Abstract
Background and aim:
IgE-mediated immune responses contribute to the pathogenesis of eosinophilic esophagitis (EoE). Interleukin (IL)-4 is a well-established cytokine involved in B cell activation, immunoglobulin (Ig) E production and isotype class switching. Earlier reports indicated that IL-15, B cells and IgE are induced in EoE pathogenesis. Therefore, we hypothesized that induced IL-15 and IgE may have a significant correlation in promoting EoE pathogenesis.
Methods:
Accordingly, we performed ELISA, qPCR, flowcytometric and immunostaining analyses to examine IgE, B cells, eosinophils and mast cells in the esophagus of IL-15 overexpressed mice following EoE induction.
Results:
Herein, we show that IL-15 overexpressed mice indeed have induced baseline IL-4, B cells, eosinophils, mast cells and IgE levels in the blood and esophagus. Further, we observed that IL-15 overexpressed mice show induction of IgE, and accumulation of degranulated eosinophils and mast cells in allergen-induced experimental EoE. Notably, despite induced blood IgE, esophageal eosinophilia is not induced in intestinal fatty acid binding protein IL-15 overexpressed gene (Fabpi-IL-15) mice. Fabpi-IL-15 transgenic mice showed IgE in the blood and intestine and intestinal eosinophilia, but no esophageal eosinophilia at baseline and comparable eosinophils in the esophagus of saline and allergen challenged Fabpi-IL-15 mice. Similarly, allergen challenged IL-15 gene-deficient mice show reduced IgE and esophageal eosinophilia in allergen-induced experimental EoE.
Conclusions:
Taken together, we for the first time provide direct evidence that tissue-specific IL-15 induced IgE mediated responses, not systemic IgE is critical in promoting EoE pathogenesis.
Keywords: B cell, eosinophils, esophagus, Immunoglobulin, Interleukin, mast cells
Introduction
IL-15 is a pleiotropic cytokine and is similar in structure to IL-2; both IL-15 and IL-2 share a number of biological activities, including the ability to stimulate the proliferation and differentiation of activated T cells 1–3. In addition, IL-15 is required in the maintenance of natural killer (NK) cells and some T-cell subsets, including their activation in an antigen-independent manner. 4, 5 This process is believed to contribute to intestinal inflammatory responses, including those found Celiac disease shares features with Eosinophilic esophagitis (EoE), such as being triggered by food antigens, the involvement of epithelial cells (although squamous epithelium in EoE), and the overexpression of NK cells activation antigens such as the MHC-like molecule MIC. 6 Notably, mice deficient in IL-15 or the IL-15 receptor (IL-15R)−/− have defective naïve and memory CD8+ T cells, intestinal intraepithelial lymphocytes and NK cells. 4, 5 Interestingly, IL-15 mRNA levels correlated with esophageal eosinophilia in human EoE and IL-15 levels were reduced in improved EoE patients. 7 The quantitative PCR analyses showed that levels of IL-15 and its receptor IL-15Rα were increased in tissues from patients with EoE, as well as in a murine model of EoE. 7 In addition to the induction of IL-15, B cells and Immunoglobulin (Ig) E expression are increased in esophageal biopsies of pediatric and adult EoE patients.8 The majority of EoE patients (90%) have evidence of food and aeroallergen hypersensitivity, and a substantial proportion (10–30%) have a history of food anaphylaxis.9 Recent literature on pediatric patients with EoE confirms that nearly all patients respond to an elemental diet with resolution of symptoms and normalization of biopsies,9 reintroduction of foods causes symptoms and esophageal eosinophilia to return, strong proof that food allergies are the main causes of EoE in humans,10 the essential role of IgE has been reported in atopic disorders; however, its role in EoE is still unclear and IgE was not found to be predictive of EoE.11 Studies also indicated that IgE-deficient or B cells-deficient mice are not protected from the induction of experimental EoE; therefore IgE may not have a critical role in experimental or human EoE. 12–14 However, the utility of IgG4 for predicting the disease cannot be ignored. Intraepithelial IgG4 deposits have been detected in EoE patients, allowing differentiation of the disease from Gastroesophageal Reflux Disease (GERD) with a 88% sensitivity and 100% specificity. 15 In addition, reports indicate that IL-15 responsive iNKTs are induced in EoE patients, 16–19 and promote esophageal eosinophilic inflammation in experimental EoE.18 iNKT cells have been shown to provide help to B-cell in both T-cell–dependent and T-cell–independent antibody responses.20,21 Therefore, it would be of great interest to know if IL-15 has a role in iNKT mediated IgE associated EoE pathogenesis. Accordingly, we generated Doxycycline (DOX)-inducible IL-15 overexpressed mice to establish that IL-15 has a role in IgE mediated EoE pathogenesis. We show DOX induced IL-15 overexpression promotes significantly higher levels of blood IgE and esophageal eosinophilia in mice compared to no- DOX exposed mice following allergen-induced experimental EoE.
Materials and Methods
Mice:
rtTA-CC-10 IL-15 bitransgenic mice were generated as per the construct shown in Figure 1, A and back cross to Balb/c background in our laboratory. IL-15 gene-deficient (IL-15−/−) Balb/c background was gifted to us by Dr. Fred D. Finkelman (Cincinnati Children’s Hospital Medical Centre, Cincinnati, OH, USA). Specific pathogen-free Balb/c mice 8–10 weeks old were obtained from the Jackson laboratory (Bar Harbor, ME, USA), used as a wild type (WT) mice. The Institutional Animal Care and Use Committee (IACUC) approved the animal protocol in accordance with National Institute of Health (NIH) guidelines. Therefore, all the experiments performed are according to animal ethics rules and regulations.
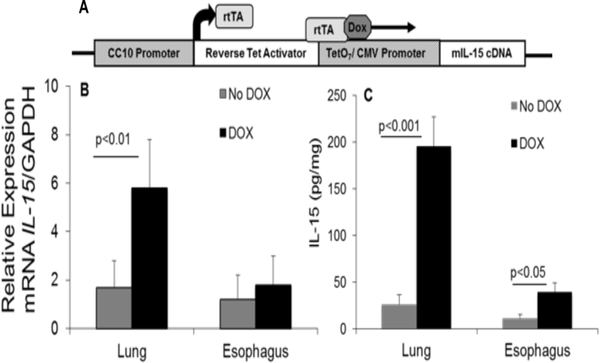
Figure 1. rtTA-CC10-IL-15 bi-transgenic mice show induced IL-15 protein in the lung and esophagus following DOX-food exposure:
The construct of (tetO)7 CMV-IL15 transgenic mice and TA-CC10 transgenic mice to generate rtTA-CC10-IL-15 bi-transgenic mice is shown (A). The rtTA-CC10-IL15 bi-transgenic mice were exposed to DOX and No-DOX food for the mouse phenotype analysis. The 3 weeks exposure to DOX food of IL-15 bi-transgenic mice induces several fold increase in IL-15 expression in the lung and esophagus of mice. The quantitative real-time PCR and ELISA analysis of IL-15 mRNA and protein levels (B, C) in the lungs and esophagus of 3 weeks DOX or No-DOX exposed or CC10-rtTA-IL-15 mice are shown. Data are expressed as mean ± SD, n =10 mice/group, p<0.05, p<0.01, p<0.001.
Generation of rtTA-CC10-IL-15 bitransgenic mice:
DOX regulated rtTA-CC10-IL-15 bitransgenic mice were generated in our laboratory. The single transgenic lines were the tetracycline reverse-trans activator transgenic mouse line CC10-rtTA (expressing rtTA cDNA under the regulation of the lung-specific rat 2.3-kb CC10 promoter) and the responder transgenic mouse line teto-IL-15 [containing germline IL-15 cDNA under the regulation of the tetracycline operator (tetO)]. These transgenic constructs were used in two single transgenic mice and were crossed to generate DOX-inducible rtTA-CC10-IL-15 bitransgenic mice for the experiments. The final constructs of IL-15 bitransgenic mice has been shown in Figure 1A.
Quantitative PCR:
Total RNA was isolated from the lung samples using TRIZOL (Invitrogen, Indianapolis, IN). The RNA samples (500 ng) were subjected to reverse transcription analyses using iScript reverse transcriptase (Bio-Rad, Hercules, CA) according to the manufacturer’s instructions. IL-15 mRNA was quantified by real-time PCR using IQ5 (Bio-Rad, Hercules, CA). Results were then normalized to mouse Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) amplified from the same cDNA mix and expressed as relative gene expression. cDNA was amplified using the primers as follows. mGAPDH, Forward 5′-ACCCAGAAGACTGTGGATGG-3′; Reverse 5′-CACATTGGGGGTA GGAACAC-3′ and mIL-15, Forward 5′-TTGCAGTGCATCTCCTTACG-3′; Reverse 5′-CCTTCCAAA CACAGCAGGAT-3′.
Induction of experimental EoE:
Experimental EoE was induced in mice following an established protocol described previously 22–24. In brief, mice were lightly anesthetized with isofluorane inhalation (methoxy-fluorane; Pittman-Moore, Mundelein, IL), and 100 μg (50 μl) of Aspergillus fumigatus (Greer Laboratories, Lenoir, NC) or 50 μl of normal saline alone was applied to the nares using a micropipette with the mouse held in the supine position. After instillation, mice were held upright until alert. After three treatments per week for 3 weeks, mice were sacrificed between 18 and 20 h after the last intranasal challenge.
Immunohistochemical analysis for eosinophils:
5-μm esophageal paraffin tissue sections of mouse esophagus were immunostained with antiserum against mouse eosinophil major basic protein (anti-MBP) as per the method described previously. 25–27 In brief, endogenous peroxidase. In. the. tissues was quenched with 0.3% hydrogen peroxide in methanol followed by non-specific protein blocking using 3 % normal goat serum. Tissue sections were then incubated with rat anti-MBP (1:6000 dilution) overnight at 4°C (purchased from Mayo Clinic, Scottsdale, AZ, USA), followed by a 1:250 dilution of biotinylated goat anti-rat IgG secondary antibody and avidin-peroxidase complex (Vector Laboratories, Burlingame, CA) for 30 minutes each. Slides were further developed with nickel diaminobenzidine-cobalt chloride solution to form a black precipitate, and counterstained with nuclear fast red. Negative controls included replacing the primary antibody with normal rabbit serum to control for endogenous biotin and peroxidase activity. Eosinophils were quantified by counting the anti-MBP positive-stained cells in each tissue section with the assistance of digital morphometry using the Metamorph Imaging System (Universal Imaging Corp, West Chester, PA) and expressed as eosinophils/mm2 tissue area as described earlier. 28, 29
Mast cell staining:
5 μm esophageal paraffin tissue sections were de-paraffinized and stained with hexazotized new fuchsin (Sigma-Aldrich, St. Louis, MO) with 4% sodium nitrite in naphthol-AS-D chloroacetate (Sigma-Aldrich, St. Louis, MO) and phosphate buffered saline solution for 30 minutes and counterstained with haematoxylin as per the method described previously. 28, 29 The histological analysis was performed using light microscopy. Mast cells were quantified by counting the positive-stained cells in each tissue section with the assistance of digital morphometry using the Metamorph Imaging System (Universal Imaging Corp, West Chester, PA) and expressed as mast cell/mm2 tissue area as described earlier. 28, 29
ELISA analysis of serum and tissue IgE and tissue cytokine (IL-4 and IL-15):
Total serum and tissue IgE levels were measured using a BD OptEIA ELISA set (BD Biosciences, San Diego, CA) as per the manufacturer’s protocol. Briefly, after blocking non-specific protein binding with 10% fetal calf serum (FCS), each mouse serum sample, esophageal cell lysate or purified mouse IgE was applied to an anti-mouse IgE monoclonal antibody coated 96-well ELISA plate (Immuron, DYNEX Technologies, Chantilly, VA). The plate was incubated for 2 hours at room temperature, and washed with 0.05% Tween-20 in PBS; biotinylated anti-mouse IgE monoclonal antibody was applied to each well followed by avidin-HRP conjugate reagent. Finally, tetramethylbenzidine (TMB) substrate solution (BD Biosciences, San Diego, CA) was added to each well, and the color was developed in the dark at room temperature. The IgE concentration of each sample was calculated by using a standard curve. Protein levels of IL-4, IL-15 were determined by using the commercially available ELISA analysis (R&D Systems, Minneapolis, MN) as per the manufacture’s protocol. Briefly, esophageal cell lysate was applied to a 96-well ELISA plate pre-coated with cytokine-specific monoclonal antibody and blocked against non-specific protein binding with 10% FCS. The plate was incubated for 2 hours at room temperature and washed with 0.05% Tween −20- PBS. Biotinylated cytokine-specific monoclonal antibody was applied to each well followed by avidin-HRP conjugate reagent. Finally, TMB,
In vitro B cell activation by IL-15:
Purified B cells were isolated from spleen and plated at 1.5 × 105/ml and cultured with various concentrations of IL-15 (0 −500 ng/ml) and IL-4 (100 ng/ml). Cells were cultured in 5 ml/well in 6 well flat-bottomed culture plates. The activation of B cells were analysed by flowcytometer.
Flow cytometric analysis:
The isolated esophageal cells were stained with cell surface molecule-specific antibodies for flow cytometric analysis. Total esophageal cells were isolated substrate solution (BD Biosciences, San Diego CA) was added to each well, the color was developed in the dark at room temperature and the optical density (OD) was read at 450 nm immediately following. The cytokine concentration of each sample was calculated by using a standard curve as described earlier.30 as per the protocol described earlier 30. The following reagents were used for specific antigen analysis: anti-CD45-APC-Cy7, anti-B220-PE-Cy7, anti-CD69-PE and respective isotype controls obtained from BD Biosciences, San Diego, CA. Live/dead cell marker 7AAD was used to exclude dead cells. The cells were incubated for specific antigens with the required combination of antibodies at 4°C for 45 minutes followed by two washes. FACS analysis was performed using a FACSCalibur (BD Biosciences, San Diego, CA) and analyzed using CellQuest software (BD Biosciences, San Diego, CA).
Statistical analysis:
For all cell counts, stained slides were analyzed randomly and in a blinded fashion. For non-parametric data, the Mann–Whitney U test was employed for comparisons between two groups, and the Kruskal–Wallis test was used for comparisons among three or more groups. Parametric data were compared using t tests or analysis of variance. Values are reported as mean ± standard deviation (SD). P values < 0.05 were considered statistically significant.
Results
rtTA-CC10-IL-15 bi-transgenic mice show induced IL-15 protein in the lung and esophagus following DOX-food exposure:
DOX-induced IL-15 bitransgenic mice were examined for IL-15 mRNA and protein in the lung and esophagus following No DOX and DOX containing food by performing real-time PCR and ELISA analysis, respectively. Our analysis indicated a significant ~ 4-fold increase in mRNA and 8-fold increase in protein levels of IL-15 in the lung of 3 weeks DOX-exposed mice compared to No DOX mice (Figure 1 B, C). Similarly, we observed a significant increase in ~ 5-fold in IL-15 protein and no increase of IL-15 mRNA in the esophagus of DOX-exposed mice compared to No-DOX mice (Figure 1 B, C).
IL-15 overexpression in the esophagus induces in vivo B cell induction, activation and IgE production.
It has been shown that in vitro IL-15 induces B cell proliferation; 31 therefore, we tested the hypothesis that IL-15 overexpression promotes IL-4 mediated B cell proliferation, activation and IgE induction. Our ELISA and flow cytometry analyses detected induced IL-4, IgE levels in the blood, as well as B cells induction and activation of B cells in the esophagus of DOX exposed mice (Figure 2 A-C) Further, analysis showed that absolute B cells and activated B cells. are increased in the esophagus of DOX exposed CC-10-IL-15 bitransgenic mice compare to the no-DOX exposed mice (Figure 2 D, E). Further, we also examined whether IL-15-induced B cell activation is independent to IL-4; therefore, the spleenocytes of WT mice B cells were cultured in the presence of various concentrations of IL-15 and IL-4. The cells were stained for B220 and CD19 for B cell marker and CD69 activation marker and further analyzed by flow cytometer. The cells were enumerated activation B cell gated population as shown in (Figure 2 F). The analysis indicated indeed B cells of WT mice show dose dependent activation of B cells. Data are expressed as mean ± SD, n = 6 mice/group.
Figure 2. IL-15 overexpression in the esophagus promotes in vivo B cell induction, activation and IgE production.
IgE and IL-4 levels in the blood of IL-15 overexpressed mice were measured by ELISA analysis (A, B). Analysis of B cells and activated B cells in the esophagus of IL-15 transgenic DOX and No-DOX exposed mice was performed by flow cytometry analyses using fluorochrome conjugated anti-CD45-APC-Cy7, anti-B220-PE-Cy7, anti-CD69-PE and respective matched isotype controls antibodies (C). The absolute and activated B cells from 3 independent experiments are shown (D, E). Further, we also examined whether IL-15-induced B cell activation is independent to IL-4; therefore, the spleenocytes of WT mice B cells were cultured in the presence of various concentrations of IL- 15 and IL-4. The activation of B cells was analyzed by flow cytometer (F). Data are expressed as mean ± SD, n = 6 mice/group. Data are expressed as mean ± SD, n = 10 mice/group, p<0.05, p<0.01, p<0.006.
Allergen-challenged DOX-exposed IL-15 overexpressed mice showed significant increase of blood and esophageal IgE levels compare to no-DOX exposed mice.
Next we tested the hypothesis that IL-15 overexpression induced IgE is associated with increased esophageal eosinophilia in experimentally induced EoE. Accordingly, CC-10-IL-15 bitransgenic mice were challenged with Aspergillus extract following the protocol describe earlier 22–24, and first examined IgE levels in the blood and esophagus. ELISA analysis indicated that saline challenged DOX exposed mice showed induced IgE levels in the blood and esophagus (Figure 3 A, B) compared to No-DOX exposed mice. In addition, we observed both blood and esophageal IgE levels is indeed significantly induced in DOX exposed compare to no DOX exposed allergen challenged mouse model of EoE (Figure 3 A-B).
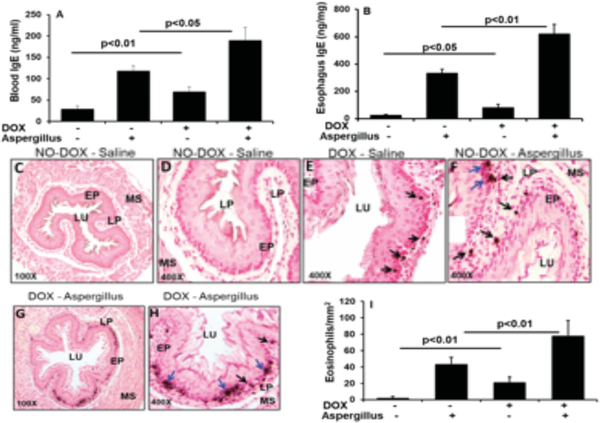
Figure 3. Allergen-challenged DOX exposed IL-15 transgenic mice showed induced IgE, esophageal eosinophilia.
ELISA analysis of blood and esophageal IgE levels in rtTA-CC10-IL-15 bitransgenic mice that were exposed to DOX and No-DOX food in experimental EoE (A, B). Tissues from allergen-challenged IL-15 transgenic mice were analyzed for eosinophils and in the esophagus. Anti-MBP immunostaining was used to analyze eosinophils in the esophageal tissue sections of allergen-challenged IL-15 transgenic mice. The no-DOX and DOX exposed saline challenged mice showed very few baseline eosinophils and limited to the esophageal lamina propria (C, D, F). In DOX and No-DOX exposed allergen challenged mice showed accumulation of a high number of eosinophils in the lamina propria and epithelial mucosa (intraepithelial eosinophilia, marked with black arrows) as well as eosinophils microabscess, epithelial layer disruption and extracellular eosinophilic granules (marked with blue arrows), in the esophageal mucosa (E, G, H). Representative photomicrographs (C and G x100 original magnifications) and D, E, F and H are x400 original magnifications are shown. EP, Epithelium; LU, Lumen; LP, Lamina propria; MS, Muscularis Mucosa. Quantitation of Anti- MBP immunostained eosinophils in the esophageal sections of DOX and No-DOX exposed rtTA-CC10-IL-15 bitransgenic mice in experimental EoE (I). Data is expressed as means ± SD pooled from 3 experiments ± SD, n=12. p<0.04, p<0.001, p<0.0001.
Esophageal eosinophilia in allergen-induced no DOX and DOX exposed IL-15 bitransgenic mice.
Next, we examined that induced IgE in DOX exposed mice is associated with the esophageal eosinophilia in allergen-induced no-DOX and DOX exposed IL-15 bitansgenic mice. The no-DOX and DOX exposed saline challenged mice showed very few baseline eosinophils and limited to the esophageal lamina propria (Figure 3 C, D, F), whereas, allergen-induced DOX exposed mice showed EoE characteristics such as accumulation of a high number of eosinophils in the lamina propria and epithelial mucosa (intraepithelial eosinophilia, marked with black arrows) as well as eosinophils microabscess, epithelial layer disruption and extracellular eosinophilic granules (marked with blue arrows), in the esophageal mucosa (Figure 3 E, G, H). These EoE characteristics are more insistently observed in DOX exposed IL-15 transgenic mice (Figure 3 G, H) compare to no No-DOX exposed allergen challenged mice (Figure 3 E). The number of eosinophils stained with anti-MBP were quantified in the esophagus of no-DOX and DOX exposed Aspergillus challenged CC-10-IL-15 transgenic mice were 43.6 ± 7.9/mm2, 78.4 ± 18.3/mm2 compare to saline challenged No-DOX and DOX exposed mice 2.4 ± 1.6, 21.4 ± 6.4, respectively (mean ± SD, n=12 mice/group) (Figure 3 I).
Mast cells analysis in the esophagus of allergen-induced no DOX and DOX exposed IL-15 bitransgenic mice.
Further, we also examined mast cell accumulation in the saline and Aspergillus challenged no-DOX and DOX exposed IL-15 bitransgenic mice. Saline challenged no-DOX and DOX exposed IL-15 bitransgenic mice showed some baseline intact mast cells, mostly restricted to the esophageal lamina propria (Figure 4 A, B), whereas induced mast cells were observed in the muscle cell mucosa of allergen-challenged DOX exposed IL-15 transgenic mice compared to No-DOX exposed Aspergillus challenged mice (marked with black arrows) (Figure 4 C, D). Of note, extracellular mast cells granules are also observed in DOX exposed IL-15 transgenic mice (marked with blue arrows). A representative extracellular chloroacetate esterase stained mast cells granule is indicated in DOX exposed allergen challenged mice (Figure 4 D). The number of mast cells stained with chloroacetate esterase were quantified in the esophagus of no-DOX and DOX exposed Aspergillus challenged CC-10-IL-15 transgenic mice were 25.3 ± 2.2/mm2, 34.5 ± 7.7/mm2 compare to saline challenged No-DOX and DOX exposed mice 8.7 ± 1.1, 20.9 ± 4.3, respectively (mean ± SD, n=12 mice/group) (Figure 4 E). The data are expressed as the mean ± SD, mice =12/group).
Figure 4. DOX exposure promotes mastcellinflammation in IL-15 transgenic allergen challenged m ice.
Tissues from allergen-challenged IL-15 transgenic mice were analyzed for mast cells in the esophagus. Chloroacetate esterase staining was performed in the esophageal tissue sections for mast cells analysis in No-DOX and DOX exposed rtTA-CC10-IL-15 bitransgenic allergen-challenged mice. No-DOX exposed mice showed few baseline mast cells compared to an induced number of mast cells in esophageal lamina propria of DOX exposed as well as the allergen challenged No-DOX and DOX treated mice (A-D). The histopathological analysis of esophageal tissue sections identified m ast cells in the lamina propria ofallergen-challenged mice.Interestingly,numberofmastcells was detected in the muscle cell mucosa of DOX exposed allergen-challenged mice compared with the No-DOX exposed mice (muscle cellmucosa mastcells,marked with black arrows)(C,D).Additionally,activated degranulating mastcells including extracellular granules were noted in the muscular mucosa of DOX exposed allergen-challenged mice (marked with blue arrows) (4, D). Quantitation of chloroacetate esterase stained mastcells in the esophagealsections ofDOX and No-DOX exposed rtTA-CC10-IL-15 bitransgenic mice in experimental EoE (4E). All representative photomicrographs are shown ×400 of original magnification. Data is expressed as means ± SD pooled from 3 experiments ± SD,n=12.p<0.04, p<0.01.
Allergen challenged IL-15 gene-deficient (IL-15−/−) mice have reduced IgE levels and esophageal eosinophilia.
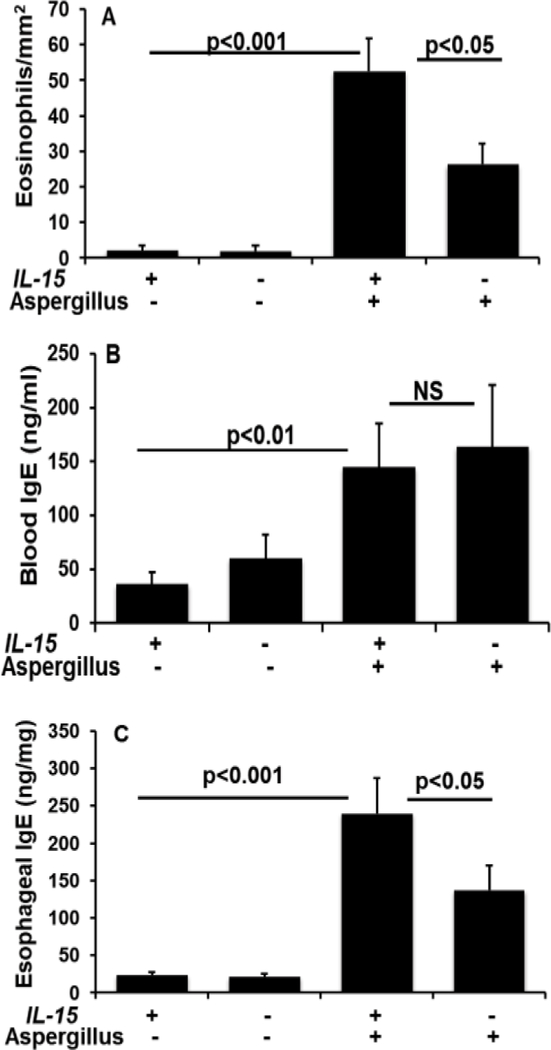
Further, to establish induced IgE association with the esophageal eosinophilia in experimental EoE, we examined allergen-challenged wild type (WT) mice and IL-15 gene-deficient (IL-15−/−) mice for IgE levels and esophageal eosinophilia. Herein, we show that allergen-induced esophageal eosinophilia is significant increase in WT mice compared to the allergen challenged IL-15−/− mice (Figure 5 A). The level of eosinophils in the esophagus of Aspergillus challenged WT mice were 52.6 ± 9.2/mm2 compared to saline WT challenged mice 2.1 ± 1.4 whereas the eosinophil levels in the allergen challenged IL-15−/− mice were 26.2 ± 5.8 compare to the saline challenged IL-15−/− mice 1.8 ± 1.6 respectively (mean ± SD, n=12 mice/group). The levels of IgE in the blood of WT mice and IL-15−/− mice were found comparable (Figure 5 B) but IgE levels in the esophagus of WT mice were significant induced following allergen challenge compared to the allergen-challenged IL-15−/− mice (Figure 5 C), indicating an association of IL-15 induced tissue IgE in promoting allergen-induced esophageal eosinophilia.
Figure 5. Allergen challenged IL-15 gene-deficient (IL-15−/−) mice have reduced IgE levels and esophageal eosinophilia.
Eosinophil quantitation was performed using anti-MBP antibody immunostaining of esophageal sections of both IL-15−/− and WT mice following allergen-induced experimental EoE (A). IgE levels in the blood and esophagus of allergen challenged IL-15−/− and WT mice were measured by ELISA analysis (B, C). Data is expressed as mean ± SD, n=3 experiments. p<0.001, p<0.05.
Intestinal-overexpression of IL-15 does not show induced esophageal IgE and esophageal eosinophilia in mice.
We were next interested in determining whether intestinal IL-15 overexpression induced blood and intestinal levels of IgE but not esophageal IgE also levels promotes blood IgE mediated esophageal eosinophilia. The intestinal IL-15 transgenic mice were developed in our laboratory using the intestinal specific Fabpi promoter as per the method described earlier 32. The induction of blood and intestine IgE compare to WT mice and comparable IgE levels in the esophagus is presented (Figure 6 A). The eosinophil analysis also shows.induced intestinal eosinophilia in Fabpi-IL-15 transgenic compared to WT mice; but no esophageal eosinophilia either wild type or Fabpi-IL-15 transgenic mice (Figure 6 B). Further, we also show that following allergen challenge both WT mice and Fabpi-IL-15 mice develops induced blood and esophageal IgE along with esophageal eosinophilia (Figure, 6 C, D). These data indicate that tissue specific IL-15-induced IgE is required for promoting EoE pathogenesis; whereas, blood IgE levels is not sufficient to induce IgE associated EoE pathogenesis in mice.
Figure 6. Intestinal-overexpression (Fabpi) of IL-15 does not induce esophageal eosinophilia.
IgE levels in the blood, esophagus and jejunum of Fabpi-IL-15 and WT mice were measured by ELISA analysis (A). Eosinophil quantitation was performed by using anti-MBP antibody immunostaining in the esophageal sections of both Fabpi-IL-15 and WT mice following the allergen- induced experimental EoE (B). IgE levels in the blood of allergen-challenged both Fabpi-IL-15 and WT mice were measured by ELISA analysis (C). Eosinophil quantification was also similarly performed by using anti-MBP antibody immunostaining in the esophageal sections of both Fabpi-IL-15 and WT mice following allergen-induced experimental EoE (D). Data is expressed as mean ± SD, n=3 experiments. p<0.001, NS (not significant).
Discussion
EoE is an established allergic disease with rising prevalence throughout the world 33. EoE is differentiated from reflux esophagitis by the enormity of mucosal eosinophilia and also on the basis of specific esophageal pathology. 34, 35 The consensus guidelines for diagnosis are recommended. 36 but novel therapy for EoE has yet to be established. IgE plays an essential role in atopic disorders; however, its role in EoE is still unclear. Recent literature on pediatric patients with EoE confirms that nearly all patients respond to an elemental diet with resolution of symptoms and normalization of biopsies. 10 However, reintroduction of foods causes symptoms and esophageal eosinophilia to reoccur, provides strong evidence that food allergies are the causes of EoE in humans. Additionally, EoE patients have evidence of food and aeroallergen hypersensitivity supports the findings that IgE is involved in EoE pathogenesis. 9 However, our current findings and previous experimental and clinical reports indicated that induced blood IgE is not critical in promoting EoE. 11–14. However, we provide evidences that esophageal induction of IL-15 activates B cells that induce tissue specific IgE and IgE isotype switching (data not shown). This tissue specific induction of IgE binds to tissue mast cells and basophils receptors, activate and degranulate these cells. The degranulation of these inflammatory cells via IL-15-induced IgE is critical in promoting EoE pathogenesis. Previously, the roles of IL-15 and IL-15 responsive iNKT cell initiated Th2-type allergic responses were implicated in promoting EoE. 12, 24, 28, 29 IL-15 induction is reported in the blood and esophageal biopsies of pediatric and adult EoE patients and IL-15 responsive iNKTs were found induced in EoE patients.16–19 Both IL-15 and iNKT cells correlate with esophageal eosinophilic inflammation in human EoE 17, 18. Several reports implicate iNKT cells to the IgE and IgG induction in innate and acquired immunity. 37, 38 This is consistent with the current data that chronic IL-15 expression induces both IgE and esophageal eosinophilia following the induction of allergen-induced EoE. The IgE association with inducing esophageal eosinophilia is further confirmed by examining allergen-challenged IL-15−/− and WT mice. The allergen challenged IL-15−/− mice showed a remarkable decrease in IgE production and significantly reduced esophageal eosinophilia compare to the allergen challenged WT mice. Earlier, we have also reported a direct link between indoor insect (cockroach and dust mites) and food (peanut and corn) allergies in the development of iNKT cell mediated experimental EoE. 17 Earlier, we have shown that ~30% of EoE patients have insect and 40–50% have peanut or corn hypersensitivity and induced. levels. of immunoglobulin (Serum IgE and antigen-specific IgG1). 17, 39 Intensive efforts have been made by the medical community to determine whether specific IgEs are causative in the pathogenesis of EoE. We show that. Induced. Baseline. Blood. and intestinal IgE levels did not promote esophageal eosinophilia in Fabpi-IL-15 transgenic mice and both WT and Fabpi-IL-15 transgenic mice show comparable esophageal and blood eosinophilia following the induction of EoE. The IgE isotype switching and utility of IgG4 deposition has been detected in EoE patients with 88% sensitivity and 100% specificity versus GERD patients. 15 Th2 cytokine induced IgG4 is considered a key marker for desensitization in immunotherapy and found induce EoE in about 2.7% of the patients undergoing this treatment. 40 Earlier, B cell induction and IgE switching have been identified in the esophageal mucosa of human EoE. 8 Earlier, IL-15 responsive iNKT cells have been shown to provide B-cell help in both T-cell–dependent and T-cell–independent antibody responses. 20, 21.
In summary, we first demonstrated that IL-15 overexpression promise B cells proliferation, activation and IgE induction. Second, we showed that an lL-15 overexpressed mouse promotes tissue-specific IgE and esophageal eosinophilia and mast cells induction and degranulation in experimental EoE. Third, we provided the critical role of IL-15 in tissue specific IgE associated esophageal eosinophilia, and lastly, we showed that systemic IgE induction has no role in the induction of experimental EoE. Taken together, the current study provides a rationale and a better understanding on the significance of the earlier findings that induce tissue-specific IL-15, IgE has an direct association in eosinophils mast cells recruitment, activation and degranulation in EoE. Most importantly, the current study identifies a novel role for IL-15 induced esophageal IgE in promoting allergen- induced EoE and provide the evidence that systemic IgE induction has no role in EoE pathogenesis.
Acknowledgements
This work was supported by the grants NIH R01 AI080581 (AM) and Edward G. Schlieder Educational Foundation for their support in our eosinophil associated diseases.
References
- 1.Giri JG, Kumaki S, Ahdieh M, et al. ’ Identification and cloning of a novel IL-15 binding protein that is structurally related the alpha chain of the IL-2 receptor. Embo J 1995;14:3654–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giri JG, Anderson DM, Kumaki S, et al. IL-15, a novel T cell growth factor that shares activities and receptor components with IL-2. J Leukoc Biol 1995;57:763–6. [DOI] [PubMed] [Google Scholar]
- 3.Anderson DM, Kumaki S, Ahdieh M, et al. Functional characterization of the human interleukin-15 receptor alpha chain and close linkage of IL15RA and IL2RA genes. J Biol Chem 1995;270:29862–9. [DOI] [PubMed] [Google Scholar]
- 4.Lodolce JP, Boone DL, Chai S, et al. IL-15 receptor maintains lymphoid homeostasis by supporting lymphocyte homing and proliferation. Immunity 1998;9:669–76. [DOI] [PubMed] [Google Scholar]
- 5.Kennedy MK, Park LS. Characterization of interleukin-15 (IL-15) and the IL-15 receptor complex. J Clin Immunol 1996;16:134–43. [DOI] [PubMed] [Google Scholar]
- 6.Kawano T, Cui J, Koezuka Y, et al. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science 1997;278:1626–9. [DOI] [PubMed] [Google Scholar]
- 7.Zhu X, Wang M, Mavi P, et al. Interleukin-15 expression is increased in human eosinophilic esophagitis and mediates pathogenesis in mice. Gastroenterology 2010;139:182–93e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vicario M, Blanchard C, Stringer KF, et al. Local B cells and IgE production in the oesophageal mucosa in eosinophilic oesophagitis. Gut 2010;59:12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spergel JM. Eosinophilic esophagitis in adults and children: evidence for a food allergy component in many patients. Curr Opin Allergy Clin Immunol 2007;7:274–8. [DOI] [PubMed] [Google Scholar]
- 10.Spergel JM, Andrews T, Brown-Whitehorn TF, et al. Treatment of eosinophilic esophagitis with specific food elimination diet directed by a combination of skin prick and patch tests. Ann Allergy Asthma Immunol 2005;95:336–43. [DOI] [PubMed] [Google Scholar]
- 11.Safroneeva E, Straumann A, Coslovsky M, et al. Symptoms Have Modest Accuracy in Detecting Endoscopic and Histologic Remission in Adults With Eosinophilic Esophagitis. Gastroenterology 2016;150:581–590e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mishra A, Schlotman J, Wang M, et al. Critical role for adaptive T cell immunity in experimental eosinophilic esophagitis in mice. J Leukoc Biol 2007;81:916–924. [DOI] [PubMed] [Google Scholar]
- 13.Noti M, Kim BS, Siracusa MC, et al. Exposure to food allergens through inflamed skin promotes intestinal food allergy through the thymic stromal lymphopoietin-basophil axis. J Allergy Clin Immunol 2014;133:1390–9, 1399e1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noti M, Wojno ED, Kim BS, et al. Thymic stromal lymphopoietin-elicited basophil responses promote eosinophilic esophagitis. Nat Med 2013;19:1005–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zukerberg L, Mahadevan K, Selig M, et al. Oesophageal intrasquamous IgG4 deposits: an adjunctive marker to distinguish eosinophilic oesophagitis from reflux oesophagitis. Histopathology 2016;68:968–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jyonouchi S, Smith CL, Saretta F, et al. Invariant natural killer T cells in children with eosinophilic esophagitis. Clin Exp Allergy 2014;44:58–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajavelu P, Rayapudi M, Moffitt M, et al. Significance of para-esophageal lymph nodes in food or aeroallergen-induced iNKT cell-mediated experimental eosinophilic esophagitis. Am J Physiol Gastrointest Liver Physiol 2012;302:G645–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rayapudi M, Rajavelu P, Zhu X, et al. Invariant natural killer T-cell neutralization is a possible novel therapy for human eosinophilic esophagitis. Clin Transl Immunology 2014;3:e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lexmond WS, Neves JF, Nurko S, et al. Involvement of the iNKT cell pathway is associated with early-onset eosinophilic esophagitis and response to allergen avoidance therapy. Am J Gastroenterol 2014;109:646–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leadbetter EA, Brigl M, Illarionov P, et al. NK T cells provide lipid antigen-specific cognate help for B cells. Proc Natl Acad Sci U S A 2008;105:8339–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lang GA, Exley MA, Lang ML. The CD1d-binding glycolipid alpha-galactosylceramide enhances humoral immunity to T-dependent and T-independent antigen in a CD1d-dependent manner. Immunology 2006;119:116–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mavi P, Niranjan R, Dutt P, et al. Allergen-induced resistin-like molecule-alpha promotes esophageal epithelial cell hyperplasia in eosinophilic esophagitis. Am J Physiol Gastrointest Liver Physiol 2014;307:G499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mavi P, Niranjan R, Dutt P, et al. Allergen-induced resistin-like molecule- promotes esophageal epithelial cell hyperplasia in eosinophilic esophagitis. Am J Physiol Gastrointest Liver Physiol 2015;309:G281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mishra A, Hogan SP, Brandt EB, et al. An etiological role for aeroallergens and eosinophils in experimental esophagitis. J Clin Invest 2001;107:83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matthews AN, Friend DS, Zimmermann N, et al. Eotaxin is required for the baseline level of tissue eosinophils. Proc Natl Acad Sci U S A 1998;95:6273–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mishra A, Hogan SP, Lee JJ, et al. Fundamental signals that regulate eosinophil homing to the gastrointestinal tract. J Clin Invest 1999;103:1719–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.NiranjaR, MavP, RayapudM, et al. Pathogenic role of mast cells in experimental eosinophilic esophagitis. Am J Physiol Gastrointest Liver Physiol 2013;304:G1087–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mishra A, Hogan SP, Brandt EB, et al. IL-5 promotes eosinophil trafficking to the esophagus. J Immunol 2002;168:2464–9. [DOI] [PubMed] [Google Scholar]
- 29.Mishra A, Rothenberg ME. Intratracheal IL-13 induces eosinophilic esophagitis by an IL-5, eotaxin-1, and STAT6-dependent mechanism. Gastroenterology 2003;125:1419–27. [DOI] [PubMed] [Google Scholar]
- 30.Zhu X, Wang M, Crump CH, et al. An imbalance of esophageal effector and regulatory T cell subsets in experimental eosinophilic esophagitis in mice. Am J Physiol Gastrointest Liver Physiol 2009;297:G550–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Armitage RJ, Macduff BM, Eisenman J, et al. IL-15 has stimulatory activity for the induction of B cell proliferation and differentiation. J Immunol 1995;154:483–90. [PubMed] [Google Scholar]
- 32.Mishra A, Hogan SP, Brandt EB, et al. Enterocyte expression of the eotaxin and interleukin-5 transgenes induces compartmentalized dysregulation of eosinophil trafficking. J Biol Chem 2002;277:4406–12. [DOI] [PubMed] [Google Scholar]
- 33.Mishra A Mechanism of eosinophilic esophagitis. Immunol Allergy Clin North Am 2009;29:29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Furuta GT, Liacouras CA, Collins MH, et al. Eosinophilic esophagitis in children and adults: a systematic review and consensus recommendations for diagnosis and treatment. Gastroenterology 2007;133:1342–63. [DOI] [PubMed] [Google Scholar]
- 35.Rothenberg ME, Mishra A, Collins MH, et al. Pathogenesis and clinical features of eosinophilic esophagitis. J Allergy Clin Immunol 2001;108:891–894. [DOI] [PubMed] [Google Scholar]
- 36.Liacouras CA, Furuta GT, Hirano I, et al. Eosinophilic esophagitis: Updated consensus recommendations for children and adults. J Allergy Clin Immunol 2011. [DOI] [PubMed] [Google Scholar]
- 37.Kim HY, Kim HJ, Min HS, et al. NKT cells promote antibody-induced joint inflammation by suppressing transforming growth factor beta1 production. J Exp Med 2005;201:41–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taniguchi M, Harada M, Kojo S, et al. The regulatory role of Valpha14 NKT cells in innate and acquired immune response. Annu Rev Immunol 2003;21:483–513. [DOI] [PubMed] [Google Scholar]
- 39.Rayapudi M, Mavi P, Zhu X, et al. Indoor insect allergens are potent inducers of experimental eosinophilic esophagitis in mice. J Leukoc Biol 2010;88:337–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lucendo AJ, Arias A, Tenias JM. Relation between eosinophilic esophagitis and oral immunotherapy for food allergy: a systematic review with meta-analysis. Ann Allergy Asthma Immunol 2014;113:624–9 [DOI] [PubMed] [Google Scholar]