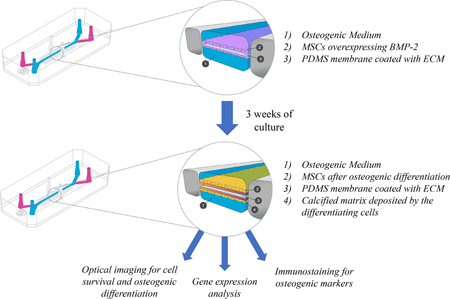
Abstract
Human organoids and organ-on-chip systems to predict human responses to new therapies and for the understanding of disease mechanisms are being more commonly used in translational research. We have developed a bone-chip system to study osteogenic differentiation in vitro, coupled with optical imaging approach which provides the opportunity of monitoring cell survival, proliferation and differentiation in vitro without the need to terminate the culture. We used the mesenchymal stem cell (MSC) line over-expressing bone morphogenetic protein-2 (BMP-2), under Tet-Off system, and luciferase reporter gene under constitutive promoter. Cells were seeded on chips and supplemented with osteogenic medium. Flow of media was started 24 h later, while static cultures were performed using media reservoirs. Cells grown on the bone-chips under constant flow of media showed enhanced survival/proliferation, comparing to the cells grown in static conditions; luciferase reporter gene expression and activity, reflecting the cell survival and proliferation, was quantified using bioluminescence imaging and a significant advantage to the flow system was observed. In addition, the flow had positive effect on osteogenic differentiation, when compared with static cultures. Quantitative fluorescent imaging, performed using the osteogenic extra-cellular matrix-targeted probes, showed higher osteogenic differentiation of the cells under the flow conditions. Gene expression analysis of osteogenic markers confirmed the osteogenic differentiation of the MSC-BMP2 cells. Immunofluorescent staining performed against the Osteocalcin, Col1, and BSP markers illustrated robust osteogenic differentiation in the flow culture and lessened differentiation in the static culture. To sum, the bone-chip allows monitoring cell survival, proliferation, and osteogenic differentiation using optical imaging.
Keywords: Organ-on-a-chip, Mesenchymal stem cells, Osteogenesis, Optical Imaging
Graphical Abstract
1. Introduction
The tissue engineering field emerged from the enormous need for enhanced tissue and organ regeneration when the self-regenerative capabilities of the human body are insufficient. Modern approaches to the development of novel tissue engineering strategies acknowledge the need to recapitulate the physiological and pathological conditions including: cell-to-cell and cell-to-ECM interactions, three-dimensional (3D) organization, shear forces, and other aspects, while retaining reasonable costs and reducing the quantities of animals used and sacrificed (Gomes et al. 2017). To close the translational gap, researchers have been intensifying their efforts to develop 3D cell culture systems known as “Organs-on-Chips” that attempt to recapitulate human physiology on a micro-scale. These micro-engineered culture systems are composed of continuously perfused compartments seeded with multi-lineage cells (Bhatia and Ingber 2014; Sung et al. 2013). We have developed a bone-chip to study osteogenic differentiation in vitro. It provides a platform for optical imaging of the bone-chip without the need to terminate the cultures, as well as the application of mechanical forces known to affect cell function, gene expression, and differentiation (Ingber 2006).
In the last few years those systems have been proven as an effective tool for the study of metabolism (Maschmeyer et al. 2015), for the prediction of drug toxicity in human tissue (Esch et al. 2015; Bhise et al. 2014), for disease modeling (Zhang et al. 2017b; Shuler 2017), and for the investigation of innovative tissue engineering approaches. (Huh et al. 2011) Hitherto, the scientific community has been introduced to a heart-on-a-chip (Zhang et al. 2016), a vessel-on-a-chip (Kim et al. 2013), a kidney-on-a-chip (Homan et al. 2016), a gut-on-a-chip (Kim et al. 2012), a liver-on-a-chip (Bhise et al. 2016), and lastly, a bone-on-a-chip (Wobma and Vunjak-Novakovic 2016; Kolesky et al. 2016). Although these systems allow fluorescent and optical microscopy imaging of cultured cells, one of the previously published systems’ limitations is that various types of analyses require termination of the chip culture. The current bone-chip design coupled with optical imaging approach addresses this limitation. Osteogenic differentiation, for example, can be imaged in vitro using several approaches; two prevalent methods are Von Kossa staining which detects mineralization based on a precipitation reaction between silver ion and the phosphate inorganic ion composing the bone mineral, widely used since the 1980s (Benayahu et al. 1989), and the alizarin red staining based on a pigment that sticks to cellular calcium deposition (Syftestad et al. 1985). Additional approach to demonstrate stem cells differentiation is to monitor marker expression, for example the presence of alkaline phosphatase that rises in the early osteoblast (Hoemann et al. 2009). Another option is to perform immunohistochemistry staining osteogenic markers e.g. Osteocalcin, Bone-sialoprotein, and Osteopontin. Unfortunately, all the aforementioned strategies require termination of the culture. Longitudinal imaging of the same culture over time can be achieved by the introduction of a genetic tag; one example is the knock-in of the luciferase gene, driven by the osteocalcin promoter, so the bioluminescent signal will reflect osteogenic cell differentiation. This can be performed using transgenic animal model (Ben Arav et al. 2012; Cohn Yakubovich et al. 2015) when cells are derived from such animals, or by stably transfected primary cells (Sheyn et al. 2011). The usage of a constitutively expressed promoter, such as the ubiquitin promotor, presents a reliable tool to monitor cell survival and/or proliferation (Sheyn et al. 2011, 2016).
In this study, we focused on non-terminal imaging of a bone-chip system. For this purpose, we used a well-established differentiating system that includes mesenchymal stem cells (MSCs) over-expressing bone morphogenetic protein-2 (BMP-2) (Moutsatsos et al. 2001; Pelled et al. 2007; Tai et al. 2008), under exogenous control of Tetracycline-off system (Moutsatsos et al. 2001), that has been previously examined in various clinically relevant models including spinal fusion (Hasharoni et al. 2005), long-bone allograft regeneration (Xie et al. 2007), radial defects (Kimelman-Bleich et al. 2009), and more. Recombinant human BMP-2 protein is approved by the FDA and widely used in the clinical setting. In the clinic, a very high dose of BMP-2 is administered, which quickly deteriorates. In a variety of experimental systems, a lower dose is required and this is achieved by nonviral gene delivery, offering sustained expression for as long as 14 days (Aslan et al. 2006), or for the cell lifetime in case of MSCs transfected with lentivirus. BMP-2 acts on MSCs via both, paracrine and autocrine signaling, transduced to the nucleus via secondary messengers including the Smad family, which enhance the expression of osteogenic transcription factors such as RUNX2, Osterix and others (Ryoo et al. 2006).
Our hypothesis for the current study is that optical imaging can be utilized to non-invasively monitor stem cell survival and differentiation while cultured in a bone-chip. Furthermore, we hypothesize that the micro-engineered environment within the bone-chip will promote cell proliferation and more efficient osteogenic differentiation and that optical imaging can be utilized to non-invasively monitor stem cell survival and differentiation while cultured in a bone-chip.
2. Methods
2.1. Generation of MSC-Tet-Off-BMP2 cell with constitutive Luc reporter gene expression
Generation of rhBMP-2 overexpressing MSCs was described previously (Moutsatsos et al. 2001). Concisely, cells from the C3H10T1/2 MSC line were stably transfected with a ptTATopBMP2 plasmid vector that encodes for a tetracycline trans-activator and rhBMP-2 (creating a tet-off system). Using the inducible human BMP-2 expression vector, ptTATop-BMP2, the expression of hBMP-2 could be shut down by the administration of doxycycline, an analogue of tetracycline, or turned on by doxycycline absence. Then, the cells were transfected with a Lenti-viral vector encoding for Luc2 reporter gene under the constitutively expressed ubiquitin promotor (Sheyn et al. 2011). Cells were cultured in 100 mm-diameter culture dishes in a complete growth medium (Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum, 2 mM L-glutamine, 100 U/mL penicillin, and 100 U/mL streptomycin (Gibco-Life Technologies, Carlsbad, CA, USA) in 5% CO2 at 37 °C. 1 mg/mL Doxycycline was added to the medium to prevent cell differentiation before seeding onto the chips.
2.2. Bone-chip system
We used a modified method previously described by Huh et al. to fabricate the chips with polydimethylsiloxane (PDMS) as previously described (Huh et al. 2013). We chose to use channel design to allow microscopical analysis of the differentiating cells under controllable flow conditions. The chips were then coated with 50 μg/mL Laminin in PBS (Sigma). Before the chip studies commenced, the cells were trypsinized and centrifuged at 300g and 4 °C for 5 min. The cells were counted using the Trypan blue exclusion test and seeded on the coated chips in 106 cells/mL concentration, total volume of 50 μL, in high glucose complete growth media supplemented with 1 μg/mL doxycycline to prevent BMP2 expression. Dead/non-adhered cells were removed after 3–6 h by flushing media through the Chip and flow was started 24 h after seeding at a rate 30 μL/h of media pulled through, using a syringe pump as previously described (Huh et al. 2013). The static cultures were performed using 200 μL media reservoirs that were replenished every other day. Once the cells reached confluence (2–3 days), the growth media was replaced by doxycycline-deficient osteogenic media (10 mM β-glycerophosphate, 50 μg L-ascorbic acid).
2.3. Cell growth and viability
Micrographs were taken twice a week for 2 weeks using optical microscopy (XL Core ThermoFisher, Waltham, MA, USA). Cell survival was monitored biweekly using bioluminescent imaging (BLI); the media was changed to media supplemented with (0.126 μg/μL) Luciferin, and the BLI signal was captured immediately using Xenogen IVIS Spectrum (PerkinElmer, Waltham, MA, USA),1 min exposure time. The image analysis was done using total influx data calculated on the same size ROI, normalized to background noise in each image.
2.4. Noninvasive osteogenic differentiation assessment (FLI, NIRI—OsteoSense650®/BoneTag800®)
The osteogenic differentiation was evaluated after 3 weeks of culture in osteogenic media using florescent probes. We compare tetracycline-deprived in static and flow culture, and used non-differentiating cells supplemented with tetracycline as a negative control. OsteoSense650® (Perkin Elmer) 0.625 nmol and B oneTag800® (LI-COR Biosciences, Lincoln, NE, USA) 2.56 nmol were introduced 24 h before the imaging, and were imaged using fluorescent imaging (FLI) and near-infrared imaging (NIRI) systems—using IVIS Xenogen for the former, O dyssey® CLx Imaging System (LI-COR Biosciences) for the later.
2.5. Gene expression analysis
Total RNA was isolated using RNAeasy isolation kit (Qiagen, Hilden, Germany) on week 3 of osteogenic differentiation. Single-stranded cDNA was created with the aid of a reverse transcription kit (Invitrogen) and employed as a template for real time-Polymerase Chain Reaction (PCR) with Taqman® gene expression assays using ABI7500 Prism (Applied Biosystems, Carlsbad, CA, USA), as previously described (Zhang et al. 2002). Quantitative RT-PCR was performed to quantify the expression of the osteogenic genes Osteopontin (Opn), Collagen 1 (Col1) and Bone-sialoprotein (BSP). The housekeeping gene 18S was used as a to normalize the data, non-differentiating cells (day 0) was used as calibrator sample to quantify the relative gene expression (RQs).
2.6. Immunofluorescence
Four weeks after seeding the cells were fixed for immunostaining using 4% formaldehyde. Nonspecific antigens were blocked by applying a blocking serum–free solution (Dako, Santa Clara, CA, USA). Chips were stained with primary antibodies against mouse BSP (1:100 Cat# MBS176061, MyBiosourse, San Diego, CA, USA), Col1 (1:250 Cat# ab21286, Abcam, Cambridge, MA, USA) and OC (1:100 Cat# PA1–85754, ThermoFisher Scientific) to examine osteogenic differentiation. The primary antibodies were applied into the chips, incubated in 4 °C overnight, and washed off using PBS; the chips were then incubated with secondary antibodies (Supplemental Table 1) for 1 h in room temperature, after which they were washed off with PBS. The chips were then stained with 4′,6-diamidino2-phenylindole dihydrochloride (DAPI, 1 μg/mL) for 5 min in the dark, followed by washing 3 times with PBS. A Vecta-Mount mounting medium (Vector Laboratories, Burlingame, CA, USA) was applied into the bone-chip. The chips were imaged using a 4-channel Laser Scanning Microscope 780 (Zeiss, Pleasanton, CA, USA) with 10 × magnification lens, z-stacking, and tile scanning. For zoom-in images, a single z-stacked image was generated using 10 × magnification. All chips were scanned using the same gain and exposure settings.
3. Results
3.1. V iability
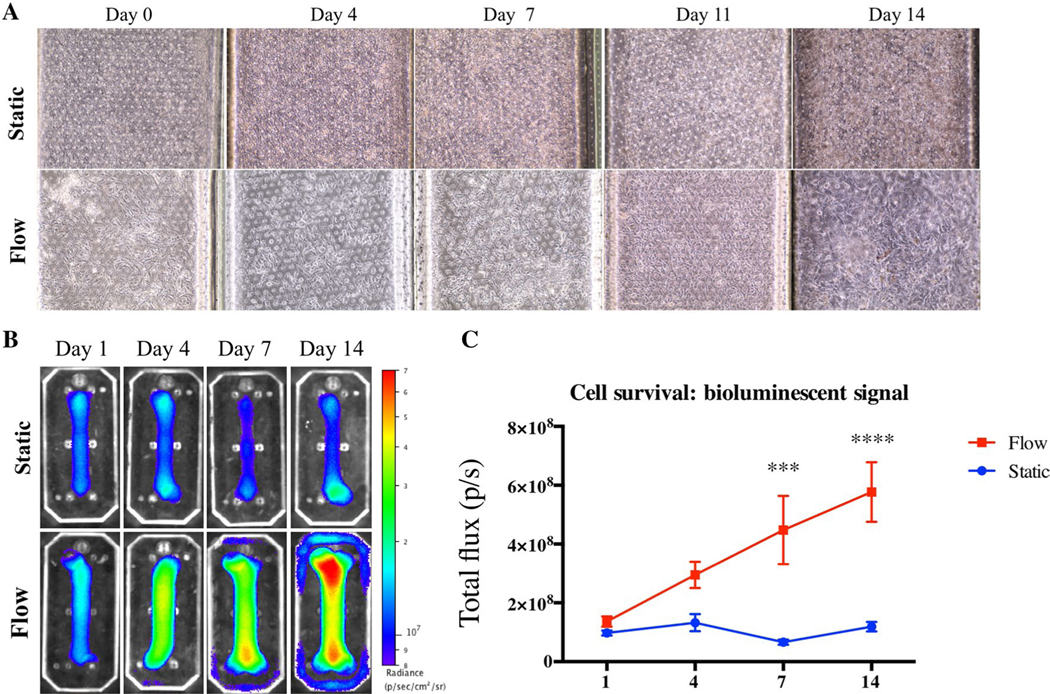
Cells growing on the bone-chips under a constant flow of media showed higher proliferation than the cells growing in static conditions. The microscopic images show proliferation of the cells under flow conditions, achieving 100% confluence 14 days after seeding. In comparison, the cells grown in static media were not as confluent and lacked the MSC characteristic fusiform shape (Fig. 1a). However, the BLI signal generated by the constitutively expressed luciferase reporter gene, reflecting the cell survival and proliferation, demonstrated a notable advantage to the flow system in qualitative analysis (Fig. 1b). Quantitative analysis showed a significantly higher signal in the flow-culture group beginning a week after cell seeding (Fig. 1c). The cells in the bone-chips maintained under continuous flow demonstrated an elevation in the measured BLI signal indicating proliferation, while the cells in the static group has yielded a constant BLI signal denoting survival, but no proliferation.
Fig. 1.
Cell survival and proliferation on the bone-chip. Micrographs of the cells in the bone-chip grown for 2 weeks were taken biweekly, in osteogenic conditions with or without flow. a Bioluminescent images taken on the same time points using IVIS imaging system. The images are relative, and the scale of the color code is depicted on the right side of each group (b). Quantitative analysis of total BLI imaging that was done for 2 weeks (c, bars indicate SD, n = 5, ***P < 0.01; ****P < 0.001)
3.2. Optical imaging of osteogenesis
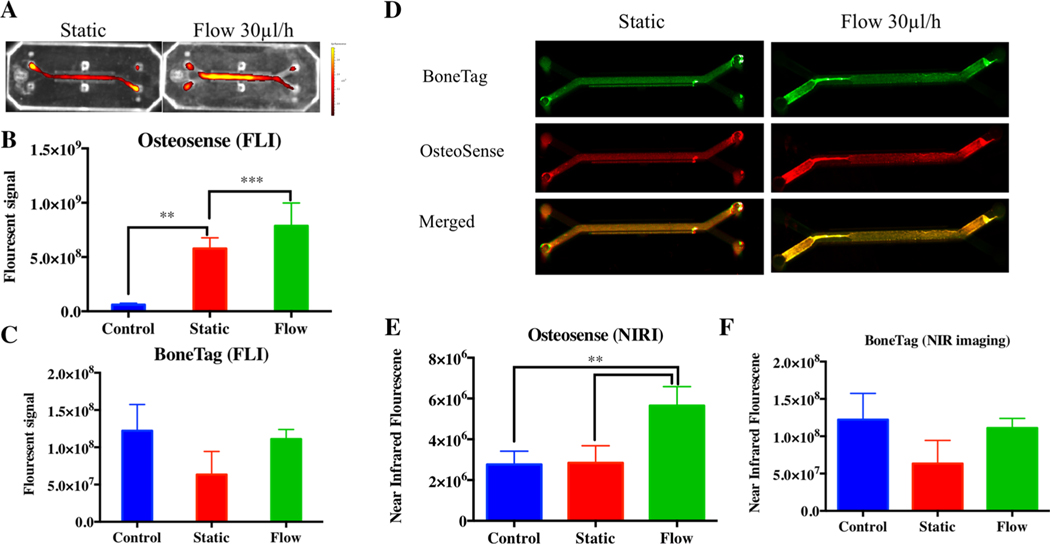
The micro-engineered environment with flow had positive effect on osteogenic differentiation compared with static cultures (Figs. 2, 3). This effect was observed in fluorescent imaging of osteogenic differentiation probes using two different systems—FLI and NIRI (Fig. 2). The probes can be detected using different wavelengths of fluorescence, therefore, both probes could be added simultaneously and imaged separately. The FLI quantification of OsteoSense 650, performed 3 weeks after seeding, showed significantly higher osteogenic differentiation of the cells growing inflow conditions, and both were significantly higher than the control, MSC-BMP2 cells in which differentiation was halted by tetracycline supplementation (Fig. 2a, b). The fluorescent signals measured in the static and the flow cultures were higher than the control: the static by the significance of P < 0.05 while the flow by the significance of P < 0.001. In response to the BoneT ag800 probe, on the other hand, most live cells produced a very low BoneTag 800 FLI signal, comparable to the signal displayed by control cells (Fig. 2c). The NIRI system is considered more sensitive and provides a more accurate quantification of signal. Here, we show that this system is capable of detecting a similar trend (Fig. 2d); the OsteoSense650 signal detected by NIRI was significantly higher in the flow culture comparing to both static culture and control (Fig. 2e), while the use of BoneTag 800 probe did not produce signal higher than the control using both imaging systems (Fig. 2f).
Fig. 2.
Live-monitoring of MSCs osteogenic differentiation on a bone-chip. The differentiation was evaluated 3 weeks after seeding with Os teoSense650 and BoneTag800 probes, that were added to the media for 24 h and washed off prior to imaging performed using two different imaging systems; fluorescent imaging system (a), the labeling was quantified using IVIS (B&C for OsteoSense650 and BoneTag800, respectively. n = 5, *P < 0.05; ***P < 0.001 bars indicate standard deviation), and near infrared imaging system (d NIRI performed using Odyssey® CLx, Li-Cor), which allowed quantification as well (e, f, n = 5, *P < 0.05; ***P < 0.001 bars indicate standard deviation)
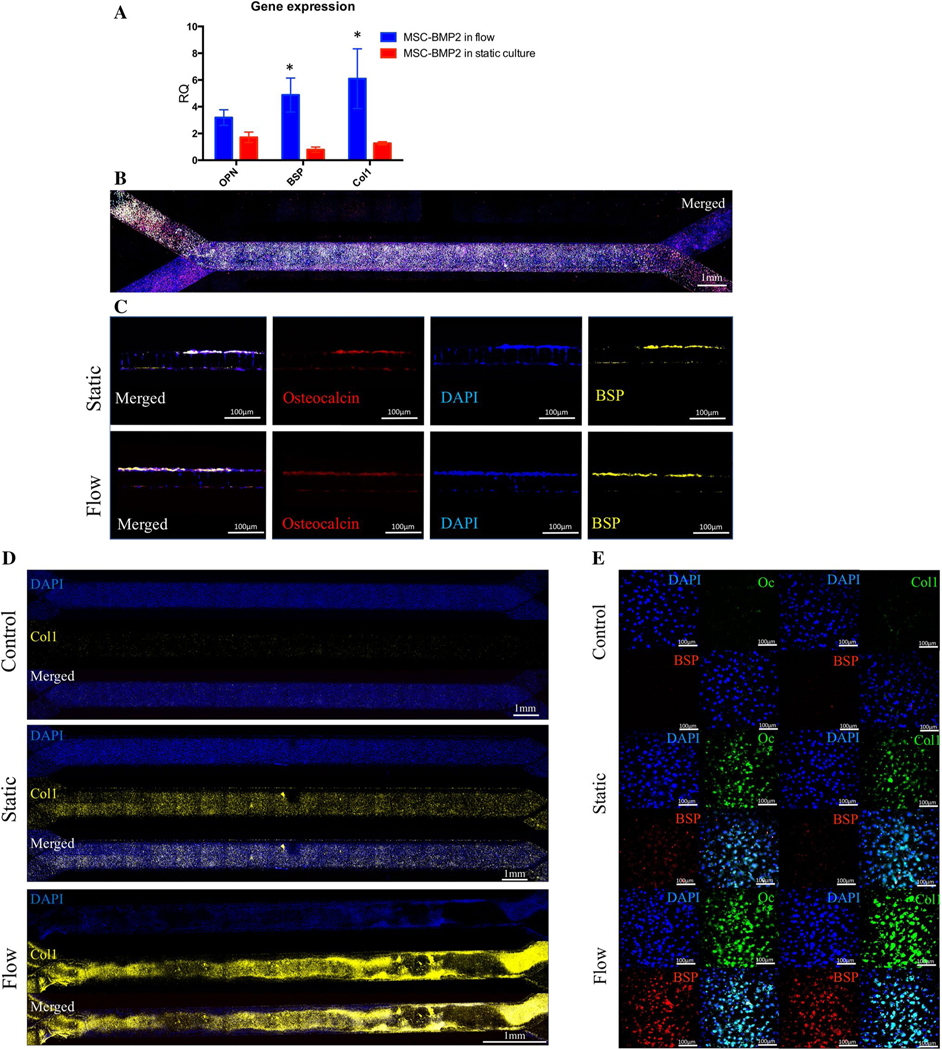
Fig. 3.
Validation of MSC-BMP2 differentiation in a bone chip. Gene expression of the osteogenic markers Osteopontin (OPN), Bone-sialoprotein (BSP) and Collagen-1 (Col1) was evaluated 3 weeks after seeding using qRT-PCR (a, n = 5, *P < 0.05; Bars indicate standard deviation). The cells were fixed, bone-chips were sectioned using vibratome across the channels and stained for Osteocalcin and BSP (b). Other chips were fixed, whole chips were fluorescently stained against the same osteogenic markers and imaged in × 10 magnification (c). Whole chips were stained for Col1 and the entire chip was imaged using confocal microscopy (d). In other sample, set staining was performed against Osteocalcin (Oc), BSP and Col1 and imaged using confocal microscopy (e)
3.3. Gene expression
The gene expression analysis was performed after 3 weeks of culture in osteogenic media, to validate the findings obtained by imaging methods. The analysis confirmed the osteogenic differentiation of the MSC-BMP2 cells in both static and flow conditions, showing overexpression of Osteopontin (OPN), Collagen type 1 and Bone Sialoprotein (BSP) in all cells, but significantly higher expression of Collagen type 1 and BSP was observed in the cells cultured in flow conditions comparing to static culture (Fig. 3a). The osteogenic genes were elevated in the static culture to 1–2 times comparing to non-differentiating cells. In the flow cultured cells, the OPN expression was tripled, the BSP expression raised by factor 5 and the Collagen-1 expression was increased by factor 6.
3.4. Immunofluorescence
To further affirm the imaging results, immunofluorescent staining was performed for the Osteocalcin and BSP markers on whole bone-chips (Fig. 3b) and transverse sections across the channels (Fig. 3c). The staining shows cells on both sides of the membrane in both conditions, but mainly in the top channel. In both conditions, there was positive staining for both markers, indicating osteogenic differentiation, being the staining more prominent in the bone-chips that were cultured underflow. When we stained against Collagen-1 in the whole chip, confocal microscopy revealed prominent expression in the flow group, while only mild expression was detected in the static cultured chip, comparing to null expression in the control group (Fig. 3d).
4. Discussion
The bone-chip channel design provides a platform for optical imaging of the cells (Huh et al. 2010; Korin et al. 2012). The cell response to environmental clues can be tested in vitro without the need to harvest the cells. Furthermore, the chips are composed of a membrane that may be coated covalently or non-covalently with any desired ECM components allowing recapitulation of the cell’s natural environment. Additional prominent advantage is the ability to control and to manipulate the flow sensed by cells, which has a known effect on cell differentiation as shown in several models (Polacheck et al. 2014; Jain et al. 2016). Measurements performed in a model similar to ours have shown that the flow of about 1–5 dynes cm2 (Huh et al. 2013) applied forces that enhanced osteogenic differentiation of MSCs, induced by BMP-2 (Liu et al. 2012; Moore et al. 2011). Previously, we have shown that microfluidic flow can be used to pattern osteogenic differentiation of MSC bearing the Tet-offBMP-2, by patterning the delivery of BMP-2 modulator doxycycline (Zhang et al. 2011), although we were able to demonstrate osteogenesis only using aforementioned imaging techniques that require termination of the culture—ALP and Von Kossa stains, calcium deposition measurement and immunohistochemistry against Bone-sialoprotein. We hypothesized that optical imaging may enable non-invasive monitoring of cell proliferation and differentiation. We could demonstrate that BLI of cells continuously expressing the luciferase gene can faithfully reflect cell proliferation and detect superior proliferation when it was present under the flow conditions (Fig. 1). Next, we showed that using FLI to discern a targeted molecular probe allows detection of osteogenic differentiation, which implies secretion of osteogenic ECM (Fig. 2). The OsteoSense650 probe is a fluorescent probe attached to a bisphosphonate, that targets the molecule to newly secreted bone-matrix as it ossifies. As expected following our previous in vivo studies (Sheyn et al. 2013), we found that the osteoid-targeted OsteoSense650 probe facilitated to monitor osteogenesis and even discerned more rigorous differentiation comparing to moderate differentiation, as confirmed by gene expression analysis and immunofluorescence (Fig. 3). The BoneTag800 which serves as a calcium chelator, could not be used to produce meaningful data, yet it might be used for cultures sustained for longer periods when the culture is expected to mature. Only the combined use of the IVIS system and the NIR imager, conceived to read out smaller samples such as western blot gels, has assembled a finer image of the differentiation process. We infer the IVIS can be used in our system to answer a yes/no question: is there any differentiation? While the NIR imager can be used to answer a more delicate question: what’s the extent of the desired differentiation?
In recent years, several other methods have been proposed for the nondestructive monitoring of differentiating in vitro systems, including microscopic magnetic resonance imaging for the monitoring of the changes in tissue stiffness during in vitro osteogenesis (Xu et al. 2006), or nuclear imaging of a tracer for the quantification of secreted osteogenic ECM (Tobias et al. 2016). In addition to the optical imaging, we also experimented with non-invasive monitoring of cell differentiation by micro-computed tomography (data not shown). However, due to low signal-to-noise ratio of the 2 layers of cells on relatively radiopaque PDMS, we concluded that it could not demonstrate the osteogenic differentiation and mineral deposition. We find our approach accessible and easy to follow since to implement it there is no need of special devices or instruments, rather widely used experimental equipment available to most of the research community. To mention a few, we believe that the optical imaging can be used not only for regenerative medicine oriented research but also to metabolism research and to disease modeling using any of the commercially available probes for hypoxia (Woolf et al. 2015), metabolism (Ocak et al. 2015), inflammation (Shanmugam et al. 2015) and others. Also, the genetic tagging by the luciferase transgene can be used as a reporter to virtually any promoter, including prokaryotic genes (Bergmann et al. 2013).
Our model is the first step in the further development to mimic the regenerating bone in finer ways. One example would be to use the bone-chip and induce different mechanical forces that are often exerted on healing fracture, such as stretch, as shear force is shown to play a key role in differentiation and gene expression (Huh et al. 2010). Different bone ECM components may be used, such as Collagen type 1, Osteopontin, Bone Sialoprotein to recreate the in vivo bone microenvironment. The flow can be used to introduce the system with chemokines that are known to play a role in in vivo fracture healing, such as TNF-alpha and IL-17 (Chang et al. 2013), or systemically administrated osteogenic therapies such as parathyroid hormone (Cohn Yakubovich et al. 2017a, b). The outflow can be collected and analyzed for the cell secretome (Wang et al. 2016). In the future, the chip may be used to study the relation of regenerating bone to adjacent tissues including tendon and cartilage. Furthermore, the molecular optical imaging may be integrated into the next generation of the multisensory-organ-on-a-chip (Zhang et al. 2017a) which will allow correlation of cell differentiation to secreted cell markers (Riahi et al. 2016).
Supplementary Material
Footnotes
Compliance with ethical standards
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s10404–019-2261–7) contains supplementary material, which is available to authorized users.
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Conflict of interest DS, DCY, SBD, SADM, VC, DG and ZG has nothing to declare. CH, NW and GAH are employees of Emulate Inc.
References
- Aslan H, Zilberman Y, Arbeli V, Sheyn D, Matan Y, Liebergall M et al. (2006) Nucleofection-based ex vivo nonviral gene delivery to human stem cells as a platform for tissue regeneration. Tissue Eng 12(4):877–889. 10.1089/ten.2006.12.877 [DOI] [PubMed] [Google Scholar]
- Ben Arav A, Pelled G, Zilberman Y, Kimelman-Bleich N, Gazit Z, Schwarz EM et al. (2012) Adeno-associated virus-coated allografts: a novel approach for cranioplasty. J Tissue Eng Regen Med 6(10):e43–e50. 10.1002/term.1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benayahu D, Kletter Y, Zipori D, Wientroub S (1989) Bone marrowderived stromal cell line expressing osteoblastic phenotype in vitro and osteogenic capacity in vivo. J Cell Physiol 140(1):1–7. 10.1002/jcp.1041400102 [DOI] [PubMed] [Google Scholar]
- Bergmann S, Rohde M, Schughart K, Lengeling A (2013) The bioluminescent Listeria monocytogenes strain Xen32 is defective in flagella expression and highly attenuated in orally infected BALB/cJ mice. Gut Pathog 5(1):19 10.1186/1757-4749-5-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia SN, Ingber DE (2014) Microfluidic organs-on-chips. Nat Biotechnol 32(8):760–772. 10.1038/nbt.2989 [DOI] [PubMed] [Google Scholar]
- Bhise NS, Ribas J, Manoharan V, Zhang YS, Polini A, Massa S et al. (2014) Organ-on-a-chip platforms for studying drug delivery systems. J Control Release 190:82–93. 10.1016/j.jconrel.2014.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhise NS, Manoharan V, Massa S, Tamayol A, Ghaderi M, Miscuglio M et al. (2016) A liver-on-a-chip platform with bioprinted hepatic spheroids. Biofabrication 8(1):014101 10.1088/1758-5090/8/1/014101 [DOI] [PubMed] [Google Scholar]
- Chang J, Liu F, Lee M, Wu B, Ting K, Zara JN et al. (2013) NF-κB inhibits osteogenic differentiation of mesenchymal stem cells by promoting β-catenin degradation. Proc Natl Acad Sci USA 110(23):9469–9474. 10.1073/pnas.1300532110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn Yakubovich D, Tawackoli W, Sheyn D, Kallai I, Da X, Pelled G et al. (2015) Computed tomography and optical imaging of osteogenesis-angiogenesis coupling to assess integration of cranial bone autografts and allografts. J Vis Exp. 10.3791/53459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn Yakubovich D, Eliav U, Yalon E, Schary Y, Sheyn D, Cook-Wiens G et al. (2017a) Teriparatide attenuates scarring around murine cranial bone allograft via modulation of angiogenesis. Bone 97:192–200. 10.1016/j.bone.2017.01.020 [DOI] [PubMed] [Google Scholar]
- Cohn Yakubovich D, Sheyn D, Bez M, Schary Y, Yalon E, Sirhan A et al. (2017b) Systemic administration of mesenchymal stem cells combined with parathyroid hormone therapy synergistically regenerates multiple rib fractures. Stem Cell Res Ther 8(1):51 10.1186/s13287-017-0502-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch EW, Bahinski A, Huh D (2015) Organs-on-chips at the frontiers of drug discovery. Nat Rev Drug Discov 14(4):248–260. 10.1038/nrd4539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes ME, Rodrigues MT, Domingues RMA, Reis RL (2017) Tissue engineering and regenerative medicine: new trends and directions—a year in review. Tissue Eng Part B Rev 23(3):211–224. 10.1089/ten.TEB.2017.0081 [DOI] [PubMed] [Google Scholar]
- Hasharoni A, Zilberman Y, Turgeman G, Helm GA, Liebergall M, Gazit D (2005) Murine spinal fusion induced by engineered mesenchymal stem cells that conditionally express bone morphogenetic protein-2. J Neurosurg Spine 3(1):47–52 [DOI] [PubMed] [Google Scholar]
- Hoemann CD, El-Gabalawy H, McKee MD (2009) In vitro osteogenesis assays: influence of the primary cell source on alkaline phosphatase activity and mineralization. Pathol Biol (Paris) 57(4):318–323. 10.1016/j.patbio.2008.06.004 [DOI] [PubMed] [Google Scholar]
- Homan KA, Kolesky DB, Skylar-Scott MA, Herrmann J, Obuobi H, Moisan A et al. (2016) Bioprinting of 3D convoluted renal proximal tubules on perfusable chips. Sci Rep 6:34845 10.1038/srep34845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE (2010) Reconstituting organ-level lung functions on a chip. Science 328(5986):1662–1668. 10.1126/science.1188302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh D, Hamilton GA, Ingber DE (2011) From 3D cell culture to organs-on-chips. Trends Cell Biol 21(12):745–754. 10.1016/j.tcb.2011.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh D, Kim HJ, Fraser JP, Shea DE, Khan M, Bahinski A et al. (2013) Microfabrication of human organs-on-chips. Nat Protoc 8(11):2135–2157. 10.1038/nprot.2013.137 [DOI] [PubMed] [Google Scholar]
- Ingber DE (2006) Cellular mechanotransduction: putting all the pieces together again. FASEB J 20(7):811–827. 10.1096/fj.05-5424rev [DOI] [PubMed] [Google Scholar]
- Jain A, van der Meer AD, Papa AL, Barrile R, Lai A, Schlechter BL et al. (2016) Assessment of whole blood thrombosis in a microfluidic device lined by fixed human endothelium. Biomed Microdevices 18(4):73 10.1007/s10544-016-0095-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Huh D, Hamilton G, Ingber DE (2012) Human gut-ona-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 12(12):2165–2174. 10.1039/c2lc40074j [DOI] [PubMed] [Google Scholar]
- Kim S, Lee H, Chung M, Jeon NL (2013) Engineering of functional, perfusable 3D microvascular networks on a chip. Lab Chip 13(8):1489–1500. 10.1039/c3lc41320a [DOI] [PubMed] [Google Scholar]
- Kimelman-Bleich N, Pelled G, Sheyn D, Kallai I, Zilberman Y, Mizrahi O et al. (2009) The use of a synthetic oxygen carrier-enriched hydrogel to enhance mesenchymal stem cell-based bone formation in vivo. Biomaterials 30(27):4639–4648. 10.1016/j.biomaterials.2009.05.027 [DOI] [PubMed] [Google Scholar]
- Kolesky DB, Homan KA, Skylar-Scott MA, Lewis JA (2016) Three-dimensional bioprinting of thick vascularized tissues. Proc Natl Acad Sci USA 113(12):3179–3184. 10.1073/pnas.1521342113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korin N, Kanapathipillai M, Matthews BD, Crescente M, Brill A, Mammoto T et al. (2012) Shear-activated nanotherapeutics for drug targeting to obstructed blood vessels. Science 337(6095):738–742. 10.1126/science.1217815 [DOI] [PubMed] [Google Scholar]
- Liu L, Yu B, Chen J, Tang Z, Zong C, Shen D et al. (2012) Different effects of intermittent and continuous fluid shear stresses on osteogenic differentiation of human mesenchymal stem cells. Biomech Model Mechanobiol 11(3–4):391–401. 10.1007/s10237-011-0319-x [DOI] [PubMed] [Google Scholar]
- Maschmeyer I, Lorenz AK, Schimek K, Hasenberg T, Ramme AP, Hübner J et al. (2015) A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip 15(12):2688–2699. 10.1039/c5lc00392j [DOI] [PubMed] [Google Scholar]
- Moore NM, Lin NJ, Gallant ND, Becker ML (2011) Synergistic enhancement of human bone marrow stromal cell proliferation and osteogenic differentiation on BMP-2-derived and RGD peptide concentration gradients. Acta Biomater 7(5):2091–2100. 10.1016/j.actbio.2011.01.019 [DOI] [PubMed] [Google Scholar]
- Moutsatsos IK, Turgeman G, Zhou S, Kurkalli BG, Pelled G, Tzur L et al. (2001) Exogenously regulated stem cell-mediated gene therapy for bone regeneration. Mol Ther 3(4):449–461. 10.1006/mthe.2001.0291 [DOI] [PubMed] [Google Scholar]
- Ocak M, Gillman AG, Bresee J, Zhang L, Vlad AM, Müller C et al. (2015) Folate receptor-targeted multimodality imaging of ovarian cancer in a novel syngeneic mouse model. Mol Pharm 12(2):542–553. 10.1021/mp500628g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelled G, Tai K, Sheyn D, Zilberman Y, Kumbar S, Nair LS et al. (2007) Structural and nanoindentation studies of stem cell-based tissue-engineered bone. J Biomech 40(2):399–411. 10.1016/j.jbiomech.2005.12.012 [DOI] [PubMed] [Google Scholar]
- Polacheck WJ, German AE, Mammoto A, Ingber DE, Kamm RD (2014) Mechanotransduction of fluid stresses governs 3D cell migration. Proc Natl Acad Sci USA 111(7):2447–2452. 10.1073/pnas.1316848111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riahi R, Shaegh SA, Ghaderi M, Zhang YS, Shin SR, Aleman J et al. (2016) Automated microfluidic platform of bead-based electrochemical immunosensor integrated with bioreactor for continual monitoring of cell secreted biomarkers. Sci Rep 6:24598 10.1038/srep24598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo HM, Lee MH, Kim YJ (2006) Critical molecular switches involved in BMP-2-induced osteogenic differentiation of mesenchymal cells. Gene 366(1):51–57. 10.1016/j.gene.2005.10.011 [DOI] [PubMed] [Google Scholar]
- Shanmugam VK, Tassi E, Schmidt MO, McNish S, Baker S, Attinger C et al. (2015) Utility of a human-mouse xenograft model and in vivo near-infrared fluorescent imaging for studying wound healing. Int Wound J 12(6):699–705. 10.1111/iwj.12205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheyn D, Kallai I, Tawackoli W, Cohn Yakubovich D, Oh A, Su S et al. (2011) Gene-modified adult stem cells regenerate vertebral bone defect in a rat model. Mol Pharm 8(5):1592–1601. 10.1021/mp200226c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheyn D, Yakubovich DC, Kallai I, Su S, Da X, Pelled G et al. (2013) PTH promotes allograft integration in a calvarial bone defect. Mol Pharm 10(12):4462–4471. 10.1021/mp400292p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheyn D, Shapiro G, Tawackoli W, Jun DS, Koh Y, Kang KB et al. (2016) PTH induces systemically administered mesenchymal stem cells to migrate to and regenerate spine injuries. Mol Ther 24(2):318–330. 10.1038/mt.2015.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuler ML (2017) Organ-, body- and disease-on-a-chip systems. Lab Chip 17(14):2345–2346. 10.1039/c7lc90068f [DOI] [PubMed] [Google Scholar]
- Sung JH, Esch MB, Prot JM, Long CJ, Smith A, Hickman JJ et al. (2013) Microfabricated mammalian organ systems and their integration into models of whole animals and humans. Lab Chip 13(7):1201–1212. 10.1039/c3lc41017j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syftestad GT, Weitzhandler M, Caplan AI (1985) Isolation and characterization of osteogenic cells derived from first bone of the embryonic tibia. Dev Biol 110(2):275–283 [DOI] [PubMed] [Google Scholar]
- Tai K, Pelled G, Sheyn D, Bershteyn A, Han L, Kallai I et al. (2008) Nanobiomechanics of repair bone regenerated by genetically modified mesenchymal stem cells. Tissue Eng Part A 14(10):1709–1720. 10.1089/ten.tea.2007.0241 [DOI] [PubMed] [Google Scholar]
- Tobias G, Uwe H, Tobias G (2016) Pantoprazol inhibits the stimulating effect for bone formation of diclofenac in vitro evaluated by the novel method of 99m-Tc-HDP-labeling in vitro. J Nucl Med 57:1239 [Google Scholar]
- Wang B, Lee WY, Huang B, Zhang JF, Wu T, Jiang X et al. (2016) Secretome of human fetal mesenchymal stem cell ameliorates replicative senescen. Stem Cells Dev 25(22):1755–1766. 10.1089/scd.2016.0079 [DOI] [PubMed] [Google Scholar]
- Wobma H, Vunjak-Novakovic G (2016) Tissue engineering and regenerative medicine 2015: a year in review. Tissue Eng Part B Rev 22(2):101–113. 10.1089/ten.TEB.2015.0535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolf EC, Curley KL, Liu Q, Turner GH, Charlton JA, Preul MC et al. (2015) The ketogenic diet alters the hypoxic response and affects expression of proteins associated with angiogenesis, invasive potential and vascular permeability in a mouse glioma model. PLoS One 10(6):e0130357 10.1371/journal.pone.0130357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Reynolds D, Awad H, Rubery PT, Pelled G, Gazit D et al. (2007) Structural bone allograft combined with genetically engineered mesenchymal stem cells as a novel platform for bone tissue engineering. Tissue Eng 13(3):435–445 [DOI] [PubMed] [Google Scholar]
- Xu H, Othman SF, Hong L, Peptan IA, Magin RL (2006) Magnetic resonance microscopy for monitoring osteogenesis in tissue-engineered construct in vitro. Phys Med Biol 51(3):719–732. 10.1088/0031-9155/51/3/016 [DOI] [PubMed] [Google Scholar]
- Zhang X, Schwarz EM, Young DA, Puzas JE, Rosier RN, O’Keefe RJ (2002) Cyclooxygenase-2 regulates mesenchymal cell differentiation into the osteoblast lineage and is critically involved in bone repair. J Clin Investig 109(11):1405–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gazit Z, Pelled G, Gazit D, Vunjak-Novakovic G (2011) Patterning osteogenesis by inducible gene expression in microfluidic culture systems. Integr Biol (Camb) 3(1):39–47. 10.1039/c0ib00053a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YS, Arneri A, Bersini S, Shin SR, Zhu K, Goli-Malekabadi Z et al. (2016) Bioprinting 3D microfibrous scaffolds for engineering endothelialized myocardium and heart-on-a-chip. Biomaterials 110:45–59. 10.1016/j.biomaterials.2016.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YS, Aleman J, Shin SR, Kilic T, Kim D, Mousavi Shaegh SA et al. (2017a) Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc Natl Acad Sci USA 114(12):E2293–E2302. 10.1073/pnas.1612906114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YS, Zhang YN, Zhang W (2017b) Cancer-on-a-chip systems at the frontier of nanomedicine. Drug Discov Today. 10.1016/j.drudis.2017.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.