJ et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proceedings of the National Academy of Sciences 110, 11982–11987 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin J-R, Fallahi-Sichani M & Sorger PK Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nature Communications 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Angelo M et al. Multiplexed ion beam imaging of human breast tumors. Nat Med 20, 436–442 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keren L et al. MIBI-TOF: A multiplexed imaging platform relates cellular phenotypes and tissue structure. Sci Adv 5, eaax5851 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levenson RM, Borowsky AD & Angelo M Immunohistochemistry and mass spectrometry for highly multiplexed cellular molecular imaging. Lab Invest 95, 397–405 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zrazhevskiy P & Gao X Quantum dot imaging platform for single-cell molecular profiling. Nat Commun 4, 1619 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Remark R et al. In-depth tissue profiling using multiplexed immunohistochemical consecutive staining on single slide. Sci Immunol 1, aaf6925 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stack EC, Wang C, Roman KA & Hoyt CC Multiplexed immunohistochemistry, imaging, and quantitation: a review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods 70, 46–58 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Zrazhevskiy P, True LD & Gao X Multicolor multicycle molecular profiling with quantum dots for single-cell analysis. Nature Protocols 8, 1852–1869 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin J-R, Fallahi-Sichani M, Chen J-Y & Sorger PK in Current Protocols in Chemical Biology 251–264 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glass G, Papin JA & Mandell JW SIMPLE: a sequential immunoperoxidase labeling and erasing method. J Histochem Cytochem 57, 899–905 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giesen C et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods 11, 417–422 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Ullal AV et al. Cancer cell profiling by barcoding allows multiplexed protein analysis in fine-needle aspirates. Sci Transl Med 6, 219ra219 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Decalf J, Albert ML & Ziai J New tools for pathology: a user’s review of a highly multiplexed method for in situ analysis of protein and RNA expression in tissue. J Pathol 247, 650–661 (2019). [DOI] [PubMed] [Google Scholar]