Abstract
3D printing (3DP), an additive manufacturing (AM) approach allowing for rapid prototyping and decentralized fabrication on-demand, has become a common method for creating parts or whole devices. The wide scope of the AM extends from organized sectors of construction, ornament, medical, and R&D industries to individual explorers attributed to the low cost, high quality printers along with revolutionary tools and polymers. While progress is being made but big manufacturing challenges are still there. Considering the quickly shifting narrative towards miniaturized analytical systems (MAS) we focus on the development/rapid prototyping and manufacturing of MAS with 3DP, and application dependent challenges in engineering designs and choice of the polymeric materials and provide an exhaustive background to the applications of 3DP in biology and chemistry. This will allow readers to perceive the most important features of AM in creating (i) various individual and modular components, and (ii) complete integrated tools.
Keywords: 3D printing, Additive manufacturing, Microfluidics, Lab-on-a-chip, DLP, SLA, SLS, MultiJet, FDM
1. Introduction
The scale of testing on time, reagent volumes, and costs are a big burden on the overall workflow in biology and chemistry. Miniaturization of experiments with microfluidic tools, such as mTAS, Lab-on-a-Chip, Lab-in-a-Trench [1] etc., has significantly cut down these scales [1,2]. Integration of better sensors to these tools has further helped in achieving equivalent or in some cases better functionality [3]. Therefore, fluidics and fluidic networks at microscale (mFN) have greatly impacted how we conduct multiplexed, robust, rapid, experimentation and data analyses. Several educational tutorials can be found for biologists to learn and understand the fundamental concepts of mF and manufacturing [1,4].
Even with all these possibilities and massive amount of funding going into the fluidics research and number of published articles [5–7] (Fig. 1) the market cost share of mFluidic tools along with its penetration into the general laboratory space as a replacement to conventional tools and techniques is still very restricted [8]. This is mostly attributed to the complex manufacturing processes and need of highly specialized and bulky engineering tools [9]. Carr has mentioned in his report “from cleanroom to makerspace”10 that this can only be changed by increasing the participation of the end-users, which in this case are biologists, chemists, and hobbiysts. This has been increasingly achieved with additive manufacturing (AM) or 3D printing (3DP) - a three-decade old technology first patented and commercialized by Chuck Hull as ‘solid imaging stereo lithography and STL file format’ that has gained momentum in past five years allowing researchers in biology and chemistry to work cross-functionally using trial and error approaches [7,11–13]. An ever expanding choice of printable materials [9–12], such as metals, polymers, powders, and ceramics, has revolutionized rapid prototyping of bio-safe and biodegradable tools.
Fig. 1.

Analytics of how research in 3DP has increased over a decade. In 2016 global 3DP healthcare market share was USD 170 million which is growing at 20% CAGR.
New precursor materials allow for creating systems of custom physical properties including tensile strength, heat resistance etc. to create complete devices with integrated features, such as membranes [13], modules, holders, fittings, and microscopy chamber [14] etc., and on few instances complete tools, such as microscopes [15]. Since AM/3DP enables printing complete tools without machining or molding, along with minimum dedicated trained operator-inputs [16] thus, is cheaper than conventional lithography methods [17]. This also allows for the personalization of printed tools with fast turnaround time for testing the tool viability w.r.t the application [18]. However, this enabler disruptive technology is limited in achieving physical resolution smaller than the X–, Y–, and Z-resolution of stage movement and of that the light source making most devices restricted in printing structures less than 50–100 microns in general [19,20] while as small as 25–50 microns through improved workflows combined with inprocess and post-process chemistries [21,22]. However, this engineering challenge is not a practical set-back for any of the chemical and biological applications.
3D printing has empowered non-engineers thus this review will have wider interest. Past 5 years this technology has emerged as a mainstay for rapid prototyping. Creating small integrated devices by the person and at the point of use has an unrealized potential to revolutionize the overall workflow as it provides freedom to create and change without prior knowledge. We have reviewed technological advancements in 3D printed fluidics for past 5 years. One of the focuses of this article is to provide an illustrious review of recent developments and their extended applications giving readers a holistic summary of AM technology. This is the only article covering fundamental concepts of fluidics w.r.t 3D printing and the USFDA regulations around creating 3D printed tools which will increase the readership base.
2. Challenges in 3D printing
Quality of the printed tools and mechanical strength of the structures has been critically assessed after an increase in the demand of 3D printed objects. This has led to the realization of several restrictions even though they are fewer than the overall benefits but may impact some applications. Printing speed, allowed printable dimensions and their resolution, stability and reproducibility of the prints, post processing requirements, and strength of printed objects are of critical importance [16,22] (Table 1).
Table 1.
Comparison of modern desktop 3D printing techniques.
| Printer | Form2 | Moonray | Carbon3D | Uniz slash+ | Uniz sltv15 | |
|---|---|---|---|---|---|---|
| Technology | Laser | DLP | CLIP | UDP w/RI Colling | ||
| XY-Resolution; μm | 150 | 100 | 75 | 75 | 89 | |
| (1280 × 800) | (2560 × 1600) | (2560 × 1600) | (3840 × 2160) | |||
| Z-Resolution; μm | 25 | 5 | 1 | 10 | 10 | |
| Solid output speed (cc/hr) | 19 | 10–30 | 60–100 | 1000 | 3000 | |
| Z-axis Speed | mm/hr | 20 | 25 | 500 | 720 | 720 |
| cc/hr | 420 | 600 | 11,600 | 16,700 | 45,200 | |
| Build size (inch) | 5.7 × 5.7 × 6.9 | 4 × 3 × 7.9 | 7.5 × 4.8 × 13 | 7.5 × 4.8 × 15 | 13 × 7.5 × 15 | |
| Price (USD) | 3500 | 4500 | 250,000 | 5000 | 7500 |
3D printing, synonymously called rapid prototyping, is other- wise faster when functional validation of printed tools is considered; however, it is very slow compared to bulk manufacturing methods [9]. Fused Deposition Modeling (FDM) is the cheapest and most common of all AM types. It is easy to operate and print with FDM but printing being rough results in weak structures and may potentially cause leakage [24] (Fig. 2a). Recently, Ho’s group has created leakage-free prints with a FDM D-Force 400 printer [25]. They have achieved it by creating plug like Legos that can be assembled into leakage-free complex fluidic structures, which were driven through capillary force and can be plugged thus. Several SLA printers have been developed and are marketed for fast print speeds and with better surface finish compared to FDM but these are still not considerably fast. FormLabs Form2 is one of the most functionally able printer in the market but laser-based printing inherently cannot print faster than 0.03 cc/min. Continuous Liquid Interface Production [23] (CLIP) is another SLA technology as marketed by Carbon3D is very fast, somewhat equivalent to mass production methods with print speeds of 4.4 cc/min. Most recently, UNIZ [26] developed Unidirectional Peel Technology (UDP), which is unprecedentedly faster and achieved speeds of 21 cc/min (~75 mm width/min; ~3.5 mm height/min). Like Carbon3D, UNIZ is also yet to go into mass production of printers. While printing speeds are getting faster the most commonly used printers are still restricted on that front. Resolution on these common printers is also a concern where complexity of applications is increasing each day for which 3D printed tools are being used. 3DP most commonly is restricted in terms of producing sizes at dimensional scales below 200 microns [20]. However, new technologies have improved printing resolution of structures as small as 10 micron, such as CLIP and UDP. New workflows, such as shrinkage cycles for achieving small features using bigger dimension molds [22], can also allow for achieving smaller features. Clogging due to trapped material within the channels having long print times is another of challenge [24]. Drain ports that can be sealed later have been printed along the channels to flush out trapped material [29]. This is not a reliable solution as these ports are hard to seal for later use. Post-processing of the printed structures, such as cleaning in isopropanol for SLA prints and in acetone for FDM prints, is tricky. Sticky patches of uncured resin in undertreated or uncured channels may causes non-specific loss of sample during processing [30], something very frequently observed with PDMS fluidic devices [31,32], while over treatment will destabilizes the structures. Print roughness and surface finish variability of 3DP increases in order of Multi Photon > UDP ~ CLIP ~ PolyJet < Laser Sintering < DLP < SLA < FDM. High roughness and finish are important considerations in microfluidics as even a small bump in surface can change the fluidic profile of the microchannel and can create undesirable eddies and vortices [33]. In some cases, the roughness of printed structures used as molds may introduce dead volumes in the molded structures [20,34,35] (Fig. 2ced). Moreover, transparency of the printed structures is a significant consideration when developing tools for immunoassays, and other biological applications [6].
Fig. 2.
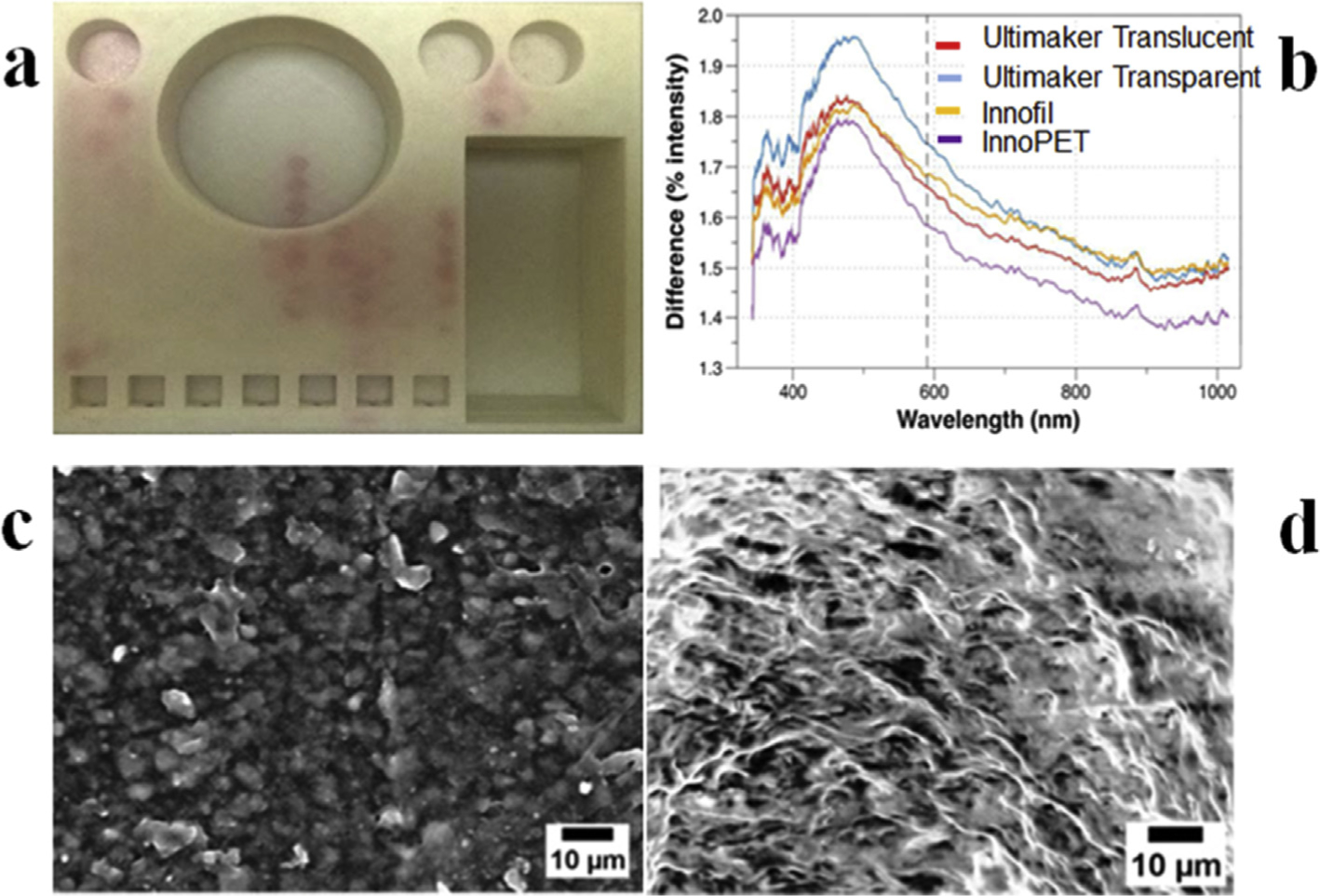
Illustration of common challenges faced with 3D printing tools. Printing fluidic networks on a single device using FDM technology usually result in reagent leakage as shown in (a)24. Carefully choosing a polymeric material is very crucial as variants of same material from different vendors may result in significant structural differences as visible in (b)27. Ultimaker Transparent, Ultimaker Translucent, Innofil, and InnoPET were employed to print optically clear cuvettes demonstrating this variation against a standard PMMA cuvette (adapted from ref [37] with permission). In (c & d) the SEM image shows effective replication of structural inaccuracies on the 3D-printed molds (c) on the PDMS casts (d).
High print to print variability due to varying vendor-specific compositions of the same polymer [27] should be another crucial point when choosing a specific 3D printing strategy (Fig. 2b). Inability to print gas permeable structures, and the potential environmental burden of these methods [36] must also be critically assessed. However, the utility of 3D printing certainly outweighs nearly all these restrictions. In the near future, we expect that these problems will be solved, and 3D printing technologies will improve further in resolution and reliability.
3. Conventional lithography and 3DP vis-a-vis
Lithography, with its deep penetration in our routines, is conventionally performed through molding, milling, imprinting, and drilling; however, at microscale it is mainly direct and indirect writing [37]. Most common approaches of manufacturing at microscale followed in conventional and 3DP lithography (Table 2) are briefly discussed in this section.
Table 2.
Summary of 3D printing technologies.
| Method | Concept | Materials | X,Y,Z (μm) | Pros | Cons | Printers |
|---|---|---|---|---|---|---|
|
I3DP SLS [54–56] |
MJM PJM SLS |
Powder, Metals, Ceramics, ABR, PP, PA, ABS |
X:38 Y:38 Z:32 |
Multiple material Rigid and smooth Soluble supports High porosity designs Bio compatible Better XY resolutions Large library of materials |
Expensive Bulky equipment |
Multijet (3D systems) Polyjet (Stratasys) Pro X SLS 500 (3D systems) |
|
SLA CLIP [19,57,58] UDP |
UVLED Laser DLP CLIP |
ABR, PDMS, Various |
X:50 Y:50 Z:25 |
User friendly Moderate to high speed printing Inexpensive. Widely used Bio-compatible resins |
Single material Manual processing Limited resolutions compared to i3DP Postprint processing |
Pico plus 27 Asiga Form 2 Miicraft Autodesk Ember Carbon 3D |
| FDM [19,58] | Extrusion | AP, ABS, PET, PLA, PETG PDMS |
X:400 Y:400 Z:200 |
Multimaterial object Simplicity Inexpensive Portable Wide range of commercial TP |
Low resolutions Build roughness lack of transparent material |
Makerbot replicator 2X Easy 3DMaker 3D Touch |
| 2 PP [59–61] | Laser | SU-8 | <1 μm | High resolutions Fast printing Hallow complex microstructures |
Additional curing Proprietary material Time consuming |
Coherent Mira 900-F) EnvisionTEC |
| 3SP [59,62] | Laser | ABS resins Acrylic resins |
X:100 Y:100 Z:25 |
Wide range of specialized materials Low surface roughness Durability Large print sizes |
Limited resolutions | ULTRA 3SP |
MJM: MultiJet Modeling; PJM: PolyJet Modeling; SLS: Selective Laser Sintering; DLP: Digital Light Process; CLIP: Continuous Liquid Interface Printing; ABR: Acrylate based resins; PP: Polypropylene; PA: Polyamide nylon; ABS: Acrylonitrile butadiene styrene; AP: Acetoxysilicone polymer; PLA: Polylactic acid; PET: Polyethylene terephthalate; PETG: Polyethylene terephthalate glycol-modified; TP: Thermoplastic.
3.1. Lithography methods for MAS development
3.1.1. Mechanical/milling
Modern machining technologies have allowed for precise replication of miniaturized microstructures up to lower μm ranges thus, creating high resolution fluidic platforms with high aspect ratios [38]. Micromachining streamlined by automated Computer Numerical Control (CNC) of XYZ axis via a CAD design file can create structures with desired dimensions by removing material in specified patterns. These methods allow better precision and reproducibility by reducing manual errors and resulting in rapid prototyping. One of the major advantages includes ability to fabricate specialized and tough materials like stainless steel with relatively smooth surface [39,40]. Although micromilling allows for precision manufacturing but direct machining of small features as small as 20 microns is still a challenge due to either unavailability of machining drill bits or mechanical strength of the bit head to sustain prolonged machining [38]. Therefore, this technology is apt to develop micromolds acting as masks or negative templates to make functional micro component platforms ranging from PDMS based devices to embossing based structures [39,41,42]. Despite its many benefits micromilling has been underutilized due to the requirement of bulky and expensive equipments along with trained operative technical expertise.
3.1.2. Negative imprinting e SU8 and epoxy
Creating structures by imprinting on a soft matrix by photopolymerization or by hot-embossing on hard plastics are most common approaches to achieve molding. SU-8 is widely used photoresist for high aspect ratio and three dimensional lithography that is biocompatible and chemical resistant to develop microstructures ranging from 1 μm up to 1.5 mm with a single spin coat [43]. Typical negative imprinting process involves spin coating of SU-8 resin, pre-exposure baking (soft bake to remove solvents), UV exposure to perform crosslinking, post exposure baking and hard baking in a clean room in presence of a template mask. This creates protruding microstructures on the negative resist which serves as a mold for PDMS casting to form microfluidic platforms. Quality of the microstructures mainly depend on soft baking time, exposure time, post baking and developing time [44,45]. Some of the challenges associated with SU-8 negative photoresist is achieving ultra-thick microstructures without multiple coats, labor intensive multistep process, and expensive clean room requirements [46]. Also, thin SU-8 structures wither after few molding cycles and are highly susceptible to breakage [47]. Epoxy resins have been used by DIY enthusiasts to develop hard molds to potentially address this drawback associated with SU8 molds [48,49]. Epoxy molds are developed sequentially via negative by casting PDMS on a SU8 mold and later using casted PDMS with imprinted microfluidic network as a template for imprinting epoxy polymer. This creates a positive epoxy mask that later is used as mold for lithography.
3.1.3. Laser
Focussed laser for creating microfluidic parts via surface ablation under extremely controlled power is an important technology in rapid prototyping. This technology obviates the use of cleanroom fabrication due to self-cleaning nature of laser allowing for simple fabrication in non-clean room settings [50–52]. Typically, a laser ablation setup involves exposure of short duration eximer (UV) laser pulses onto the polymeric substrates, leading to a bond breaking of long chains of the polymeric molecules in presence of a patterned mask or an aperture. Ablation area and depth is defined by beam width, patterning, exposure time, and laser repetition rates [50]. Under ideal laser beam energy distribution with specified laser pulse rates and XY movement of substrate the resulted microstructures can be created with minimal thermal defects. Besides ability to work with multiple polymeric materials like poly styrene, PMMA, nitrocellulose etc., laser ablation is challenged by need for technical expertise, costs of the laser systems, post patterning bonding for enclosed fluidics and thermal related fabrication defects like surface roughness, cracking etc.
3.2. 3DP methods for MAS development
3.2.1. Material extrusion
This approach, also known as Fused Filament Manufacturing (FDM) is another type of additive manufacturing where polymers are added in layers each at a time by melting polymeric filaments and depositing them over layer-by-layer. This approach is very simple yet efficient in creating tools and provides a wide range of materials to choose from Ref. [53]; however, the main concern while printing microfluidic structures is poor print precision, high roughness of the fluidic components that may introduce undesirable eddies, and inability to print transparent tools, specifically for application that may require optical or visual monitoring [27,63–66]. Yet, with all those restriction there are a great number of publications based on FDM which are mainly attributed to its low cost.
3.2.1.1. Polymeric materials.
Commonly employed materials for FDM are styrene-(Acrylonitrile Butadiene Styrene:ABS; High Impact Polystyrene:HIPS), ethylene-(High density Polyethylene:HDPE), nylon-, polycarbonate-, polylactic acid-based [53]. There are several new materials have been developed in last two years. The USAFDA approved 3D-printable versions of medical polymers, such as Dioxaprene (dioxanone polymer), Lactoprene (polylactic acid), Maxprene (poly glycolid-co-lactide), and caproprene (95% caprolactone 5% glycolide) [67].
Reinforced polymers constitute another new class of materials reported recently [68]. Due to growing interests and applications in developing integrated membranes and sensors, new composite materials, such as carbon nanotubes, graphene, ceramics, and magnetic materials have been used as components in highly integrated devices [5,64,69,70]. Diels-Alder reversible thermoset (DART) materials are another novel group which has been promoted for minimal reduction (~4%) of material strength upon deposition and lowest shrinkage [71].
3.2.1.2. Printing methods
3.2.1.2.1. Fused Deposition Modeling (FDM).
Fused deposition modeling (FDM) is an approach where a thermoplastic filament is melted while passing through a hot outlet/nozzle and is deposited in layer over layer onto a stage thus generating structures in XYZ planes. This type of printing follows the STL-based design input like most other 3D printing approaches [69]. FDM 3D printing is versatile due to the availability of several thermoplastics along with composites with peculiar physical properties.
While this research has laid the foundation for cheap 3D printing of integrated microfluidic devices, there are several associated potential challenges. Since the heated thermoplastic in FDM is added layer by layer, the layers may not fuse well resulting in weak structures that are highly susceptible to compressive stress fracture [5,20]. Certain newer machines, such as QIDI Tech’s 3DP-QDA 16–01 dual extruder printer, mitigate poor layer-to-layer fusion by printing of either heated platforms or build structures in heated chambers. This problem was efficiently addressed by modifying the print head on a HE3D DLT180 printer to accommodate three channels in a single extruder [72]. This allowed for printing three different materials via modulating their injection to achieve multimaterial prints with better finish than conventional FDM. Also, heating plastic filaments create potentially hazardous fumes requiring additional ventilation to be used. In an additional technological advancement, heating filaments can be replaced by liquid polymer precursors thus alleviating health hazards and improving layer fusion [73]. DART process may address these drawbacks with new class of materials that instead causes minimum burning of polymer during melting. Extent of crosslinking in DART polymers can be easily manipulated through de-crosslinking and re-crosslinking of polymers by regulating temperature around a critical point. It also poses low risk of health or environmental hazard while at the same time addresses other challenges, such as creating smooth surfaces at fairly high strengths [71].
3.2.1.2.2. Modified FDM for printing PDMS.
Polydimethylsiloxane, being the primary choice for routine microfluidics due to its high transparency and compressibility, has been 3D printed for creating structures as smooth and transparent as those conventionally made by PDMS. Researchers have switched the basic filament dispensing system of a FDM machine with a syringe and needle. This allowed them to avoid modifications in the stage, software, or the printing unit. However, they have to employ a gel matrix to print the PDMS structures until these are heat-cured (Fig. 3). Hydrophilic support bath via freeform reversible embedding was used by Hinton and group to extrude PDMS within the hydrophilic Carbopol gel which was later cured via heating in two rounds [74]. They have managed to create perfusable manifolds using this technique. In another attempt Li and collaborators printed wax instead of PDMS by replacing syringe with a glass nozzle [75]. Later, they have employed these wax prints as molds for casting PDMS. This approach was better than direct printing PDMS as it obviates the need of employing a complicated gel matrix-based printing. Also, it creates smoother surfaces as wax reduces the process-introduced roughness.
Fig. 3.
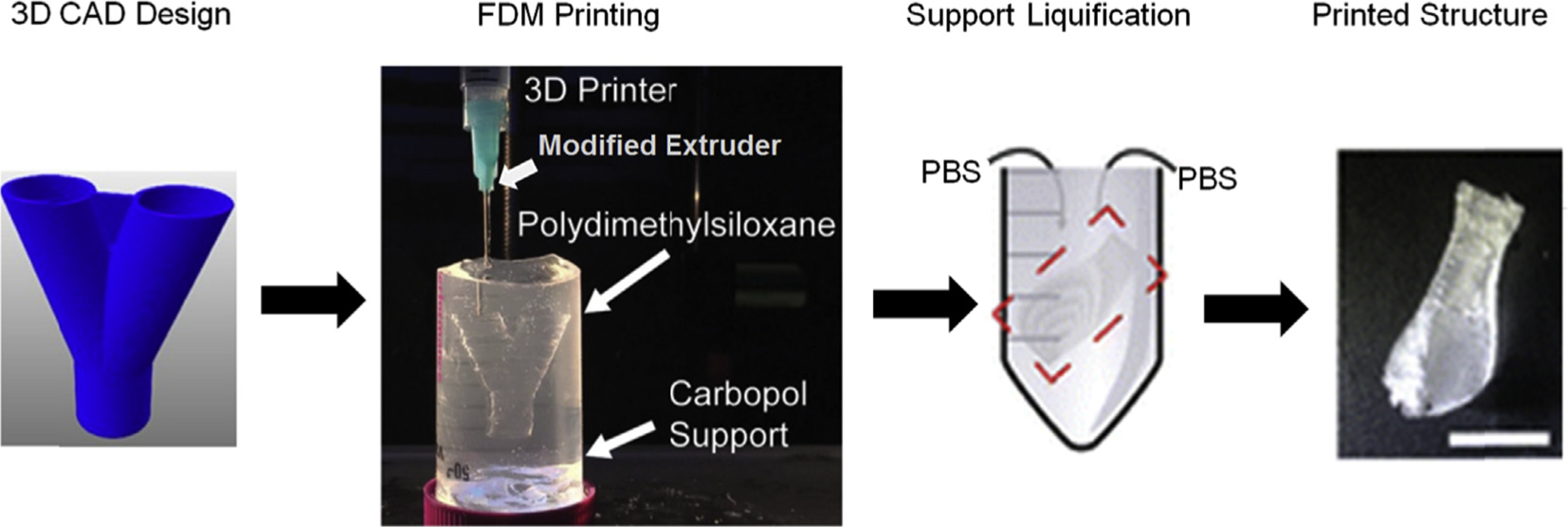
Illustrates the workflow associated to print PDMS with filament extrusion using a modified syringe-based extrusion head.
3.2.2. Stereolithogaphy
Central workflow for 3D printing involves designing the device in computer-aided design (CAD) software available in paid or open license formats. The CAD designs are transformed into ‘.STL’ format which changes the three dimensional design into triangulated sub-structural dimensions allowing printers to designate files in XYZ directions. This workflow simplifies the overall printing process with fewer steps in a simple time efficient manner [23,76]. The additive manufacturing aspect of 3D printing is based on sequentially assembling layers of specific materials one over the other with instrument-limited spatial resolution [57]. Thus, to achieve a high degree of robustness and integrity of the sub-millimeter features only a few techniques are useful. These are laser sintering (SLS), fused deposition modeling (FDM)/filament deposition manufacturing (FFM), inkjet printing of photopolymers/microjetting (PolyJet or Multijet), stereolithography (SLA) [5] (Fig. 4). Despite all these methods being available, most reports on microfluidic device fabrication are focused on printing with FDM and SLA.
Fig. 4. (a).
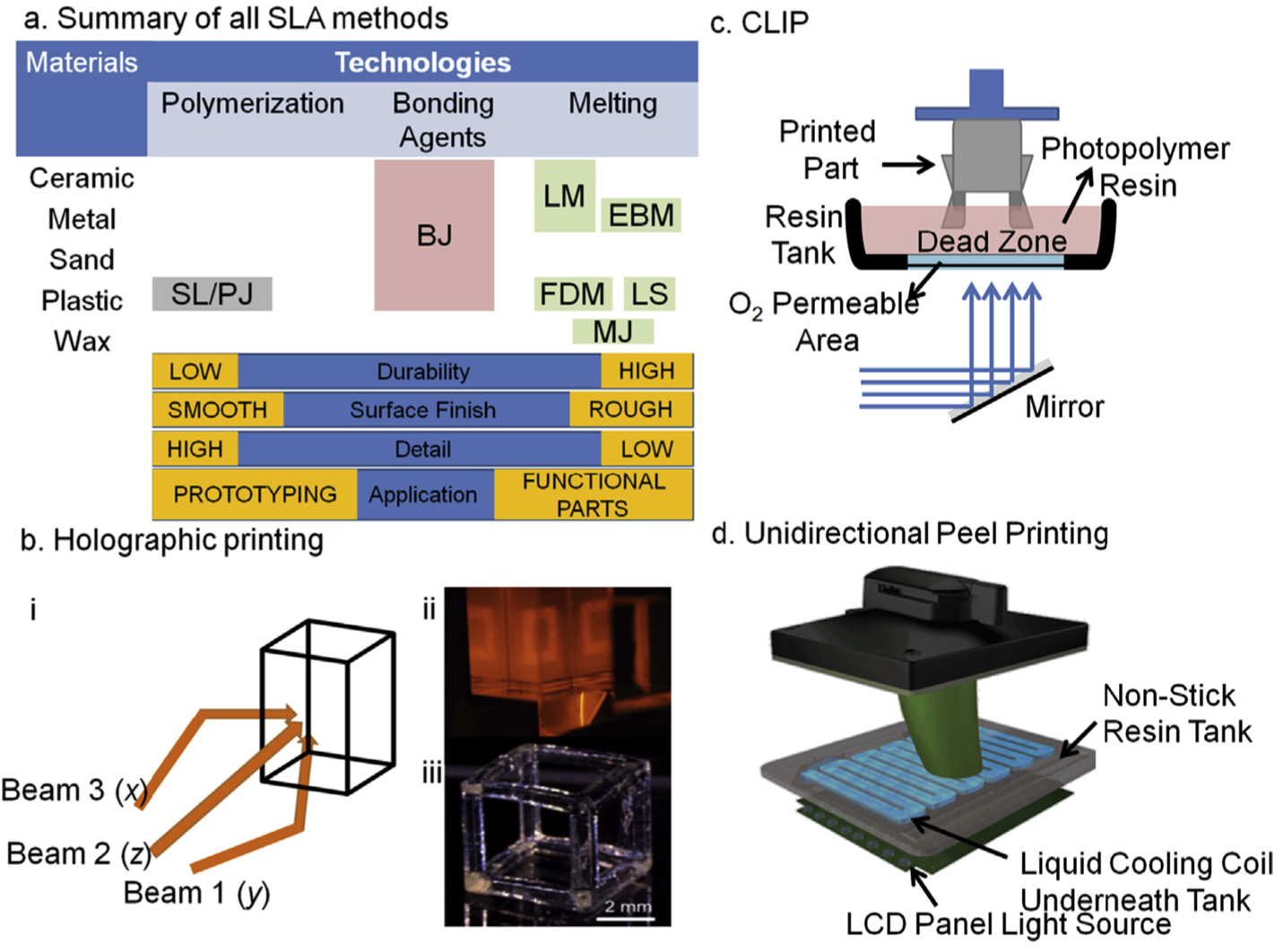
Illustrates a summary of different approaches for 3D printing with their print qualities. Binder Jetting: BJ; Electron Deposition Modeling: EDP; Fused Deposition Modeling: FDM; Hybrid Process: HP; Laser Melting: LM; Laser Sintering; Material Jetting: MJ; Photopolymer Jetting: PJ; Stereolithography: SL. (b) An overview of 3 dimensional light projection scheme in Holographic 3D printing set up (i) while in panel (ii) actual light projections of squares can be seen from all three planes that will create a hollow cube as depicted in a single projection of light as shown in panel (iii), a video related to this technology can be find at (https://www.youtube.com/watch?v=00H-hXufpQE). (c) Depicts Continuous Liquid Interface Production scheme, which uses a combination of oxygen-based quenching of the reactive photoinitiator, allowing rapid printing within few minutes (15–20 min). (d) A set up of recently developed UDP method by Uniz Technologies, which is basically the fastest 3D printer in all segments. This is an arrangement of peeling in only z-direction by introducing a cooling coil in the otherwise non-sticky resin vat. The cooling causes peeling of the cured parts which also minimizes polymer shrinking due to hot-cold cycles.
3.2.2.1. Photopolymers.
This is the class of materials which when exposed to specific wavelength of light undergo polymerization. In 3DP mostly UV photopolymerization is employed and polymer precursors with UV polymerization ability are typically used. Main component in the recipe of these photopolymers are photo-curable prepolymer/monomer/oligomer, binder, initiator, colorant, and some cases plasticizer [9,77]. Methacrylate monomers, Thiol-Ene, Thiol-Yne, epoxide, and ether-based systems are broad categories of monomeric chemistries for photopolymerization in AM (Table 3). Acrylate polymerization is rapid compared to epoxies and is commonly used for 3D printing. However, epoxies with high strength, low crosslinking-associated shrinkage, heat resistance, and optical transparency serve as better alternatives to acrylates [77]. Although all the components are essential to achieve a stable polymer but initiator plays a crucial role in initiating the polymerization process and enhancing the rates. A balance between the interplay of light wavelength and polymerization process is also controlled by initiator to some extent. Most common initiators are radical systems, photoacid generators, and two-photon initiators which are usually added as reactive solvents (Table 3). Several new variants are reported recently. The most interesting of those is biotinylated photopolymer [78]. Due to the extent of applications of biotin in biological and biochemical analysis, this biotinylated polymer can open up a whole new area of applications.
Table 3.
Common SLA-specific photopolymers.
| Class | Polymer | Initiator |
|---|---|---|
| Acrylates [9,85] | Diacrylate + Methacrylate + N-vinyl-pyrrolidone (reactive diluent) | Darocur 1173, Irgacure 183 |
| Radical-mediated | Urethane acrylate or Bisphenol A diglycidyl ether + dipropyleneglycol diacrylate (reactive diluent) or + pentaerythritol tetraacrylate (reactive diluent) | |
| Thiol-Ene [86–90] | Dinorbornene + trimethyloylpropane tris-(mercaptopropionate) | 6-benzothiazol-2-yl (2-naphthyl) diphenylamine |
| Thiol-Yne | NOA 61/72 (mercapto esters) + tetrahydrofuran methacrylate | |
| Radical-mediated | ||
| Epoxides Cation-mediated | 3,4-epoxycyclohexylmethyl-3,4-epoxycyclohexanecarboxylate Soy bean oil (ESBO) |
3.2.2.2. Printing methods
3.2.2.2.1. Stereolithography (SLA).
Stereolithography is fabricating layers via fusion which is facilitated by photopolymerization of a liquid polymer precursor. In a typical SLA set-up, polymer precursor in a vat is polymerized with UV light, as was the case in the first patented technology by Charles W Hull 1986, and in current technology with highly focused laser or diode light sources 5.
The common workflow for SLA printing is generating a CAD-based design followed by conversion to ‘.STL’ format which is basically extracted geometric information [79]. A computer program interface, often referred to as slicer program, is required to change the STL design into slices/layers. These layers then serve as a path guide for moving the light source to photopolymerize a precursor layer [80]. The polymer precursor type and light source govern the dimensional limits of the printed objects. For a light wavelength designated for efficient polymerization of a thin precursor like Formlab’s clear resin, a thicker pre-polymer, such as flexible resin by Formlabs, will have smaller light penetration thus resulting in either no structures or anomalous build up. Therefore, light penetration depth controls how much energy is transferred upon exposure to effect the polymerization, and hence controls the integrity and boundary sharpness of the printed structures. Several new polymer materials are being introduced as well as improving the existing polymer versions are ongoing to address these challenges [81–83]. Based on the laser type used, limit of the printed structures can typically reach 100 microns in the XY direction and may have print-to-print variations. Therefore, SLA methods, although better than FDM in terms of surface roughness, material choices, and structural reproducibility [22] have dimensional limits for some types of resin and light source [65]. Recently, Shusteff and colleagues have reported a novel approach of SLA printing but volumetrically [84] in dimension. Unlike conventional SLA, material is exposed to light as a holographic image instead of a layer at a time thus reducing all whole procedure to few seconds (Fig. 4b). Light is exposed from x-, y-, and z-planes with appropriate prism mirrors. When light from all three directions coincide at a point equals the energy required for photoinitiating the reaction. However, controlling exposure times is very critical while printing hollow structures as overexposing will cure resin in the supposed to be hollow areas. Most of these limitations can be addressed by employing the Digital Light Projection approach.
3.2.2.2.2. Digital light processing (DLP).
Rather than polymerizing a single layer by moving a laser along the STL design path, light for a whole layer or slice is projected as one single event, thus making this printing method faster than laser-based SLA. In addition, since light is projected as pixels, the dimensional limit can be decreased to 20–30 microns along XYZ planes, which is 3-to 5-fold lower than typical laser-based approaches. However, this restricts the dimensional resolution to the printable area. Since light is projected as pixels the biggest area that can be projected without losing the resolution is quite small, viz. 2 cm 2 cm, and as this area is increased the pixel density decreases thus drastically affecting the spatial resolution of the printed structures [28,91]. Additionally, there might be shrinkage with DLP SLA printing [92].
There is another new approach, namely continuous liquid interface production (CLIP) that has been developed as a proof of concept study but is not yet open for commercial manufacturing.
3.2.2.2.3. Continuous liquid interface production (CLIP).
Tumbleston et al. described another variant of laser SLA which incorporates an oxygen-permeable membrane between the optical window and the cured part of the device [93,94] thus, creating a dead zone that acts a mask between the cured part and the continuously renewing liquid precursor. This has allowed them to print continuously for which it is called Continuous Liquid Interface Production (CLIP; Fig. 3c). This is similar to the robot making process as shown in Terminator movies! Therefore, this process not only addresses all of the above challenges but also significantly reduces the printing time to only few minutes.
3.2.2.2.4. Unidirectional peel printing (UDP).
Although CLIP has presented solutions to most of the 3DP-related restrictions, continuous technological improvement has yet led to UDP [26]. Unlike CLIP, it has neither pure oxygen-based dead zone nor has an exotic separation material, as re-iterated by its CEO Dr. Houmin Li. Instead of using a conventional up-and-down peel method along XY- and Z-axes, in UDP peeling is performed by cooling the cured parts under continuous cooling that allows fast peeling along the Z-direction and thus, printing as high speeds (Fig. 4d). It is achieved by introducing a cooling zone at the bottom of the photopolymer vat. This technology also uses a LCD light source for achieving better resolution and quality surface finish.
3.2.2.2.5. Modified SLA for PDMS printing.
PDMS is reportedly has been printed in hydrogels using modified extrusion heads in FDM [74]. However, dispensing PDMS through print head produced similar effect as with filament extrusion; non-uniform structures were obtained although they functioned as intended. Therefore, recently researchers have developed new chemistries with which they can print PDMS directly in SLA format using laser-based [95] or DLP-based [96] approaches. Prints were highly uniform and structures with resolution as low as 100 μm were obtained. However, in order to track the PDMS polymerization, an ink is to be added due to which the trasparency of the printed device is partially lost [95,96]. The technology is not yet mature and will require further research to fabricate soft-lithography-like smooth and transparent prints.
3.3. Other approaches
3.3.1. Inkjet printing
Typical of an inkjet printer, the ink is dispensed in specific droplet sizes on to a solid support as a jet. This technique was modified for the first time by Objet, an Israeli company, to print in 3D with a single jet print head. In current incarnations, several print heads are incorporated to allow printing multiple lines of same or different materials and thus called MultiJet or PolyJet printing. This approach is highly sought after for printing biological materials [97] due to its ability to print liquids that can accommodate bio-molecules and cells, or tissues. However, biocompatible inks are covered through trade secrets and are expensive at the same time. It is also possible to print photopolymers by selective printing and sequentially curing the photopolymers. Few researchers have claimed printing microvalves [98] and mF networks using this method [99].
3.3.2. Laser sintering
Unlike inkjet printing, this is a powder printing technology where 3D printed structures are built out of powders by selective laser exposure. These powders may consist of metal alloys or polymers and can be printed through selective laser melting (SLM) and selective laser sintering (SLS), respectively [54]. This method is mainly employed for printing superior quality prosthetics [56]. However, it can also be employed for microfluidics and lab chip printing but such reports are not common [6]. Also, this method is deemed not suitable for bioprinting because of the exposure of precursor materials, such as powders, to lasers.
4. Tools printed with 3DP
Tools, components, modules have reportedly seen a shift from conventional lithography to 3D printing. There are several 3D printed devices and tools developed in the past five years, which are summarized in most recent review articles [5,6]. Here we selectively review a few developments in this field.
4.1. Molds and scaffolds
3D printing was used in mF fabrication of molds and scaffolds that serve as an indirect approach to develop fluidic networks. The ease of rapid prototyping and ability to change forms and factors of the molds with 3D printing allows for developing tools with various physical parameters [100,101]. Kamei et al. reported fabricating 3D printed molds used to perform soft lithography for PDMS-based complex fluidic networks [102]. They employed these for creating concentration gradients of several growth factors. In another study, Chan et al. created a method for single step molding of complex structures using 3D printed masters [103]. They demonstrated several key 3D microfluidic molds, such as basket weaved networks, 3D chaotic advective mixers, peristaltic pumps, injection-on-demand device etc. Researchers have fabricated complete LOC devices using 3D printed templates and molds [35,104]. Comina and colleagues developed an integrated microfluidic mixer by templating PDMS on 3D fabricated mold, which they described for on-chip glucose detection [104]. Their templates were smooth allowing for leakage-free PDMS-glass bonding and were reusable at the same time. In a recent attempt, Villegas and group were able to create highly smooth PDMS surfaces using cheap low-resolution 3D printed molds by creating an omniphobic lubricant-infused coating that covered surface roughness of the mold rendering it smooth [105]. Geometries unattainable with existing lithography approaches were successfully demonstrated with 3D printed molds, such as, Hwang and his team fabricated multidimensional device geometries including variable channel cross-sectional areas and diameters in a single device [106]. In recent attempts, complex capillary networks [35] and multilayered channel structures [107] in PDMS were also demonstrated. A novel single-layer two-sided molding method developed by Glick and associates was employed to fabricate a multilayer device of PDMS with unique approach to bonding and alignment, thus allowing for rapid assembly [107]. They developed two channel levels along with membranes, valves, interconnects between fluidic levels, and integrated sample inlets and outlets.
Simple scaffolding and template removal is yet another interesting approach if multilevel, complicated fluidic networks are needed. Acrylonitrile butadiene styrene (ABS) scaffolds printed by FDM were demonstrated by Saggiomo and Velders, which is a very simple, cheap yet very effective approach to soft lithography [108]. Once PDMS is cast over the ABS scaffold and cured, the scaffold was removed using acetone, which only causes minimal swelling and structural defects. As a further improvement, Chan et al. created complex dodecahedron and trifurcated complex network scaffolds using SLA-printed acrylate-based masters with extremely smooth surfaces [109]. 3D alginate structures were developed using this approach for applications in cell biology due to the fact that these complex structures are perfusable.
4.2. Direct printing of complex structures and devices
With continuously improving 3D printing technologies direct printing has progressed as an independent approach for complete mF device fabrication. Inkjet-printed microvalves [98], MultiJet printed multimaterial microfluidic valves [110], and DLP-SLA printed microvalves, pumps, and multiplexers [111] have been reported recently. In an interesting approach, Bhargava and colleagues have discussed fabricating modular style fluidic components that can be integrated to develop a complete device [112]. However, most striking feature of recent 3D printing methods is ability to create complete devices with integrated components.
A supercapacitor-powered immunoarray was developed by Kadimisetty et al. by assembling a graphite-based antibody array and 3D printed fluidic module integrated with electrodes for detecting four prostate-specific protein biomarkers in electrochemiluminescence (ECL) format [29]. In another attempt to create electrode-integrated fluidic devices via 3D printing, Bishop et al. printed flow cells for performing electrochemistry and ECL based analysis. In one study they developed a chip with SLA printing to analyze the effect of DNA concentration on the ECL of ruthenium bipyridyl [113]. In another report they developed single channel fluidic chips with FDM to synthesize Prussian Blue nanoparticles and for amperometry to quantify hydrogen peroxide [114]. Lego®-like modular microfluidic tools printed using 3DP methods are recently reported by He group [25].
5. Selective applications of MAS
Microfluidics has positively impacted the development of new chemical and biological applications. To understand where and how 3D printing has the potential to revolutionize chemical and biological analyses, and how much the 3D printing technology has penetrated into the field, we must first look into the plethora of applications that are principally based on microfluidic tools developed via conventional lithography.
5.1. Conventional lithography-based MAS
Cell biology in standard practice is a cumbersome laborious science requiring regular monitoring. In conventional cell biology-based techniques, experimentation extending up to several days with sample volumes of several hundred milliliters faces challenges, such as poor analytical cut-offs, that may be in the form of experimental sensitivities or may be confluence population of cultured cells, and poor reproducibility. mF tools have addressed all these challenges by minimizing sample size and enhancing the reaction kinetics governed by improved mass flow properties [4,115,116] thus allowing continuous monitoring of samples, often with on-board sensing.
Cell culture is the core method in cellular biology that allows for sustained growth of cells outside their site of origin. The first few reports on culture on a chip were reported over a decade ago [117–119] with chips performing basic functions of holding cells in designated chambers with culture supplies and facilitating the proliferation. However, the complexity of such chips kept increasing with integrated functions. A few of these chips introduced biosensors within the chip to monitor cellular physiology [120]. Lei et al. developed an on-chip impedimetric system to monitor cell migration and chemosensitivity [121]. Nguyen and colleagues developed a chip with an impedance biosensor for single cancer cell migration. Recently, a chip was developed with an integrated microheater for growing cultures outside the regular incubator [122]. In another report an integrated infrared spectrometer was fabricated on the chip to monitor cell densities [123]. Cell differentiation of primary preadipocytes into adipose tissues [124], 3D cell cultures [125,126], and tissue/organs on a chip, such as bone marrow-on-a-chip [127] are a few notable extended applications of cell culture chips.
Advancements in mFs has strengthened our ability to cultivate cells in time-limited settings at significantly low volumes allowing scientists to create homogeneous cell populations either in cultures or out of a tissue biopsy sample with integrated tools [128,129] allowing for identification, separation, and enrichment of a specific cell population type [130–132] as determined with (a) integrated sensors that monitor the cellular morphology, such as size and shape [133–135], mechanical properties [136], and asymmetry [137], and (b) label-free cell isolation, which is mainly achieved on chip by either hydrodynamic focusing [138] or via inertial flow and Dean forces [139]. Rare cells, such as circulating tumor cells (CTC) which are typically found in very low frequency equivalent to ~1 cell per 7.5 mL of whole blood, can be isolated and enriched using mF facilitation [139–142]. Although advanced mF platforms are now regularly being used in research labs for rare cell isolation and identification, applications also extend to many other cell types, such as stem cells [143], specifically separation and identification of blood-based cell [144] and plasma components [145].
These tools have facilitated applications which were beyond our perspective a decade back. Surface biomarker analysis [131,146] and cell-cell communication at either single cell resolution [2,147,148] or single cell level [149,150] are a few high precision applications that are revolutionizing cancer biology, biomolecular methods, and diagnostics. These developments are attributed to our ability to now perform on-chip lysis of cells that enables analysis of sub-cellular organelles and molecular components. Main strategies for cell lysis are based on optothermally induced bubbles [151,152], mechanical disruption with on-chip pump [153], and chemical and electrochemical disruption [119,154]. These techniques when integrated with appropriate microfluidic modules can also allow for applications in omics [150,155–158], molecular biology [159–161], and disease diagnostics [162–164].
5.2. 3DP-based MAS
Analytical applications of 3D-printed devices are revolutionizing the way prototype experimentations are done in the wet lab. Devices with a wide range of abilities have been developed, from simple fluidics to perform singleplex analyses to complicated on-chip microfluidic networks for multiplexed analyses. Cell and tissue printing on planar surfaces to printing 3D scaffolds and complete organs with significantly high precision are a few other interesting applications of 3D printing. The number of applications is on a rising trajectory in almost all fields of analytical biology and chemistry [165] (Table 2).
5.2.1. In cell biology
The largest number of new applications has been in the area of cell biology covering a majority or experimental activities, including cell culture, cell identification and separation, cellular communication, cell lysis, and cellular biomarkers [165]. Cell isolation has been core to cell biomarker analysis and cell sorting has been a central technology behind this. Cell sorters rely upon a high speed jet for de-clustering and focusing cells through a thin opening. However, a major restriction to this approach is exposure of cells to a massive shear force which may change the whole physiology of the cells and compromising the results. Microfluidics has changed this approach completely and we now have devices to focus and separate cells without the need to expose cells to shearing forces.
A 3D printed standalone mini-hydrocyclone device for flow focusing microalgae [166] exemplifies how rapid prototyping has made the complete workflow very sustainable and reproducible and in this case with a medium to low energy input. With this mini-hydrocyclone, Maira and colleagues were able to achieve ~7-fold more concentrated samples within only 11 min without encountering problems typical to planar mF tools, such as clogging, low throughput, associated costs and operational difficulties (Fig. 5a). This type of active cell sorters holds tremendous value to research and industrial operations. However, these may not be viable when either the sampling volume is very low or cells are in a highly viscous medium, such as whole blood. Inertial flow holds the answer to these problems. 3D printed standalone and modular inertial cell sorters have been developed to achieve high degree of cell sorting and isolation. Detection of pathogenic bacteria [169], isolation of Chinese Hamster Ovary cells in membrane-less micro-filtration [170], and reconfigurable generic separators [171] have been reported. Spivey et al. has generated a continuous flow culture device that also serves as a single cell isolator [167]. They demonstrated culturing yeast on-chip. In this device yeast cells were captured in transverse channels that serve as connectors for two parallel fluidic channels with higher width than the connector. The trapped yeast was allowed to grow while outgrowths that reached main channels were shear cut by the flow of the liquid. With this they demonstrated a powerful tool to study cellular aging and its genetic regulation (Fig. 5b).
Fig. 5.
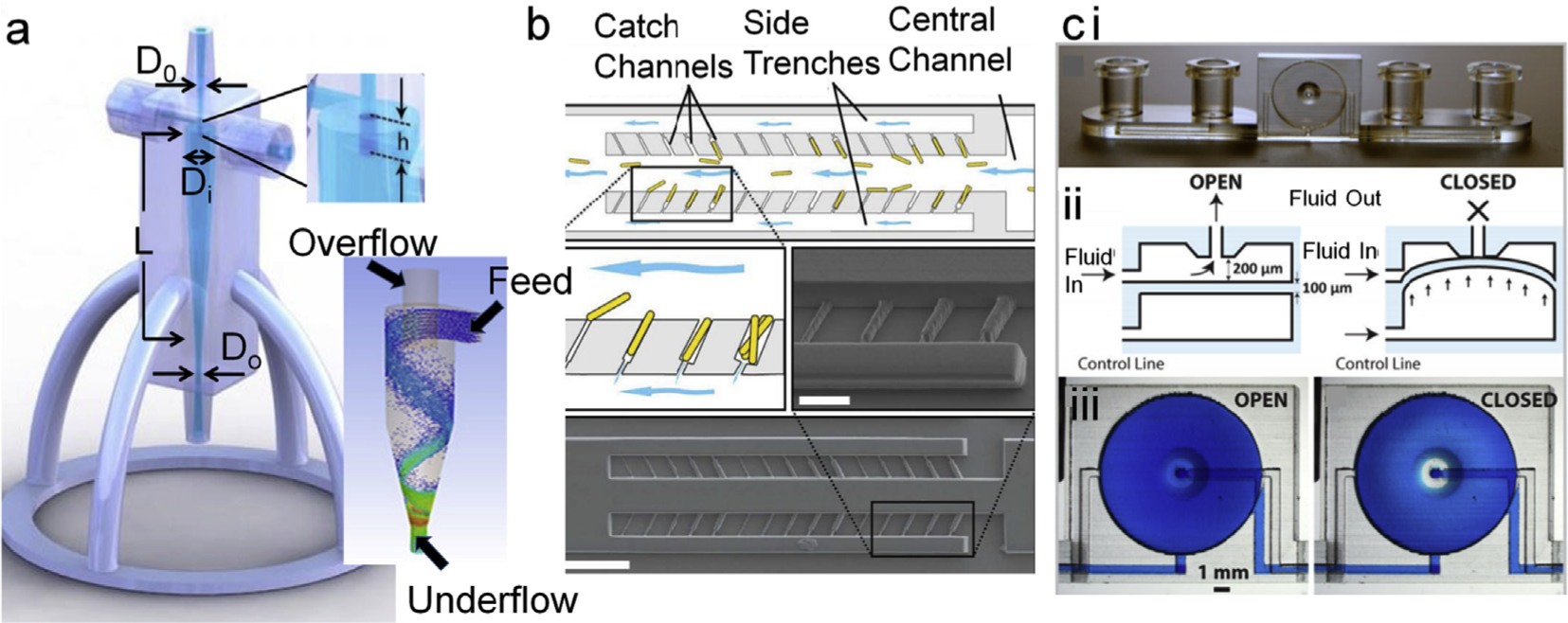
a. Minihydrocyclone for cell separation and concentration. Basic components in designing an efficient concentrator are diameters of inlet D0, Inlet diameter (Di), inlet overlap length (h), Length of the concentrator cylinder (L), and outlet diameter (Do) which controls the Feed and overflow for size-dependent focusing of cells as depicted in the bottom panel of ‘a’ [166]. Adapted from ref 164. Copyright (2017) The Royal Society of Chemistry. b. illustrates cell sorting and capturing at single cell level. Cells were exposed to fluidic sheer at the opening thus yeast buds cleaved, which authors have used for studying genetic makeup and growth physiology [167]. Adapted with permission from ref. 168. Copyright (2014) American Chemical Society. c. Illustration of highly integrated chip with on-chip valves (i) that can be manipulated on demand via appropriate airflow in the channel underneath of the fluidic channel (ii, iii). The complex functioning of on-chip integration was successfully demonstrated by culturing CHO cells that also has gradient generator on-board [168]. Reproduced from Ref. 169 with permission from The Royal Society of Chemistry.
Another application of rapid mF prototyping in cell biology is cell culture, associated workflow and modular fittings. Au and colleagues printed a highly integrated device with on-chip valves, pumps, and culture chamber [168] (Fig. 5c). They cultured CHOeK1 cells in this lab-on-a-chip (LOC) and analyzed calcium ion transportation within live cells under different conditions. Although their main focus was novel engineering, their demonstration practically opened up multiple dimensions where 3D printed integrated mF LOCs can be developed and employed. Podwin’s group has employed a completely opposite approach and developed a multi-level co-culture device via modular printing of separate levels individually, followed by integrating and assembling them with appropriate membranes and fluidic components [172]. They have employed this tower device to culture yeast and euglenas in such a way that CO2 gas permeated from ethanol fermented by yeast culture is harvested and used for euglenas which in turn generated O2 gas. Although this device is simple in composition, it could have impactful applications in unraveling biological cascade pathways. In a much simpler approach Brennan et al. fabricated a modular print with fluidic networks arranged in a desired sequence [173]. They used this as a gas perfusion and gradient generation system integrated cell culture plate.
The 3D printed mF devices are strongly competing with conventional fluidics in the field of cell biology. The only recent challenge seems to be in the form of restriction of the choice of precursor print material as currently tested materials may not be suitable for several cell based applications. However, this restriction seems to be insignificant when it comes to other applications of 3D printed fluidic tools in biomarker discovery and diagnostics. Our capability to 3D print glass will revolutionize further applications of 3D printing in cell and molecular biology applications [10,174].
5.2.2. In biomarker analysis/diagnostics
Molecular diagnostics is another area where smaller sample volume and rapid results are always sought after. Microfluidics in general has addressed these challenges to their core but 3D printing has enabled the scientists to achieve similar results faster, at much lower cost, and with a high degree of integration and complexity, although with few reservations. In a recent study, Singh and colleagues developed a microfluidic tool using 3D printing aligned in the shape of an organ with an ability to perform biomarker profiling and analysis from a whole organ [175]. They demonstrated this application using a porcine kidney and have analyzed clinically relevant metabolic and pathophysiological biomarkers including heat shock protein 70 and kidney injury molecule 1 down to 409 and 12 ng/mL concentrations, respectively. These measurements facilitate organ screening for transplantation. This is the kind of potential 3D printing has to offer. In one such study, 3D printed microfluidics integrated with biosensors for glucose and lactate was developed with a demonstrated application as an online sub-cutaneous microdialysate analyzer [62]. There are several applications of 3D printed mF tools but few high impact are designed tools for immunodetection of liver cancer cells [176], integrated multiplexed detection of cancer biomarkers [177], analysis of blood components [178], and plasma separation [179].
Interestingly, few of these tools are highly integrated and automated, which is needed for strong commercialization potential. One such tool was recently reported by Kadimisetty et al. that integrates a supercapacitor power source, a microprocessor-controlled micro-pumping system, and electrode-based microarray to perform multiplexed detection of prostate cancer biomarker proteins [29] (Fig. 6). Kadimisetty and Rusling and their group have developed other integrated tools that were employed in electrochemiluminescence (ECL)-based studies of genotoxicity chemistry [180]. From the same group, another device was developed for performing multiplexed prostate cancer-specific protein biomarker detection [177] (Fig. 6). They have validated this tool by testing patient samples and have obtained strong correlation to convention ELISA detection, but at much lower cost and higher sensitivity.
Fig. 6.
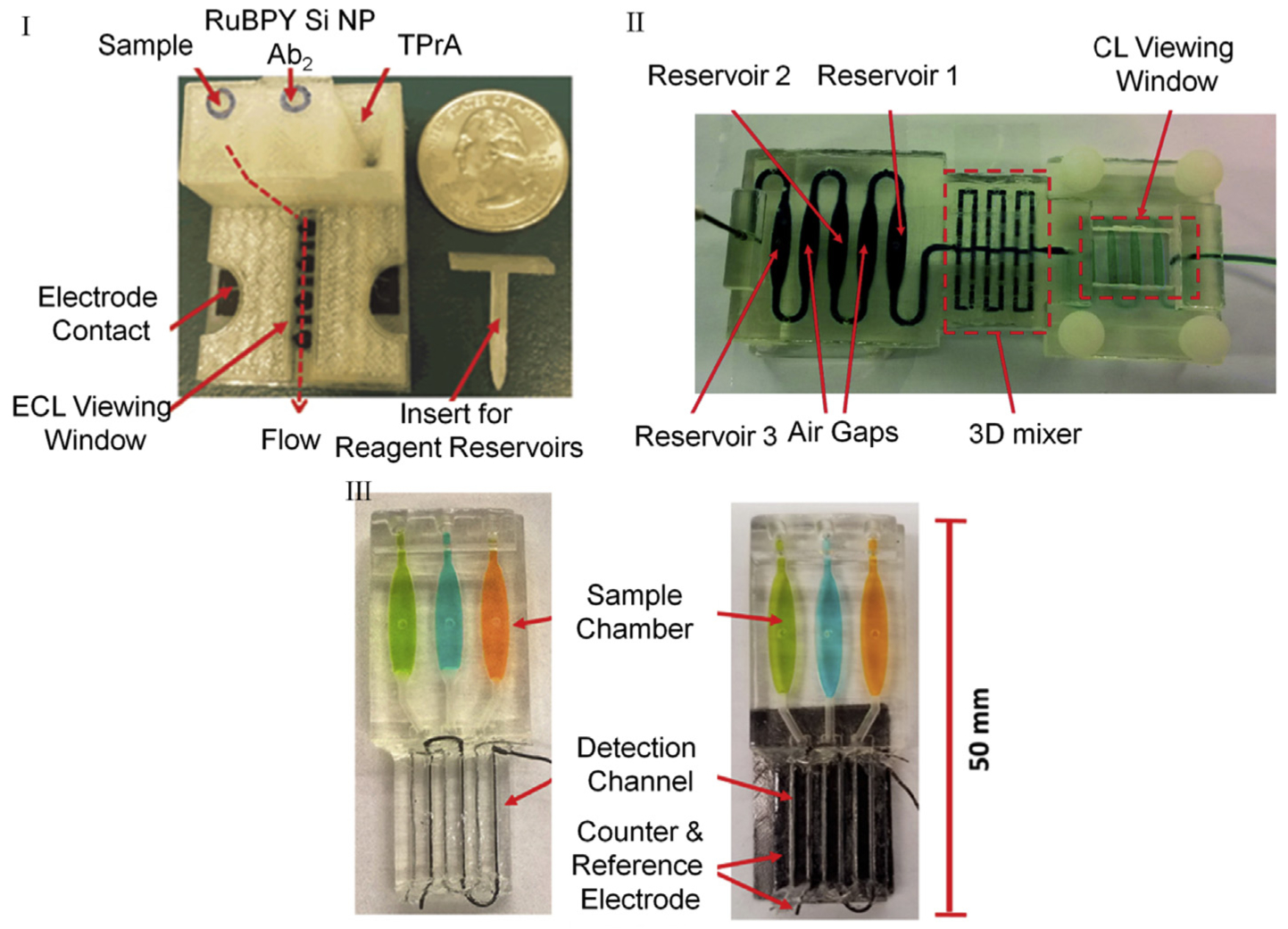
3D-printed tools to perform ECL (I, III) and CL (II) for detecting prostate cancer-specific panel of biomarkers. I. FDM printed motor-free device with integrated reagent reservoirs, supercapacitor driven pumps, and electrodes. Reprinted from ref 79. Copyright (2016), with permission from Elsevier. II. SLA printed transparent single body chip with integrated mixers. Reproduced from Ref. 152 with permission from The Royal Society of Chemistry. III. SLA printed transparent chip with integrated three electrode system on-chip for biomarker detection. Reprinted from ref. 87. Copyright (2017) American Chemical Society.
True point-of-care (POC) devices that can be used remotely without requiring any complicated machinery have also been 3D printed. For example, a 3D-printed tool was created with integrated pumps and valves that operates manually [181] (Fig. 7). This device also integrates a cartridge-type detection chamber and cell phone camera mount allowing it to detect disease-specific biomarkers in an incorporated colorimetric assay format at POC. In other work, Plevniak et al. have developed a 3D printed POC anemia detection and severity profiling chip with integrated micromixer [182,183] which was validated using 5 mL whole blood samples from patients. They concluded that clinically their chip was 50% in agreement with current hemoglobin assays and further optimizations may be required to add clinical relevance. In an attempt to further achieve true capability of getting to sample to insight in resource limited setting and POC, Jue and colleagues developed a chip for detecting Chlamydia and Neisseria with integrated sample processing, nucleic acid extraction and amplification units along with the capacity of meter mixing [184]. Chan and colleagues have critically reviewed applications of 3D printing in POC design and development [185]. Recently, Liu’s group created a highly integrated reactor for molecular diagnostics and detected N. meningitidis as low as 50 CFU and P. falciparum down to femtograms, which are comparable to routine methods [186]. They have developed this device through integrating printed structures with membranes and coating fluidic channels with static coating for improved biocompatibility.
Fig. 7.
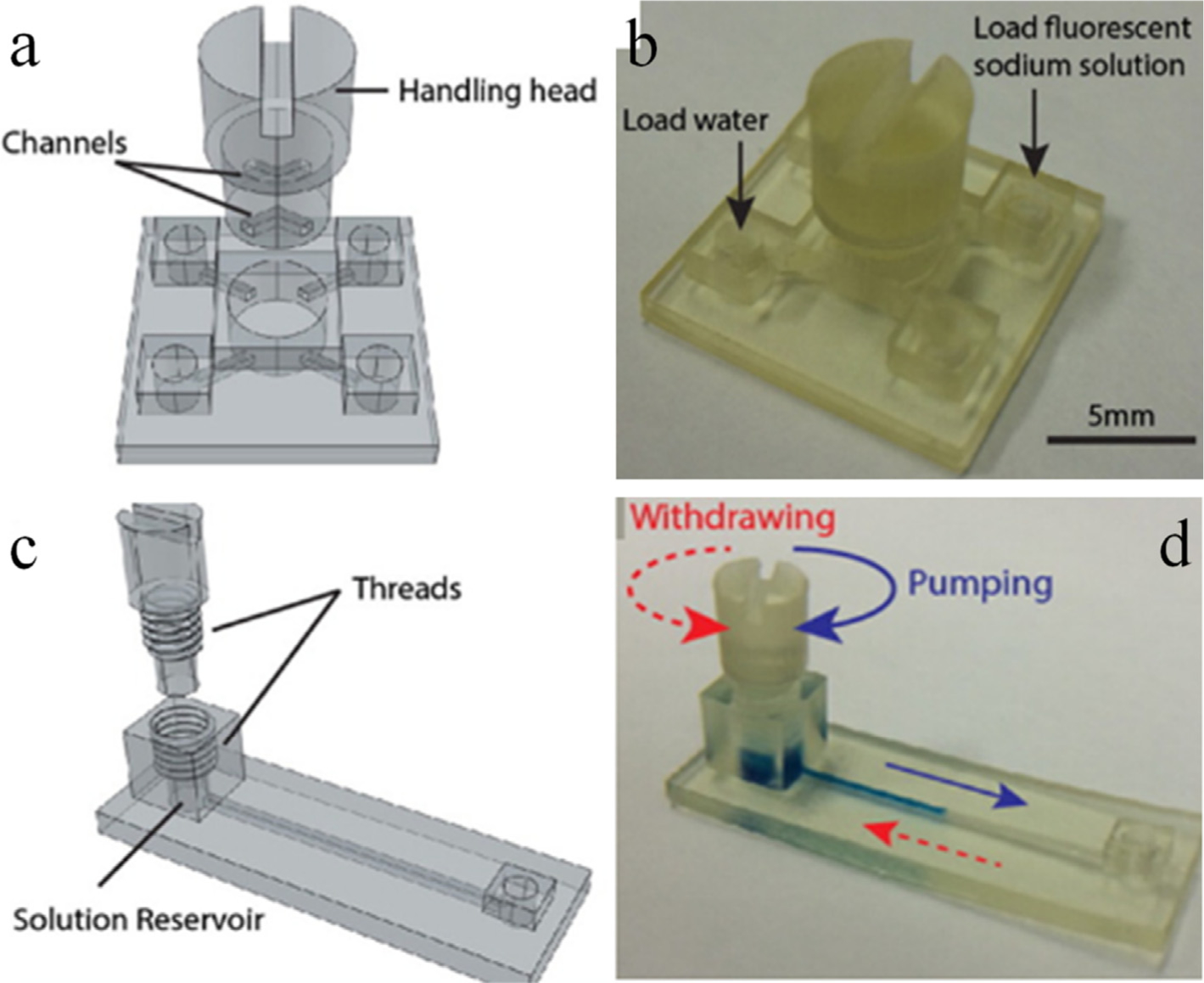
Modular valves (a, b) and pumps (c, d) are part of a bigger design challenge. These modules were combined together, and an integrated device was fabricated. The performance of the individual modules and the tool was demonstrated by detecting urinary total protein using colorimetric approach. Adapted with permission from ref. 88. Copyright (2016) American Chemical Society.
5.2.3. In chemical analysis
3D printed microfluidics (mFs) has allowed us to perceive and propose rapid solutions to several complex problems in almost all the domains of biological sciences. However, it is not only restricted to the analytical life sciences. Chemistry has also seen a massive number of applications with conventional and 3D printed mF tools [187]. Miniaturized reactors for performing chemical reactions, probing chemical species, drug testing are few.
5.2.3.1. Chemical reactors.
Performing complex chemical reactions in bulk for proof of concept studies is a big challenge for reasons such as cost, amount of waste, and the labor involved. Small volume mF chemical reactors not only address these bottlenecks but also allow for improving the reaction kinetics by controlling the law of mass action. One such device was fabricated by Li and colleagues where they integrated a membrane and reagents, such as zinc and Griess reagent, for reducing nitrate to nitrite [70]. Similarly, Su et al. developed a reactor for analyzing glucose and lactate from rat brain microdialysate with preimmobilized oxidase enzymes for glucose and lactate monitoring [188]. They detected 0.06 mM of glucose and 0.059 mM of lactate with this device. 3D printing was also used to create microfluidic reactionware for complex chemical reactions [189].
In addition to increasing reaction rates using simple fluidic formats, more complex reactors were designed for complicated reactions. Jonsson et al. created a microfluidic cartridge type tool to generate peptide fragments for mass spectrometry (MS) analysis [190]. They employed off-stoichiometric thiol-ene as a bulk material and a monolithic stationary phase with immobilized pepsin enzyme and used it in line for cleaving proteins and analyzing them with MS. Several recent developments are summarized in Table 4.
Table 4.
Summary of the applications of 3D printed tools.
| Printing Approach | Device type | Degree of Integration | Analytical Application | Signal Type |
|---|---|---|---|---|
| In Biology | ||||
| SLA [173] | μFN | Module for microwell plate integration | Controlled Oxygen delivery in microplates | EC |
| SLA [62] | μF chip | Integrated biosensing material holders | Microdialysates including glucose and lactate | EC |
| SLA [181] | μF chip | Pumps and Valves | Urinary proteins | C |
| SLA [201] | μF chip | Integrated serpentine micromixer | Cell encapsulation in hydrogel | F |
| SLA [113] | μF chip | Simple flow chip with integrated electrodes | Nucleic Acid | ECL |
| SLA [202] | OMT | Modular | Immunoassay for anti-viral antibody detection | C |
| FDM [203] | μF chip | Simple fluidics with integrated T-junction and serpentine mixers | Assembling linear DNA into complex circular DNA for biological applications, such as transfection | O/S |
| FDM [29] | μF chip | Multi reagent fluidic network in a single chip format | PSA, PSMA, PF4 | ECL |
| FDM [64] | μF chip | Integrated T-junction | Dental pulp stem cell encapsulation in alginate droplets | Optical Imaging |
| FDM [204] | LOC | Integrated standalone tool | Drug-resistant Staphylococcus bacterial harvest, culture, lysis, PCR gene amplification | C gold NPs |
| FDM [205] | LOC | Microfluidic total analysis system | Influenza virus | EC |
| Microjet Printing [166] | μF chip | Integrated T-junction and luer connectors | Microalgae separation using hydrocyclone type feed reactor | BFM |
| Microjet Printing [172] | LOC | Integrated membranes and biosensors | Bioreactor for multicellular organism co-culture; model organisms yeast and euglena | BFM |
| Inkjet Printing [179] | μF chip | Modular; Integrated post-based filtration unit | Plasma separation <10s time | Optical |
| Multiphoton Lithography [167] | μF chip | Simple fluidics with integrated multiple flow trenches | Single cell capture device | Optical |
| In Chemistry/Biochemistry | ||||
| SLA [180] | μF chip | Multi reagent fluidic network in a single chip format | DNA damage potential of E-cigarette smoke components | ECL |
| SLA [206,207] | LOC | Integrated standalone tool with on-chip pumps, valves, references and optics | Chemical testing of glucose and hydrogen peroxide (H2O2) | C and F |
| SLA [206,207] | μF chip | Integrated preconcentration unit | Trace Elements in sea water: Mn, Ni, Cu, Zn, Cd, Pb | MS |
| FDM [190] | μF chip | Microreactor fluidic interface module as a part of integrated chip | Peptide mapping analysis | UPLC-MS |
| FDM [199] | μF chip | Simple fluidics | Droplet Splitter for chemical titrations | F |
| FDM [195] | μF chip | Integrated solid-phase capsule | Solid phase extraction | CG |
| FDM [114] | μF chip | Simple fluidic channel type reactor | H2O2 | EC |
| FDM [208] | μF pad | Integrated channels and T-junction | Nitrite | C |
| FDM [209] | LOC | Integrated chip with sample processing cartridge, reaction chamber, attachments for detection device | Lactate | CL |
| FDM [27] | μF chip | Simple fluidics | Chemical testing of glucose and hydrogen peroxide (H2O2) | C |
| FDM [210] | μF chip | Simple fluidics | Low volume chemical reactor | Various methods |
| DMP [211,212] | LOC | Standalone device with integrated micromixer, gradient generator, droplet extractor, and isotachophoresis | Nitrate and Glucose | C |
| Microjet Printing [194] | μF chip | Simple fluidic channel type reactor with integrated metal electrodes | Dopamine and Nitric Oxide | EC |
| Microjet Printing [13] | μF chip | Integrated on-chip membranes | Linezolid, levofloxacin, and ciprofloxacin transportation for pharmacokinetic applications | MS |
| Inkjet Printing [179] | μF chip | Modular; Integrated post-based filtration unit | Plasma separation <10s time | Optical |
| Multiphoton | μF chip | Simple fluidics with integrated multiple flow trenches | Single cell capture device | Optical |
| Lithography [167] | ||||
| In Chemistry/Biochemistry | ||||
| SLA [180] | μF chip | Multi reagent fluidic network in a single chip format | DNA damage potential of E-cigarette smoke components | ECL |
| SLA [206,207] | LOC | Integrated standalone tool with on-chip pumps, valves, references and optics | Chemical testing of glucose and hydrogen peroxide (H2O2) | C and F |
| SLA [206,207] | μF chip | Integrated preconcentration unit | Trace Elements in sea water: Mn, Ni, Cu, Zn, Cd, Pb | MS |
| FDM [190] | μF chip | Microreactor fluidic interface module as a part of integrated chip | Peptide mapping analysis | UPLC-MS |
| FDM [199] | μF chip | Simple fluidics | Droplet Splitter for chemical titrations | F |
| FDM [195] | μF chip | Integrated solid-phase capsule | Solid phase extraction | CG |
| FDM [114] | μF chip | Simple fluidic channel type reactor | H2O2 | EC |
| FDM [208] | μF pad | Integrated channels and T-junction | Nitrite | C |
| FDM [209] | LOC | Integrated chip with sample processing cartridge, reaction chamber, attachments for detection device | Lactate | CL |
| FDM [27] | μF chip | Simple fluidics | Chemical testing of glucose and hydrogen peroxide (H2O2) | C |
| FDM [210] | μF chip | Simple fluidics | Low volume chemical reactor | Various methods |
| DMP [211,212] | LOC | Standalone device with integrated micromixer, gradient generator, droplet extractor, and isotachophoresis | Nitrate and Glucose | C |
| Microjet Printing [194] | μF chip | Simple fluidic channel type reactor with integrated metal electrodes | Dopamine and Nitric Oxide | EC |
| Microjet Printing [13] | μF chip | Integrated on-chip membranes | Linezolid, levofloxacin, and ciprofloxacin transportation for pharmacokinetic applications | MS |
| Microjet Printing [21] | Open | Simple chip with integrated microstructure-based | Separation of phenol red and bromothymol blue | C and F |
| Fluidic chip | capillary | Separation of Lysozyme and myoglobin | ||
| Microjet Printing [198] | Fluidic chip | Modular with integrated modules namely thin layer flow cell and flow-field shaped solid electrodes | Pb+2 | EC |
| Microjet Printing [197] | μF chip | Simple chip with integrated screen printed electrodes | Cd(II) and Pb(II) | EC |
SLA: Stereolithography; OMT: Optomechanical tool; FDM: Fused Deposition Manufacturing; DMP: Digital Mask Projection; LOC: Lab-on-a-Chip; ECL: Electrochemiluminescence; PSA: Prostate-Specific Antigen; PSMA: Prostate-Specific Membrane Antigen; PF4: Platelet Factor 4; MS: mass Spectrometry; NP: Nanoparticle; Electrochemistry: EC; Colorimetry: C; Fluorescence: F; Optical: O; Spectrophotometry: S; Chromatography: CG; Chemiluminescence: CL; Bright-field microscopy: BFM.
5.2.3.2. Drug testing.
Applications of 3D printed mFs have been extended to other domains of chemical science that include simple fluidics for drug and chemical testing. Testing the effect of drugs, such as cytotoxicity analysis, is another domain where integrated devices are revolutionizing the field. Sweet and colleagues reported a 3D printed device fabricated with multijet printing for determining effective activity of up to three drugs at various concentrations [191]. They were able to study antagonistic, synergistic, and suppressive interactions of drugs on bacteria using this complex fluidic network. In another effort, Anderson et al. created a fluidic device with integrated membranes for studying effective transport of drugs across membranes and their potency [13]. Several other highly integrated tools were developed for studying the effector functions of the drugs enabling parallel in-vitro pharmacokinetic (PK) profiling of drugs [192]. Lockwood’s group in this work have demonstrated the efficiency of their device by performing PK studies of levofloxacin as model drug. These studies were mainly focused on analyzing drug effects on living cells or drug transportation. In a futuristic approach, researchers have studied drug penetration and their metabolism in complex cell and tissue samples grown outside of the chip. LaBonia and colleagues have studied the penetration effects of chemotherapeutic irinotecan drug into HCT 116 colon spheroids and the metabolites were later studied with MALDI-MS [193].
5.2.3.3. Chemical testing.
Among several integrated tools, summarized in Table 4, researchers have developed 3D printed mF devices for measuring biological chemicals, such as dopamine, nitric oxide, and ATP [194] while others have successfully demonstrated devices for analyzing aerosols in the petroleum industry [195], and trace measurements of heavy metals [73,196–200].
6. Regulatory guidelines
The US FDA has recognized the importance of 3D printing in drug related research and development thus has set-up guidelines for developing such devices. As per their guidelines, the basic steps to develop medical devices with 3D printing are (i) device design for creating and validating digital models, (ii) software workflow for interfacing desired design to the printer, (iii) material controls to ensure biocompatibility and consistency, (iv) printing, (v) post-processing for cleaning up the residual substances and confirming with initial design along with sterilization if necessary, (vi) post-process validation and verification to confirm the geometries, and appropriate functioning adhering to the specifications, and (vii) device testing methods to demonstrate that device meets regulatory requirements [213].
Medical devices are typically cleared or approved by the US FDA while materials are evaluated as part of the finished device and its intended use. Devices employing new materials don’t necessarily require rigorous premarket approval, known as PMA (a process used by the US FDA to check if the product manufacturing adheres to the laid out stringent regulations); though these may be cleared/approved through a 510(k) premarket notification process. Further details of the regulatory guidelines related to 3D printed products in medical sciences can be found on the US FDA website [213].
7. Basic fluidics theory for MAS designing
In order to address the readership of this article that primarily targets enthusiasts and a wider scientific community from non-engineering and non-physics background working in the field of microfluidic manufacturing we are briefing few fundamental concepts of fluidics. These fundamentals govern flow fields, mixing, diffusion, and reaction kinetics that are important concepts for chemical and biological workflow. These fundamentals are derived as dimensionless numbers from the concepts of the conservation of mass, energy, and momentum. These dimensionless parameters become important because each of these numbers correspond to an important physical phenomenon and while designing fluidic components these numbers are useful to characterize the physics of the device in question. Simply put, the fundamental processes that could be complex to derive using dimensions, such as lengths, can easily be quantified using their representative dimensionless parameters without the need for complicated mathematical calculations [4]. For example, Tang and colleagues have developed a chip with single channel multi-reagent delivery, 3D mixer, and immunoassay-based biomarker detection using electrochemiluminescence integrated in a series (Fig. 6 II) [177]. Now for clocked delivery of reagents, knowledge of linear flow speed is necessary but the shape and volumetric hold of the oval delivery channels is also equally important. Similarly, for mixing reagents in the 3D mixer we must know the nature of flow in terms of laminarity or turbulence and how many mixer units will be required for desired degree of mixing. When in the detection module of the chip, diffusion of analyte on to the antibody-bound surface becomes clearly of great significance as it will govern the binding kinetics that is eventually responsible for limits of detection. To summarize, chip developed by Tang et al. require a prior knowledge of fluid flow rate, channel dimensions, nature of flow (laminarity/turbulence), diffusion between the fluid layers for enhancing reaction rates. Observing carefully there are several mathematical variables that will control the summarized physical parameters.
Considering the above example, most important of these parameters, which are also summarized in Table 5, is to determine the nature of flow in terms of laminarity. Reynolds Number (Re) [1,4] is the dimensionless entity, which is the most primary of all, that characterizes forced convection and quantifies the extent of flow laminarity in the system. Typically, a value of Re≪1 is characteristic of laminar flow, and indicates consistent fluid flow while Re 1 conversely indicates turbulence through chaotic mixing, vortices etc. From this we would know that in the single channel reagent delivery module Re must be several orders smaller than 1 while in the mixing module it must be significantly greater than 1. Based on this we would be able to characterize the quality of the mixing and number of units required to achieve the extent of mixing. Now, in the detection module of Tang’s unibody chip, diffusivity and extent of mass flow are important for achieving higher analyte detection sensitivity. This can be easily characterized with another dimensionless quantity called Peclet Number (Pe). It signifies convection transport rate for heat and diffusion transport rate for mass and relates mass diffusivity to Re. Other important dimensionless numbers that we encounter routinely in biological and chemical analyses in microfluidic chips are (i) the Grashof Number that defines natural convection in terms of length scales; (ii) Viscous forces, based on the applications, are described by Capillary Number and Weissenberg Number; while (iii) buoyancy is described by Richardson Number [4]. There are many other dimensionless quantities that help in understanding the fluidics at micron level to which details can be found elsewhere [1,4].
Table 5.
Few important Dimensionless Numbers in fluidics and effect of 3DP on them.
| Dimensionless number | Details | Effect of 3D printing |
|---|---|---|
| Reynolds Number | Inertial force/Viscous force convective momentum/viscous momentum Forced Convection |
Print finish and surface roughness affects the magnitude of Re drastically. Printing homogeneity must be characterized and accounted for. |
| Grashof Number (heat) | Natural convection buoyancy force/Viscous force Used to calculate Re for buoyant flow |
Ditto as above |
| Grashof Number (mass) | Controls the lengthscale to natural convection boundary layer thickness Natural Convection |
|
| Richardson Number | Buoyancy/Flow gradient | Ditto as above |
| Prandtl Number (heat) | Momentum/Species diffusivity Used to determine fluid or heat or mass transfer boundary layer thickness | Structural print limit imposed by 3DP restricts the extent of diffusivity that can be achieved. 3DP tools with surface finish similar to conventional fluidic tools can only achieve the mixing or mixing-controlled reaction kinetics only by a fraction to that of conventional fluidics |
| Prandtl-Schmidt Number (mass) | ||
| Rayleigh Number (heat) | Natural convection/Diffusive heat or mass transport Used to determine the transition to turbulence | |
| Rayleigh Number (mass) | ||
| Capillary Number | Viscous forces/Interfacial forces | Raw material viscosity could impact the measurement of these quantities. Uncured resin trapped within the channels could affect fluid transport phenomenon |
| Elasticity Number | Elastic effects/Inertial effects | |
| Weissenberg Number | Viscous forces/Elastic forces |
8. Perspective and conclusion
In past several years 3D printing has evolved as a genuine competitor to fluidics developed via conventional lithography approaches [214]. This is due in large part to the relatively recent commercial availability of very low cost ($500–3000), high quality 3D printers. Advances in the printing technology that allow developing integrated structures that were once out of the scope for soft lithography emphasize the fact that we are in the early stages of a manufacturing revolution [215]. 3D printing has already been established as a standard approach in die and casting industry. This is mainly facilitated by the availability of an array of print materials, such as tough polymers that can withstand high temperatures. Emphasis must now be given to engineering solutions, and to design new polymers/materials that can be readily printed. With the advent of CLIP technology, we can now further develop devices with z-resolution as low as 1 μm [93]. Although, xy-resolutions are still comparable to DLP but high print speed is a big buzz factor for 3DP market. UDP by UNIZ has taken over all other 3DP approaches in terms of speed by achieving unprecedented print rates, several fold faster than CLIP, of ~800 mm/h or ~73,000 cc/hr with z-resolutions as low as 10 mm. Now, with new 3D printing technologies on the horizon we are able to print complex structures and multi-level fluidics, such as curved or serpentine channels and mixers that can be routinely printed using SLA printers [83], which were presumably very challenging in the past as described in previous review articles [57,81,160]. With all the engineering advances researchers were still able to fabricate highly integrated tools by improving the workflows. PDMS 3D printing was considered an unimaginable task few years earlier. However, PDMS has been successfully printed with FDM and SLA printers [74,75,95,96]. It is achieved due to innovations in the workflow, such as modifying or replacing commercial pin heads with needles or glass capillaries. We have moved several paces forward to create complex 3D fluidic networks that are being employed in advanced applications extending from cell separation to trace element detection. This technology has advanced from printing functional modular tools to membrane or sensor integrated multimaterial devices [5,94,168]. Advancements in polymer technology have significantly complemented the engineering aspects of 3DP. New polymers with various customizable physico-chemical properties, such as tensile strength, heat resistance, and chemical resistance etc, have facilitated the development of tools for a plethora of biochemical applications. Also, glass printing is a new area under investigation that will further increase the application range of 3DP in microscale sciences [174,216,217].
Although 3D printing has enabled us to develop complex devices rapidly, it is often used as complementary to soft lithography even when a device of equivalent quality could be directly 3D printed. In addition, the research to market reach for 3D printed fluidic devices is non-existent. A consortium approach may be beneficial with commercial partners who have the capability and infrastructure to look into regulatory clearances and commercialization aspects of newly developed mF platforms. Recent developments have laid a clear path to an increased market share of 3D printed mF technology with the potential to revolutionize various scientific domains, but in particular personalized healthcare and medical research devices.
Acknowledgements
This work was supported financially by grant Nos. EB016707 from the National Institute of Biomedical Imaging and Bioengineering (NIBIB), NIH.
References
- [1].Dixit CK, Kaushik A (Editors), Microfluidics for Biologists e Fundamentals and Applications, Dixit j Chandra, first ed., Springer, Springer International Publishing, Switzerland, 2016. [Google Scholar]
- [2].O’Connell TM, King D, Dixit CK, O’Connor B, Walls D, Ducrée J, Lab Chip 14 (2014) 3629–3639. [DOI] [PubMed] [Google Scholar]
- [3].Luka G, Ahmadi A, Najjaran H, Alocilja E, DeRosa M, Wolthers K, Malki A, Aziz H, Althani A, Hoorfar M, Sensors 15 (2015) 30011–30031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Dixit CK, in: Dixit CK, Kaushik A (Editors), Microfluidics for Biologists: Fundamentals and Applications, Springer International Publishing, Cham, 2016, pp. 1–32. [Google Scholar]
- [5].Au AK, Huynh W, Horowitz LF, Folch A, Angew. Chem. Int. Ed 55 (2016) 3862–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Amin R, Knowlton S, Hart A, Yenilmez B, Ghaderinezhad F, Sara Katebifar M. Messina, Khademhosseini A, Tasoglu S, Biofabrication 8 (2016) 022001. [DOI] [PubMed] [Google Scholar]
- [7].Ho CMB, Ng SH, Li KHH, Yoon Y-J, Lab Chip 15 (2015) 3627–3637. [DOI] [PubMed] [Google Scholar]
- [8].Volpatti LR, Yetisen AK, Trends Biotechnol. 32 (2014) 347–350. [DOI] [PubMed] [Google Scholar]
- [9].Ligon SC, Liska R, Stampfl J, Gurr M, Mülhaupt R, Chem. Rev 117 (2017) 10212–10290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Nguyen DT, Meyers C, Yee TD, Dudukovic NA, Destino JF, Zhu C, Duoss EB, Baumann TF, Suratwala T, Smay JE, Dylla-Spears R, Adv. Mater 29 (2017), 1701181. [DOI] [PubMed] [Google Scholar]
- [11].Bertana V, Potrich C, Scordo G, Scaltrito L, Ferrero S, Lamberti A, Perrucci F, Pirri CF, Pederzolli C, Cocuzza M, Marasso SL, J. Vac. Sci. Technol. B Nanotechnol. Microelectron. Mater. Process. Meas. Phenom 36 (2017) 01A106. [Google Scholar]
- [12].Wang J, McMullen C, Yao P, Jiao N, Kim M, Kim J-W, Liu L, Tung S, Microfluid. Nanofluidics 21 (2017) 105. [Google Scholar]
- [13].Anderson KB, Lockwood SY, Martin RS, Spence DM, Anal. Chem 85 (2013) 5622–5626. [DOI] [PubMed] [Google Scholar]
- [14].Alessandri K, Andrique L, Feyeux M, Bikfalvi A, Nassoy P, Recher G, Sci. Rep (2017). 10.1038/srep42378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rawat S, Komatsu S, Markman A, Anand A, Javidi B, Appl. Opt 56 (2017) D127–D133. [DOI] [PubMed] [Google Scholar]
- [16].Beauchamp MJ, Nordin GP, Woolley AT, Anal. Bioanal. Chem 409 (2017) 4311–4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Au AK, Lee W, Folch A, Lab Chip 14 (2014) 12942–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bhargava KC, Thompson B, Malmstadt N, Proc. Natl. Acad. Sci 111 (2014) 15013–15018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Waheed S, Cabot JM, Macdonald NP, Lewis T, Guijt RM, Paull B, Breadmore MC, Lab Chip 16 (2016) 1993–2013. [DOI] [PubMed] [Google Scholar]
- [20].Chen C, Mehl BT, Munshi AS, Townsend AD, Spence DM, Martin RS, Anal. Methods 8 (2016) 6005–6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Macdonald NP, Cabot JM, Smejkal P, Guijt RM, Paull B, Breadmore MC, Anal. Chem 89 (2017) 3858–3866. [DOI] [PubMed] [Google Scholar]
- [22].Sun M, Xie Y, Zhu J, Li J, Eijkel JCT, Anal. Chem 89 (2017) 2227–2231. [DOI] [PubMed] [Google Scholar]
- [23].Bhattacharjee N, Urrios A, Kang S, Folch A, Lab Chip 16 (2016) 1720–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bauer M, Kulinsky L, Micromachines 9 (2018) 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Nie J, Gao Q, Qiu J, Sun M, Liu A, Shao L, Fu J, Zhao P, He Y, Biofabrication 10 (2018) 035001. [DOI] [PubMed] [Google Scholar]
- [26].3D Printers, Desktop, Professional and Industrial, Uniz, https://uniz3d.com/.
- [27].Tothill AM, Partridge M, James SW, Tatam RP, J. Micromech. Microeng 27 (2017) 035018. [Google Scholar]
- [28].Gong H, Beauchamp M, Perry S, Woolley AT, Nordin GP, RSC Adv 5 (2015) 106621–106632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kadimisetty K, Mosa IM, Malla S, Satterwhite-Warden JE, Kuhns TM, Faria RC, Lee NH, Rusling JF, Biosens. Bioelectron 77 (2016) 188–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Rogers CI, Qaderi K, Woolley AT, Nordin GP, Biomicrofluidics (2015). 10.1063/1.4905840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Gokaltun A, Yarmush ML, Asatekin A, Usta OB, Technology 05 (2017) 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wong I, Ho C-M, Microfluid. Nanofluidics 7 (2009) 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Woldemariam M, Filimonov R, Purtonen T, Sorvari J, Koiranen T, Eskelinen H, Chem. Eng. Sci 152 (2016) 26–34. [Google Scholar]
- [34].Mohanty S, Larsen LB, Trifol J, Szabo P, Burri HVR, Canali C, Dufva M, Emnéus J, Wolff A, Mater. Sci. Eng. C 55 (2015) 569–578. [DOI] [PubMed] [Google Scholar]
- [35].Olanrewaju AO, Robillard A, Dagher M, Juncker D, Lab Chip 16 (2016) 3804–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Sharma A, Mondal S, Mondal AK, Baksi S, Patel RK, Chu W-S, Pandey JK, Int. J. Precis. Eng. Manuf.-Green Technol 4 (2017) 323–334. [Google Scholar]
- [37].Aassime A, Hamouda F, Mod. Technol. Creat. Thin-Film Syst. Coat (2017). 10.5772/66028. [DOI]
- [38].Dimov N, in: Dixit CK, Kaushik A (Editors), Microfluidics for Biologists: Fundamentals and Applications, Springer International Publishing, Cham, 2016, pp. 85–102. [Google Scholar]
- [39].Lee LJ, Madou MJ, Koelling KW, Daunert S, Lai S, Koh CG, Juang Y-J, Lu Y, Yu L, Biomed. Microdevices 3 (2001) 339–351. [Google Scholar]
- [40].Chen P-C, Pan C-W, Lee W-C, Li K-M, Int. J. Adv. Manuf. Technol 71 (2014) 1623–1630. [Google Scholar]
- [41].Wilson ME, Kota N, Kim Y, Wang Y, Stolz DB, LeDuc PR, Ozdoganlar OB, Lab Chip 11 (2011) 1550–1555. [DOI] [PubMed] [Google Scholar]
- [42].Mecomber JS, Hurd D, Limbach PA, Int. J. Mach. Tool Manufact 45 (2005) 1542–1550. [Google Scholar]
- [43].Lee H, Lee K, Ahn B, Xu J, Xu L, Oh KW, J. Micromech. Microeng 21 (2011) 125006. [Google Scholar]
- [44].Anhoj TA, Jorgensen AM, Zauner DA, Hübner J, J. Micromech. Microeng 16 (2006) 1819. [Google Scholar]
- [45].Zhang J, Tan KL, Hong GD, Yang LJ, Gong HQ, J. Micromech. Microeng 11 (2001) 20. [Google Scholar]
- [46].Pinto VC, Sousa PJ, Cardoso VF, Minas G, Micromachines 5 (2014) 738–755. [Google Scholar]
- [47].Kung Yu-Chun, Huang Kuo-Wei, Fan Yu-Jui, Chiou Pei-Yu, Lab Chip 15 (2015) 1861–1868. [DOI] [PubMed] [Google Scholar]
- [48].Estévez-Torres A, Yamada A and Wang L, An inexpensive and durable epoxy mould for PDMS e Chips and Tips, http://blogs.rsc.org/chipsandtips/2009/04/22/an-inexpensive-and-durable-epoxy-mould-for-pdms/.
- [49].Kamande JW, Wang Y, Taylor AM, Biomicrofluidics (2015). 10.1063/1.4922962. [DOI] [PMC free article] [PubMed]
- [50].Suriano R, Kuznetsov A, Eaton SM, Kiyan R, Cerullo G, Osellame R, Chichkov BN, Levi M, Turri S, Appl. Surf. Sci 257 (2011) 6243–6250. [Google Scholar]
- [51].Mohammed MI, Alam MNHZ, Kouzani A, Gibson I, J. Micromech. Microeng 27 (2017) 015021. [Google Scholar]
- [52].Hsieh Y-K, Chen S-C, Huang W-L, Hsu K-P, Gorday KAV, Wang T, Wang J, Polymers 9 (2017) 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Tanikella NG, Wittbrodt B, Pearce JM, Addit. Manuf 15 (2017) 40–47. [Google Scholar]
- [54].Shirazi SFS, Gharehkhani S, Mehrali M, Yarmand H, Metselaar HSC, Kadri NA, Osman NAA, Sci. Technol. Adv. Mater 16 (2015) 033502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Olakanmi EO, Cochrane RF, Dalgarno KW, Prog. Mater. Sci 74 (2015) 401–477. [Google Scholar]
- [56].Chia HN, Wu BM, J. Biol. Eng 9 (2015) 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Bhushan B, Caspers M, Microsyst. Technol 23 (2017) 1117–1124. [Google Scholar]
- [58].Ambrosi A, Pumera M, Chem. Soc. Rev 45 (2016) 2740–2755. [DOI] [PubMed] [Google Scholar]
- [59].Zhou X, Hou Y, Lin J, AIP Adv 5 (2015) 030701. [Google Scholar]
- [60].Göring G, Dietrich P-I, Blaicher M, Sharma S, Korvink JG, Schimmel T, Koos C, Hölscher H, Appl. Phys. Lett 109 (2016) 063101. [Google Scholar]
- [61].Kumi G, Yanez CO, Belfield KD, Fourkas JT, Lab Chip 10 (2010) 1057–1060. [DOI] [PubMed] [Google Scholar]
- [62].Gowers SAN, Curto VF, Seneci CA, Wang C, Anastasova S, Vadgama P, Yang G-Z, Boutelle MG, Anal. Chem 87 (2015) 7763–7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Ertay DS, Yuen A, Altintas Y, Addit. Manuf 19 (2018) 205–213. [Google Scholar]
- [64].Morgan AJL, Jose LHS, Jamieson WD, Wymant JM, Song B, Stephens P, Barrow DA, Castell OK, PLoS One 11 (2016) e0152023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Li Y, Linke BS, Voet H, Falk B, Schmitt R, Lam M, CIRP J Manuf. Sci. Technol 16 (2017) 1–11. [Google Scholar]
- [66].McIlroy C, Olmsted PD, Rheol J. 61 (2017) 379–397. [Google Scholar]
- [67].Mohseni M, Hutmacher DW, Castro NJ, Polymers 10 (2018) 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Brenken B, Barocio E, Favaloro A, Kunc V, Pipes RB, Addit. Manuf 21 (2018) 1–16. [Google Scholar]
- [69].Salentijn GI, Oomen PE, Grajewski M, Verpoorte E, Anal. Chem 89 (2017) 7053–7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Li F, Smejkal P, Macdonald NP, Guijt RM, Breadmore MC, Anal. Chem 89 (2017) 4701–4707. [DOI] [PubMed] [Google Scholar]
- [71].Yang K, Grant JC, Lamey P, Joshi-Imre A, Lund BR, Smaldone RA, Voit W, Adv. Funct. Mater 27 (2017) (n/a-n/a). [Google Scholar]
- [72].Serex L, Bertsch A, Renaud P, Micromachines 9 (2018) 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Gross B, Lockwood SY, Spence DM, Anal. Chem 89 (2017) 57–70. [DOI] [PubMed] [Google Scholar]
- [74].Hinton TJ, Hudson A, Pusch K, Lee A, Feinberg AW, ACS Biomater. Sci. Eng 2 (2016) 1781–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Li Z, Yang J, Li K, Zhu L, Tang W, RSC Adv 7 (2017) 3313–3320. [Google Scholar]
- [76].Mao M, He J, Li X, Zhang B, Lei Q, Liu Y, Li D, Micromachines 8 (2017) 113. [Google Scholar]
- [77].Carve M, Wlodkowic D, Micromachines 9 (2018) 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Caterina Credi, Gianmarco Griffini, Marinella Levi, Stefano Turri, Small 14 (2017) 1702831. [Google Scholar]
- [79].Baumann FW, Wellekötter J, Roller D, Bonten C, Comput.-Aided Des. Appl 14 (2017) 486–497. [Google Scholar]
- [80].Heinrich A, Rank M, Suresh Nair S, Bauckhage Y, Maillard P, Organic Photonic Materials and Devices XIX Proceedings Volume 10101, San Francisco, California, United States, 2017, p. 1010118. [Google Scholar]
- [81].Stansbury JW, Idacavage MJ, Dent. Mater 32 (2016) 54–64. [DOI] [PubMed] [Google Scholar]
- [82].Wang X, Jiang M, Zhou Z, Gou J, Hui D, Compos. B Eng 110 (2017) 442–458. [Google Scholar]
- [83].Lee J-Y, An J, Chua CK, Appl. Mater. Today 7 (2017) 120–133. [Google Scholar]
- [84].Shusteff M, Browar AEM, Kelly BE, Henriksson J, Weisgraber TH, Panas RM, Fang NX, Spadaccini CM, Sci. Adv (2017). 10.1126/sciadv.aao5496. [DOI] [PMC free article] [PubMed]
- [85].United States Patent, US7211368B2, 2007.
- [86].Sycks DG, Wu T, Park HS, Gall K, J. Appl. Polym. Sci 135 (2018) 46259. [Google Scholar]
- [87].United States Patent, US20170291357A1, 2017.
- [88].Ortiz-Gutiérrez M, Olivares-Pérez A, Alvarado-Méndez E, Trejo-Durán M, Salgado-Verduzco MA, J. Appl. Res. Technol 13 (2015) 561–565. [Google Scholar]
- [89].Hoffmann A, Leonards H, Tobies N, Pongratz L, Kreuels K, Kreimendahl F, Apel C, Wehner M, Nottrodt N, J. Tissue Eng 8 (2017), 2041731417744485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Park HY, Kloxin CJ, Scott TF, Bowman CN, Macromolecules 43 (2010) 10188–10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Wu C, Yi R, Liu YJ, He Y, Wang CCL, in: 2016 IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS), 2016, pp. 2155–2160. [Google Scholar]
- [92].Kovalenko I, Verron S, Garan M, Šafka J, Moučka M, Open Eng. (2017). 10.1515/eng-2017-0016. [DOI]
- [93].Tumbleston JR, Shirvanyants D, Ermoshkin N, Janusziewicz R, Johnson AR, Kelly D, Chen K, Pinschmidt R, Rolland JP, Ermoshkin A, Samulski ET, DeSimone JM, Science 347 (2015) 1349–1352. [DOI] [PubMed] [Google Scholar]
- [94].Yang L, Hsu K, Baughman B, Godfrey D, Medina F, Menon M, Wiener S, in: Additive Manufacturing of Metals: the Technology, Materials, Design and Production, Springer, Cham, 2017, pp. 1–31. [Google Scholar]
- [95].Palaganas J, de Leon AC, Mangadlao J, Palaganas N, Mael A, Lee YJ, Lai HY, Advincula R, Macromol. Mater. Eng 302 (2017), 1600477. [Google Scholar]
- [96].Sivashankar S, Agambayev S, Alamoudi K, Buttner U, Khashab N, Salama KN, Micro & Nano Lett 11 (2016) 654–659. [Google Scholar]
- [97].Li J, Chen M, Fan X, Zhou H, J. Transl. Med 14 (2016) 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Walczak R, Adamski K, Lizanets D, J. Micromech. Microeng 27 (2017) 047002. [Google Scholar]
- [99].Sochol RD, Sweet E, Glick CC, Venkatesh S, Avetisyan A, Ekman KF, Raulinaitis A, Tsai A, Wienkers A, Korner K, Hanson K, Long A, Hightower BJ, Slatton G, Burnett DC, Massey TL, Iwai K, Lee LP, Pister KSJ, Lin L, Lab Chip 16 (2016) 668–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Bonyár A, Sántha H, Varga M, Ring B, Vitéz A, Harsányi G, Int. J. Material Form 7 (2014) 189–196. [Google Scholar]
- [101].Yuen PK, Biomicrofluidics 10 (2016) 044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Kamei K, Mashimo Y, Koyama Y, Fockenberg C, Nakashima M, Nakajima M, Li J, Chen Y, Biomed. Microdevices 17 (2015) 36. [DOI] [PubMed] [Google Scholar]
- [103].Chan HN, Chen Y, Shu Y, Chen Y, Tian Q, Wu H, Microfluid. Nanofluidics 19 (2015) 9–18. [Google Scholar]
- [104].Comina G, Suska A, Filippini D, Lab Chip 14 (2013) 424–430. [DOI] [PubMed] [Google Scholar]
- [105].Villegas M, Cetinic Z, Shakeri A, Didar TF, Anal. Chim. Acta 1000 (2018) 248–255. [DOI] [PubMed] [Google Scholar]
- [106].Hwang Y, Paydar OH, Candler RN, Sens. Actuators Phys 226 (2015) 137–142. [Google Scholar]
- [107].Glick CC, Srimongkol MT, Schwartz AJ, Zhuang WS, Lin JC, Warren RH, Tekell DR, Satamalee PA, Lin L, Microsyst. Nanoeng 2 (2016), 201663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Saggiomo V, Velders AH, Adv. Sci 2 (2015), 1500125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Chan HN, Shu Y, Tian Q, Chen Y, Chen Y, Wu H, Mater. Horiz 3 (2016) 309–313. [Google Scholar]
- [110].Keating SJ, Gariboldi MI, Patrick WG, Sharma S, Kong DS, Oxman N, PLoS One 11 (2016), e0160624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Gong H, Woolley AT, Nordin GP, Lab a Chip 16 (2016) 2450–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Bhargava KC, Ermagan R, Thompson B, Friedman A, Malmstadt N, Micromachines 8 (2017) 137. [Google Scholar]
- [113].Bishop GW, Satterwhite-Warden JE, Bist I, Chen E, Rusling JF, ACS Sens. 1 (2016) 197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Bishop GW, Satterwhite JE, Bhakta S, Kadimisetty K, Gillette KM, Chen E, Rusling JF, Anal. Chem 87 (2015) 5437–5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Xiong B, Ren K, Shu Y, Chen Y, Shen B, Wu H, Adv. Mater 26 (2014) 5525–5532. [DOI] [PubMed] [Google Scholar]
- [116].Zhang C, Bioanalysis 9 (2017) 329–331. [DOI] [PubMed] [Google Scholar]
- [117].Tourovskaia A, Figueroa-Masot X, Folch A, Lab Chip 5 (2005) 14–19. [DOI] [PubMed] [Google Scholar]
- [118].Hung PJ, Lee PJ, Sabounchi P, Lin R, Lee LP, Biotechnol. Bioeng 89 (2005) 1–8. [DOI] [PubMed] [Google Scholar]
- [119].Nevill JT, Cooper R, Dueck M, Breslauer DN, Lee LP, Lab Chip 7 (2007) 1689–1695. [DOI] [PubMed] [Google Scholar]
- [120].Lei KF, Wu M-H, Hsu C-W, Chen Y-D, Biosens. Bioelectron 51 (2014) 16–21. [DOI] [PubMed] [Google Scholar]
- [121].Nguyen TA, Yin T-I, Reyes D, Urban GA, Anal. Chem 85 (2013) 11068–11076. [DOI] [PubMed] [Google Scholar]
- [122].Nieto D, McGlynn P, de la Fuente M, Lopez-Lopez R, O’connor GM, Colloids Surfaces B Biointerfaces 154 (2017) 263–269. [DOI] [PubMed] [Google Scholar]
- [123].Gröger M, Lange M, Rennert K, Kaschowitz T, Plettenberg H, Hoffmann M, Mosig AS, Eng. Life Sci 17 (2017) 585–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Tanataweethum N, Yang F, Zelaya A, Karnik S, Cohen RN, Brey E, Bhushan A, Faseb. J 31 (2017), 886.8–886.8. [Google Scholar]
- [125].Duval K, Grover H, Han L-H, Mou Y, Pegoraro AF, Fredberg J, Chen Z, Physiology 32 (2017) 266–277. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- [126].Klingelhutz AJ, Gourronc F, Chaly A, Ankrum J, Faseb. J 31 (2017), 88.1–88.1. [Google Scholar]
- [127].Sieber S, Wirth L, Cavak N, Koenigsmark M, Marx U, Lauster R, Rosowski M, Tissue Eng J. Regen.Med (2018) 479–489. [DOI] [PubMed] [Google Scholar]
- [128].Kido H, Micic M, Smith D, Zoval J, Norton J, Madou M, Colloids Surfaces B Biointerfaces 58 (2007) 44–51. [DOI] [PubMed] [Google Scholar]
- [129].Cors JF, Kashyap A, Khartchenko AF, Schraml P, Kaigala GV, PLoS One 12 (2017), e0176691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Lee W, Tseng P, Carlo DD, in: Microtechnology for Cell Manipulation and Sorting, Springer International Publishing, Cham, 2017, pp. 1–14. [Google Scholar]
- [131].Huang L, Bian S, Cheng Y, Shi G, Liu P, Ye X, Wang W, Biomicrofluidics 11 (2017) 011501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Al-Faqheri W, Thio THG, Qasaimeh MA, Dietzel A, Madou M, Al-Halhouli A, Microfluid. Nanofluidics 21 (2017) 102. [Google Scholar]
- [133].Rollo E, Tenaglia E, Genolet R, Bianchi E, Harari A, Coukos G, Guiducci C, Biosens. Bioelectron 94 (2017) 193–199. [DOI] [PubMed] [Google Scholar]
- [134].Girault M, Kim H, Arakawa H, Matsuura K, Odaka M, Hattori A, Terazono H, Yasuda K, Sci. Rep 7 (2017) srep40072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Chen Y-C, Ingram PN, Fouladdel S, McDermott SP, Azizi E, Wicha MS, Yoon E, Sci. Rep 6 (2016) srep27301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Antfolk M, Kim SH, Koizumi S, Fujii T, Laurell T, Sci. Rep 7 (2017) srep46507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Au SH, Edd J, Stoddard AE, Wong KHK, Fachin F, Maheswaran S, Haber DA, Stott SL, Kapur R, Toner M, Sci. Rep 7 (2017) 2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Torino S, Iodice M, Rendina I, Coppola G, Biophys 4 (2017) 178–191. [Google Scholar]
- [139].Aguirre GR, Efremov V, Kitsara M, Ducrée J, Microfluid. Nanofluidics 18 (2015) 513–526. [Google Scholar]
- [140].Antfolk M, Laurell T, Anal. Chim. Acta 965 (2017) 9–35. [DOI] [PubMed] [Google Scholar]
- [141].Park M-H, Reátegui E, Li W, Tessier SN, Wong KHK, Jensen AE, Thapar V, Ting D, Toner M, Stott SL, Hammond PT, J. Am. Chem. Soc 139 (2017) 2741–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Karabacak NM, Spuhler PS, Fachin F, Lim EJ, Pai V, Ozkumur E, Martel JM, Kojic N, Smith K, Chen P, Yang J, Hwang H, Morgan B, Trautwein J, Barber TA, Stott SL, Maheswaran S, Kapur R, Haber DA, Toner M, Nat. Protoc 9 (2014) 694–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Menachery A, Kumawat N, Qasaimeh M, Trac Trends Anal. Chem 89 (2017) 1–12. [Google Scholar]
- [144].Tak For Yu Z, Guan H, Ki Cheung M, McHugh WM, Cornell TT, Shanley TP, Kurabayashi K, Fu J, Sci. Rep 5 (2015) 11339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Liu C, Liao S-C, Song J, Mauk MG, Li X, Wu G, Ge D, Greenberg RM, Yang S, Bau HH, Lab Chip 16 (2016) 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Sinkala E, Sollier-Christen E, Renier C, Rosás-Canyelles E, Che J, Heirich K, Duncombe TA, Vlassakis J, Yamauchi KA, Huang H, Jeffrey SS, Herr AE, Nat. Commun 8 (2017) ncomms14622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Lin Y-S, Liu W, Hu C, J. Chromatogr. Sep. Tech 8 (2017) 345. [Google Scholar]
- [148].Vu TQ, de Castro RMB, Qin L, Lab Chip 17 (2017) 1009–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Solanki S, Pandey CM, in: Dixit CK, Kaushik A (Editors), Microfluidics for Biologists - Funcdamentals and Applications, first ed., Springer International Publishing; Switzerland, 2016, pp. 191–221. [Google Scholar]
- [150].Prakadan SM, Shalek AK, Weitz DA, Nat. Rev. Genet 18 (2017) 345–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Shehadul Islam M, Aryasomayajula A, Selvaganapathy PR, Micromachines 8 (2017) 83. [Google Scholar]
- [152].Fan Q, Hu W, Ohta AT, Micromachines 8 (2017) 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Cheng Y, Wang Y, Wang Z, Huang L, Bi M, Xu W, Wang W, Ye X, Biomicrofluidics 11 (2017) 024112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Lim D, Yoo JC, Appl. Biochem. Biotechnol (2017) 1–10. [DOI] [PubMed] [Google Scholar]
- [155].Wollman AJM, Shashkova S, Welkenhuysen N, Hedlund EG, Hohmann S, Leake MC, Biophys. J 112 (2017) 313a.28122217 [Google Scholar]
- [156].Peter H, Wienke J, Guest PC, Bistolas N, Bier FF, in: Proteomic Methods in Neuropsychiatric Research, Springer International Publishing, Cham, 2017, pp. 339–349. [Google Scholar]
- [157].Su Y, Shi Q, Wei W, Proteomics 17 (2017) 1600267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Jie M, Mao S, Li H, Lin J-M, Chin. Chem. Lett 28 (2017) 1625–1630. [Google Scholar]
- [159].Marioni JC, Arendt D, Annu. Rev. Cell Dev. Biol 33 (2017) 537–553. [DOI] [PubMed] [Google Scholar]
- [160].Chiu DT, deMello AJ, Di Carlo D, Doyle PS, Hansen C, Maceiczyk RM, Wootton RCR, Inside Chem. 2 (2017) 201–223. [Google Scholar]
- [161].Lu C, Verbridge SS (Editors), Microfluidic Methods for Molecular Biology, first ed., Springer International Publishing; Switzerland, Cham, 2016. [Google Scholar]
- [162].Tomazelli Coltro WK, Cheng C-M, Carrilho E, de Jesus DP, Electrophoresis 35 (2014) 2309–2324. [DOI] [PubMed] [Google Scholar]
- [163].Papadakis G, Friedt JM, Eck M, Rabus D, Jobst G, Gizeli E, Biomed. Microdevices 19 (2017) 16. [DOI] [PubMed] [Google Scholar]
- [164].Becker H, Gärtner C, in: Taly V, Viovy J-L, Descroix S (Editors), Microchip Diagnostics, Springer, New York, 2017, pp. 3–21. [Google Scholar]
- [165].Zhang Y, Ge S, Yu J, Trac Trends Anal. Inside Chem 85 (2016) 166–180. [Google Scholar]
- [166].Syed MS, Rafeie M, Henderson R, Vandamme D, Asadnia M, Warkiani ME, Lab Chip 17 (2017) 2459–2469. [DOI] [PubMed] [Google Scholar]
- [167].Spivey EC, Xhemalce B, Shear JB, Finkelstein IJ, Anal. Chem 86 (2014) 7406–7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Au AK, Bhattacharjee N, Horowitz LF, Chang TC, Folch A, Lab Chip 15 (2015) 1934–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Lee W, Kwon D, Choi W, Jung GY, Au AK, Folch A, Jeon S, Sci. Rep 5 (2015) srep07717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Warkiani ME, Tay AKP, Guan G, Han J, Sci. Rep 5 (2015) srep11018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Hahn YK, Hong D, Kang JH, Choi S, Micromachines 7 (2016) 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Podwin A, Dziuban JA, J. Micromech. Microeng (2017). 10.1088/1361-6439/aa7a72. [DOI]
- [173].Brennan MD, Rexius-Hall ML, Eddington DT, PLoS One 10 (2015), e0137631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Kotz F, Arnold K, Bauer W, Schild D, Keller N, Sachsenheimer K, Nargang TM, Richter C, Helmer D, Rapp BE, Nature 544 (2017) 337–339. [DOI] [PubMed] [Google Scholar]
- [175].Singh M, Tong Y, Webster K, Cesewski E, Haring AP, Laheri S, Carswell B, O’Brien TJ, Aardema CH, Senger RS, Robertson JL, Johnson BN, Lab Chip 17 (2017) 2561–2571. [DOI] [PubMed] [Google Scholar]
- [176].Damiati S, Küpcü S, Peacock M, Eilenberger C, Zamzami M, Qadri I, Choudhry H, Sleytr UB, Schuster B, Biosens. Bioelectron 94 (2017) 500–506. [DOI] [PubMed] [Google Scholar]
- [177].Tang CK, Vaze A, Rusling JF, Lab Chip 17 (2017) 484–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Chen C, Wang Y, Lockwood SY, Spence DM, Analyst 139 (2014) 3219–3226. [DOI] [PubMed] [Google Scholar]
- [179].Bilatto SER, Adly NY, Correa DS, Wolfrum B, Offenhäausser A, Yakushenko A, Biomicrofluidics 11 (2017) 034101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Kadimisetty K, Malla S, Rusling JF, ACS Sens. 2 (2017) 670–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Chan HN, Shu Y, Xiong B, Chen Y, Chen Y, Tian Q, Michael SA, Shen B, Wu H, ACS Sens. 1 (2016) 227–234. [Google Scholar]
- [182].Plevniak K, Campbell M, He M, in: 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), 2016, pp. 267–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Plevniak K, Campbell M, Myers T, Hodges A, He M, Biomicrofluidics 10 (2016) 054113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Jue E, Schoepp NG, Witters D, Ismagilov RF, Lab Chip 16 (2016) 1852–1860. [DOI] [PubMed] [Google Scholar]
- [185].Chan HN, Tan MJA, Wu H, Lab Chip 17 (2017) 2713–2739. [DOI] [PubMed] [Google Scholar]
- [186].Kadimisetty K, Song J, Doto AM, Hwang Y, Peng J, Mauk MG, Bushman FD, Gross R, Jarvis JN, Liu C, Biosens. Bioelectron 109 (2018) 156–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Bishop GW, Satterwhite-Warden JE, Kadimisetty K, Rusling JF, Nano-technology 27 (2016) 284002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Su C-K, Yen S-C, Li T-W, Sun Y-C, Anal. Chem 88 (2016) 6265–6273. [DOI] [PubMed] [Google Scholar]
- [189].Kitson PJ, Glatzel S, Chen W, Lin C-G, Song Y-F, Cronin L, Nat. Protoc 11 (2016) 920–936. [DOI] [PubMed] [Google Scholar]
- [190].Jönsson A, Svejdal RR, Bøgelund N, Nguyen TTTN, Flindt H, Kutter JP, Rand KD, Lafleur JP, Anal. Chem 89 (2017) 4573–4580. [DOI] [PubMed] [Google Scholar]
- [191].Sweet EC, Chen JCL, Karakurt I, Long AT, Lin L, in: 2017 IEEE 30th International Conference on Micro Electro Mechanical Systems (MEMS), 2017, pp. 205–208. [Google Scholar]
- [192].Lockwood SY, Meisel JE, Monsma FJ, Spence DM, Anal. Chem 88 (2016) 1864–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].LaBonia GJ, Lockwood SY, Heller AA, Spence DM, Hummon AB, Proteomics 16 (2016) 1814–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Erkal JL, Selimovic A, Gross BC, Lockwood SY, Walton EL, McNamara S, Martin RS, Spence DM, Lab Chip 14 (2014) 2023–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Kataoka ÉM, Murer RC, Santos JM, Carvalho RM, Eberlin MN, Augusto F, Poppi RJ, Gobbi AL, Hantao LW, Anal. Chem 89 (2017) 3460–3467. [DOI] [PubMed] [Google Scholar]
- [196].Su C-K, Peng P-J, Sun Y-C, Anal. Chem 87 (2015) 6945–6950. [DOI] [PubMed] [Google Scholar]
- [197].Hong Y, Wu M, Chen G, Dai Z, Zhang Y, Chen G, Dong X, ACS Appl. Mater. Interfaces 8 (2016) 32940–32947. [DOI] [PubMed] [Google Scholar]
- [198].Sun Q, Wang J, Tang M, Huang L, Zhang Z, Liu C, Lu X, Hunter KW, Chen G, Anal. Chem 89 (2017) 5024–5029. [DOI] [PubMed] [Google Scholar]
- [199].Hutama TJ, Oleschuk RD, Lab Chip 17 (2017) 2640–2649. [DOI] [PubMed] [Google Scholar]
- [200].Calderilla C, Maya F, Cerdá V, Leal LO, Talanta 184 (2018) 15–22. [DOI] [PubMed] [Google Scholar]
- [201].Knowlton S, Yu CH, Ersoy F, Emadi S, Khademhosseini A, Tasoglu S, Biofabrication 8 (2016) 025019. [DOI] [PubMed] [Google Scholar]
- [202].Berg B, Cortazar B, Tseng D, Ozkan H, Feng S, Wei Q, Chan RY-L, Burbano J, Farooqui Q, Lewinski M, Di Carlo D, Garner OB, Ozcan A, ACS Nano 9 (2015) 7857–7866. [DOI] [PubMed] [Google Scholar]
- [203].Patrick WG, Nielsen AAK, Keating SJ, Levy TJ, Wang C-W, Rivera JJ, Mondragón-Palomino O, Carr PA, Voigt CA, Oxman N, Kong DS, PLoS One 10 (2015), e0143636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Chudobova D, Cihalova K, Skalickova S, Zitka J, Rodrigo MAM, Milosavljevic V, Hynek D, Kopel P, Vesely R, Adam V, Kizek R, Electrophoresis 36 (2015) 457–466. [DOI] [PubMed] [Google Scholar]
- [205].Krejcova L, Nejdl L, Rodrigo MAM, Zurek M, Matousek M, Hynek D, Zitka O, Kopel P, Adam V, Kizek R, Biosens. Bioelectron 54 (2014) 421–427. [DOI] [PubMed] [Google Scholar]
- [206].Comina G, Suska A, Filippini D, Lab Chip 14 (2014) 2978–2982. [DOI] [PubMed] [Google Scholar]
- [207].Comina G, Suska A, Filippini D, Angew. Chem. Int. Ed 54 (2015) 8708–8712. [DOI] [PubMed] [Google Scholar]
- [208].He Y, Wu Y, Fu J, Gao Q, Qiu J, Electroanalysis 28 (2016) 1658–1678. [Google Scholar]
- [209].Roda A, Guardigli M, Calabria D, Calabretta MM, Cevenini L, Michelini E, Analyst 139 (2014) 6494–6501. [DOI] [PubMed] [Google Scholar]
- [210].Symes MD, Kitson PJ, Yan J, Richmond CJ, Cooper GJT, Bowman RW, Vilbrandt T, Cronin L, Nat. Chem 4 (2012) 349–354. [DOI] [PubMed] [Google Scholar]
- [211].Shallan AI, Smejkal P, Corban M, Guijt RM, Breadmore MC, Anal. Chem 86 (2014) 3124–3130. [DOI] [PubMed] [Google Scholar]
- [212].Zhang H-Z, Zhang F-T, Zhang X-H, Huang D, Zhou Y-L, Li Z-H, Zhang X-X, Anal. Chem 87 (2015) 6397–6402. [DOI] [PubMed] [Google Scholar]
- [213].3D Printing of Medical Devices, https://www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/3DPrintingofMedicalDevices/default.htm.
- [214].Yazdi AA, Popma A, Wong W, Nguyen T, Pan Y, Xu J, Microfluid. Nanofluidics 20 (2016) 50. [Google Scholar]
- [215].Xu Y, Wu X, Guo X, Kong B, Zhang M, Qian X, Mi S, Sun W, Sensors 17 (2017) 1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Rapp BE, Kotz F, Keller N, Sachsenheimer K, Kirschner N, Nargang T, Richter C, Advanced fabrication technologies for micro/nano optics and photonics XI, Int. Soc. Optic. Photon 10544 (2018) 1054414. [Google Scholar]
- [217].Destino Joel F, Dudukovic Nikola A, Johnson Michael A, Nguyen Du T, Yee Timothy D, Egan Garth C, Sawvel April M, Steele William A, Baumann Theodore F, Duoss Eric B, Suratwala Tayyab, Dylla-Spears Rebecca, Adv. Mater. Technol 0 (2018), 1700323. [Google Scholar]


