Abstract
The genome organization and expression strategy of the newly identified severe acute respiratory syndrome coronavirus (SARS-CoV) were predicted using recently published genome sequences. Fourteen putative open reading frames were identified, 12 of which were predicted to be expressed from a nested set of eight subgenomic mRNAs. The synthesis of these mRNAs in SARS-CoV-infected cells was confirmed experimentally. The 4382- and 7073 amino acid residue SARS-CoV replicase polyproteins are predicted to be cleaved into 16 subunits by two viral proteinases (bringing the total number of SARS-CoV proteins to 28). A phylogenetic analysis of the replicase gene, using a distantly related torovirus as an outgroup, demonstrated that, despite a number of unique features, SARS-CoV is most closely related to group 2 coronaviruses. Distant homologs of cellular RNA processing enzymes were identified in group 2 coronaviruses, with four of them being conserved in SARS-CoV. These newly recognized viral enzymes place the mechanism of coronavirus RNA synthesis in a completely new perspective. Furthermore, together with previously described viral enzymes, they will be important targets for the design of antiviral strategies aimed at controlling the further spread of SARS-CoV.
Keywords: nidovirus, genome organization, subgenomic mRNA synthesis, replicase, RNA processing
Abbreviations: SARS-CoV, severe acute respiratory syndrome coronavirus; ORF, open reading frame; sg, subgenomic; BCoV, bovine coronavirus; EToV, equine torovirus; HCoV, human coronavirus; MHV, mouse hepatitis coronavirus; PL1pro, papain-like proteinase 1; IBV, avian infectious bronchitis coronavirus; SUD, SARS-CoV unique domain; TRS, transcription-regulating sequence; XendoU, poly(U)-specific endoribonuclease; ExoN, 3′-to-5′ exonuclease; 2′-O-MT, S-adenosylmethionine-dependent ribose 2′-O-methyltransferase; ADRP, adenosine diphosphate-ribose 1″-phosphatase; CPD, cyclic phosphodiesterase; snoRNA, small nucleolar RNA
1. Introduction
Severe acute respiratory syndrome (SARS) is a life-threatening form of atypical pneumonia that recently emerged in Guangdong Province, China. A previously unknown coronavirus was isolated from SARS patients1., 2., 3. and is considered the cause of this emerging respiratory disease. In an extraordinary effort, the full-length genome sequence of the SARS-coronavirus (SARS-CoV) was elucidated within weeks after the identification of this novel pathogen and published by the Michael Smith Genome Sciences Center (Vancouver, Canada,4 Entrez Genomes accession number NC_004718 (AY274119)), the Centers for Disease Control and Prevention (Atlanta, USA,5 GenBank accession number AY278741), and others. The SARS-CoV genome is ∼29.7 kb long and contains 14 open reading frames (ORFs) flanked by 5′ and 3′-untranslated regions of 265 and 342 nucleotides, respectively (Figure 1). Homologs of proteins conserved in all coronaviruses are encoded by the overlapping ORFs 1a and 1b, and by ORFs 2, 4, 5, 6 and 9a (Figure 1; Table 1, Table 2) .
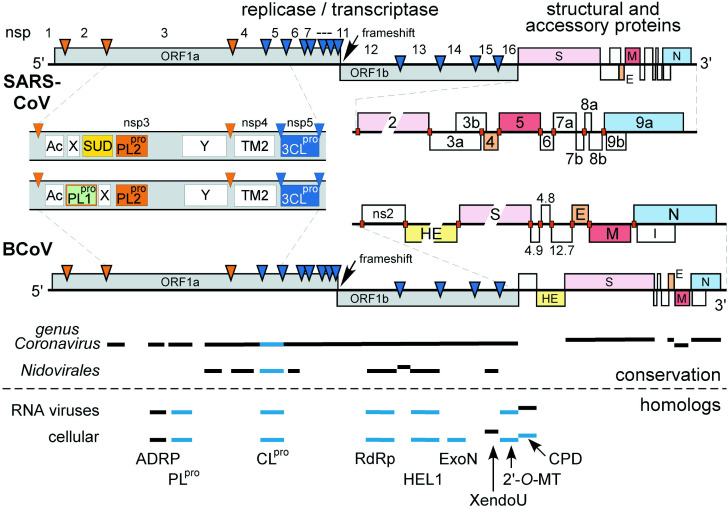
Figure 1.
Overview of the SARS-CoV genome organization and expression. Comparison of the genome organizations of SARS-CoV and bovine coronavirus (BCoV). The replicase genes are depicted, with ORF1a, ORF1b, and ribosomal frameshift site indicated. Arrows represent sites in the corresponding replicase polyproteins that are cleaved by papain-like proteinases (orange) or the 3C-like cysteine proteinase (blue). Cleavage products are provisionally numbered nsp1–nsp16 (see also Table 1). In the 3′-terminal part of the genomes, homologous structural protein genes are indicated in matching colors. Close-ups of two regions with major differences are shown (and see the text). In the N-terminal half of replicase ORF1a, SARS-CoV lacks one of the PLpro domains (indicated in orange/green in BCoV) and contains a unique insertion (SUD). In the region with structural and accessory protein genes, the location of the body TRSs involved in subgenomic RNA synthesis are indicated with red boxes (see Figure 3 and Hofmann et al.76). The bottom part of the Figure illustrates which parts of the genome are conserved in the genus Coronavirus and in the order Nidovirales (the ORF1a sequence of toroviruses, which largely remains to be sequenced, could not be included). Furthermore, it is indicated for which domains homologs have been identified in other RNA viruses and the cellular world. Enzymes for which structural data are available are shown in blue. SUD, SARS-CoV unique domain; PLpro, papainlike cysteine proteinase; 3CLpro, 3C-like cysteine proteinase; TM, transmembrane domain; ADRP, adenosine diphosphate-ribose 1″-phosphatase; ExoN, 3′-to-5′ exonuclease; CLpro, chymotrypsin-like proteinase; RdRp, RNA-dependent RNA polymerase; HEL1, superfamily 1 helicase; XendoU, (homolog of) poly(U)-specific endoribonuclease; 2′-O-MT, S-adenosylmethionine-dependent ribose 2′-O-methyltransferase; CPD, cyclic phosphodiesterase. Domains Ac, X, and Y are described by Ziebuhr et al.32 and Gorbalenya et al.47
Table 1.
Predicted SARS-CoV replicase cleavage products and their mode of expression
| Protein ordera in polyproteins pp1a/pp1ab | Position in polyproteins pp1a/pp1ab (amino acid residues)b | Protein size (amino acid residues) | Associated putative functional domain(s)c | Predicted mode of expression and release from polyproteinsd |
|---|---|---|---|---|
| nsp1-pp1a/pp1ab | 1Met-Gly180 | 180 | ? | TI+PL2pro |
| nsp2-pp1a/pp1ab | 181Ala-Gly818 | 638 | ? | PL2pro |
| nsp3e-pp1a//pp1ab | 819Ala-Gly2740 | 1922 | Ac, X, PL2pro, Y (TM1), ADRP | PL2pro |
| nsp4-pp1a/pp1ab | 2741Lys-Gln3240 | 500 | TM2 | PL2+3CLpro |
| nsp5-pp1a/pp1ab | 3241Ser-Gln3546 | 306 | 3CLpro | 3CLpro |
| nsp6-pp1a/pp1ab | 3547Gly-Gln3836 | 290 | TM3 | 3CLpro |
| nsp7-pp1a/pp1ab | 3837Ser-Gln3919 | 83 | ? | 3CLpro |
| nsp8-pp1a/pp1ab | 3920Ala-Gln4117 | 198 | ? | 3CLpro |
| nsp9-pp1a/pp1ab | 4118Asn-Gln4230 | 113 | ? | 3CLpro |
| nsp10-pp1a/pp1ab | 4231Ala-Gln4369 | 139 | GFL | 3CLpro |
| nsp11-pp1a | 4370Ser-Val4382 | 13 | ? | 3CLpro+TT |
| nsp12-pp1ab | 4370Ser-Gln5301 | 932 | RdRp | RFS+3CLpro |
| nsp13-pp1ab | 5302Ala-Gln5902 | 601 | ZD, NTPase, HEL1 | RFS+3CLpro |
| nsp14-pp1ab | 5903Ala-Gln6429 | 527 | Exonuclease (ExoN homolog) | RFS+3CLpro |
| nsp15-pp1ab | 6430Ser-Gln6775 | 346 | NTD, endoRNase (XendoU homolog) | RFS+3CLpro |
| nsp16-pp1ab | 6776Ala-Asn7073 | 298 | 2′-O-MT | RFS+3CLpro+TT |
Predictions are based on the SARS-CoV sequences published by Michael Smith Genome Sciences Centre (Vancouver, Canada; Entrez Genomes accession number NC_004718 (AY274119)4) and the Centers for Disease Control and Prevention (Atlanta, USA; GenBank accession number AY2787415) and an alignment of SARS-CoV with previously characterized coronavirus sequences as summarized in Refs. 11., 18., 32..
For convenience, replicase cleavage products were provisionally numbered non-structural protein (nsp) 1–16 according to their position in the polyproteins.
Amino acids of replicase proteins pp1a and pp1ab were numbered assuming that, as in other coronaviruses, a −1 ribosomal frameshift occurs; use of the slippery sequence UUUAAAC10 is predicted to yield a peptide bond between Asn4378 and Arg4379 in pp1ab.
Abbreviations: PL2pro, papain-like proteinase 2; ADRP, adenosine diphosphate-ribose 1″-phosphatase; TM, transmembrane domain; 3CLpro, 3C-like cysteine proteinase; GFL, growth factor-like domain; RdRp, RNA-dependent RNA polymerase; ZD, putative Zinc-binding domain; HEL1, superfamily 1 helicase; NTD, nidovirus conserved domain; ExoN, 3′-to-5′ exonuclease; 2′-O-MT, S-adenosylmethionine-dependent ribose 2′-O-methyltransferase. Domains Ac, X, and Y are described in Refs 32., 47..
Indicated are the SARS-CoV proteinases predicted to be involved in cleavage of the N- and/or C-termini of the cleavage products; TI, translation initiation; TT, translation termination; RFS, ORF1a/ORF1b ribosomal frameshift.
Table 2.
Predicted SARS–CoV proteins expressed from subgenomic mRNAs 2 to 9
| ORF numbera | Protein size (amino acid residues) | Subgenomic mRNA predicted to be used for expressiona | Protein name/function |
|---|---|---|---|
| 2 | 1255 | 2 | Spike (S) protein |
| 3a | 274 | 3 | ? |
| 3bb | 154 | 3 | ? |
| 4 | 76 | 4 | Envelope (E) protein |
| 5 | 221 | 5 | Membrane (M) protein |
| 6 | 63 | 6 | ? |
| 7a | 122 | 7 | ? |
| 7bc | 44 | 7 | ? |
| 8ad | 39 | 8 | ? |
| 8b | 84 | 8 | ? |
| 9a | 422 | 9 | Nucleocapsid (N) protein |
| 9be | 98 | 9 | ? |
Predictions are based on the SARS-CoV sequences published by Michael Smith Genome Sciences Centre (Vancouver, Canada; Entrez Genomes accession number NC_004718 (AY274119)4) and the Centers for Disease Control and Prevention (Atlanta, USA; GenBank accession number AY2787415).
ORF3b (462 nucleotides) overlaps with the 3′ half of ORF3a, the RNA4 body TRS and the 5′ end of ORF4. It is the fifth largest reading frame downstream of ORF1b (after ORFs 2, 3a, 5 and 9a) making it a likely candidate to be expressed. Since its translation initiation codon is the 13th AUG codon in mRNA3, ORF3b expression should involve a mechanism like internal ribosomal entry (as previously suggested for some other coronavirus ORFs; Ref. 78) or the synthesis of an as yet undetected additional subgenomic mRNA.
The translation termination codon of ORF7a and translation initiation codon of ORF7b overlap. The absence of any other upstream AUG codons (with the exception of that of ORF7a) and good context for translation initiation of the ORF7b AUG codon suggest that ORF7b may be expressed from subgenomic RNA7 by “leaky scanning” of ribosomes.
The putative ORF8a start codon is in a good context for translation initiation and immediately follows the body TRS involved in mRNA8 transcription, making it likely that ORF8a is expressed from mRNA8. The mechanism used to express the larger downstream ORF8b is more puzzling, since its (putative) translation initiation codon appears to have a poor context for translation initiation and two additional AUG codon are present in the region between the putative start codons of ORFs 8a and 8b. Recently, some SARS-CoV isolates from human and civet cat origin (L.L.M.P. and Y.G., unpublished results) were reported to contain a 29 nucleotides insertion that results in the in-frame fusion of ORFs 8a and 8b. Consequently, ORF8b in the Frankfurt-1 and HKU-39849 isolates used in this study may be translationally silent.
A functional “internal” open reading frame, overlapping with the N protein gene, has been described for other group 2 coronavirus, e.g. BCoV;77 ORF9b appears to occupy a corresponding position and may be expressed following “leaky scanning” by ribosomes.
Coronaviruses6., 7. are enveloped, positive-stranded RNA (+RNA) viruses, with a single-stranded genome of between 27 kb and 31.5 kb, the largest among known RNA viruses. The genomes of coronaviruses and related viruses in the order Nidovirales8., 9. are polycistronic and are expressed through a sophisticated combination of poorly understood regulatory mechanisms.6., 7. Coronavirus genome expression starts with the translation of two large replicase ORFs (1a and 1b; Figure 1), whose coding capacity is about twice that of the average complete +RNA virus genome. Via a −1 ribosomal frameshift,10 the ORF1a polyprotein (pp1a; >4000 amino acid residues) can be extended with ORF1b-encoded sequences to yield a >7000 amino acid residue pp1ab polyprotein. Replicase polyprotein processing is carried out by two or three ORF1a-encoded viral proteinases.11 The processing products are a group of largely uncharacterized (putative) replicative enzymes, including an RNA-dependent RNA polymerase, an RNA helicase that is fused to a complex N-terminal Zn-finger, and a Zn-ribbon-containing papain-like proteinase.12., 13., 14., 15. The replicase subunits are thought to assemble into a viral replication complex that is targeted to cytoplasmic membranes by various membrane-associated viral proteins.16., 17., 18. In addition to genome replication, the coronavirus replicase complex mediates the synthesis of an extensive nested set of subgenomic (sg) mRNAs (transcription) to express all ORFs downstream of ORF1b, which encode a variety of structural and accessory proteins.6., 7., 8., 9. The number and composition of these 3′-proximal ORFs vary greatly among coronaviruses, but they always include genes for the structural proteins S, M, E and N, which drive cytoplasmic virus assembly. The mechanisms underlying the synthesis of genomic and subgenomic RNAs are poorly understood. To explain the composite structure of the sg mRNAs, which are both 5′ and 3′-coterminal with the viral genome, several models have been put forward,6., 9. of which the one postulating the discontinuous synthesis of negative-stranded sg templates for sg mRNA synthesis19 has received wide support recently.
On the basis of antigenic cross-reactivity, coronaviruses were originally classified into three groups (termed groups 1, 2, and 3). Subsequently, the phylogeny-based clustering of coronaviruses proved at first (almost) identical with that based on antigenic cross-reactivity.6., 7. The same three clusters were evident upon analysis of the replicase region20., 21., 22. which does not contribute to virion antigenicity. This indicated that different regions of the coronavirus genome have indeed co-evolved and that intergroup recombination has not played a prominent role in coronavirus evolution.23 However, the agreement between the two classifications is not perfect, as some coronaviruses are sufficiently different to not have antigenic cross-reactivity with the established groups,24 but close enough to cluster with one of them (group 1) on the basis of sequence comparisons.7 Consequently, these viruses were placed into (the expanded) group 1. Here, we refer to coronavirus groups as evolutionary clusters that unite viruses not necessarily having antigenic cross-reactivity.
Using the recently published SARS-CoV genome sequences,4., 5. we provide insight into the evolution, organization and expression of SARS-CoV. The SARS-CoV genome and proteome were compared with those of other coronaviruses, distantly related nidoviruses, and databases, and several of our predictions were verified experimentally.
2. Results and Discussion
2.1. SARS-CoV represents a lineage that has split off from the group 2 branch relatively late in coronavirus evolution
To optimize our understanding of the SARS-CoV genome, we sought to infer the phylogenetic position of the novel agent relative to known coronaviruses. Recent phylogenetic analyses of different SARS-CoV proteins using unrooted trees consistently showed that SARS-CoV does not segregate into any of the three currently established coronavirus groups.4., 5. These results were interpreted as support for the classification of SARS-CoV as the prototype of a novel, fourth group of coronaviruses.4., 5. However, in our opinion, the evidence leading to this conclusion was inconclusive and alternative interpretations, with SARS-CoV being an outlier in one of the established groups, remained possible. This uncertainty can be resolved only through the reconstruction of coronavirus evolution from its origin using a rooted phylogenetic tree, which is most reliable when an outgroup is included in the analysis. The closest known outgroup for coronaviruses are the toroviruses, which form a separate genus in the same virus family.8., 25. The ORF1b part of the replicase and the two virion proteins S and M are homologous in coronaviruses and toroviruses.26., 27., 28. Unfortunately however, the level of conservation of the S and M protein genes is so low that we consider only the phylogenetic analysis of replicase ORF1b to be truly informative.
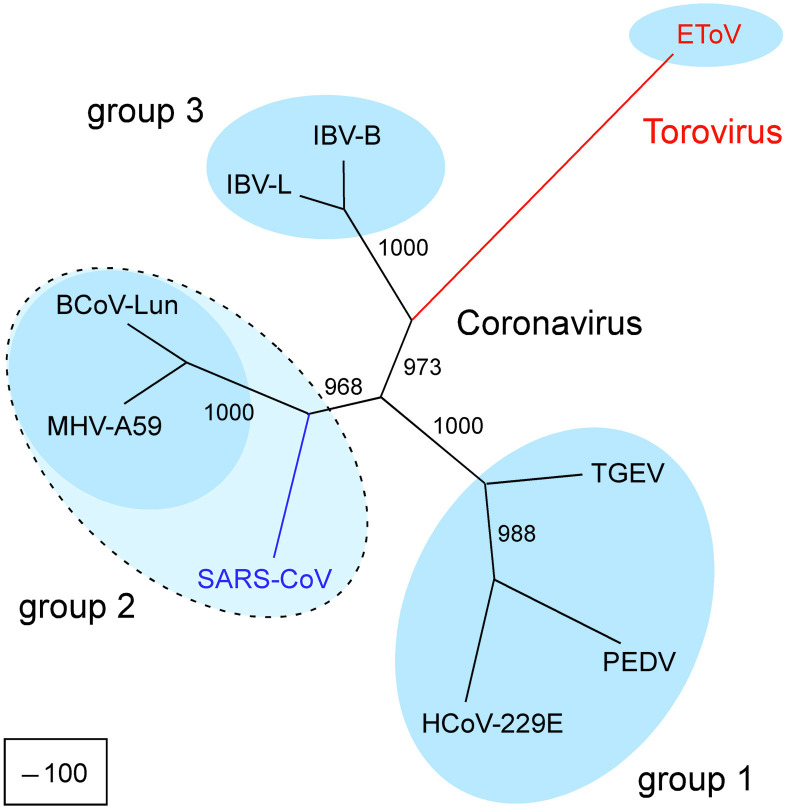
Consequently, to resolve the phylogenetic position of SARS-CoV, the equine torovirus (EToV25) was included in our analysis, which was limited to replicase ORF1b,26 the most conserved part of the genome. It should be noted, however, that the size of this genome segment (∼5500 nucleotides) approximates the combined size of the genes encoding the four virion-associated proteins S, M, E, and N. A fully resolved tree was obtained, with all branches supported in more than 960 out of 1000 bootstrap trials (Figure 2). The topology of this tree suggests strongly that the SARS-CoV lineage was an early split-off from the group 2 branch, which occurred after the two bifurcations that gave rise to the three major coronavirus groups (Figure 2). Accordingly, in two regions of the replicase ORF1a polyprotein, nsp1 and one of the nsp3 domains, which differentiate the three coronavirus groups, SARS-CoV contains orthologs of domains that are unique for group 2 coronaviruses (see Figure S1 of the Supplementary Material ). The published unrooted trees for the virion proteins and 3CLpro are also compatible with this phylogeny,4., 5. although formally we cannot exclude the occurrence of recombination with other coronaviruses in very limited regions. In this respect, we would like to stress that the differences in the composition and arrangement of ORFs in the 3′-proximal region of the genome (downstream of ORF1b; see Figure 1) between SARS-CoV and established group 2 coronaviruses does not contradict the above results. Group 1 coronaviruses also differ in this region through the presence of unique so-called “accessory non-structural protein genes”.6., 7. Some of these genes have been found to be dispensable for virus reproduction in tissue culture and/or animals.6., 7., 29. The fact that, apparently, they can be acquired or lost easily in the course of evolution indicates that these genes can not be considered reliable group markers.
Figure 2.
Phylogenetic analysis of coronavirus replicase genes. SARS-CoV replicase ORF1b amino acid sequences (Entrez Genomes accession number NC_004718 (AY274119)) were compared with those from viruses representing the three coronavirus subgroups and the genus Torovirus. Group 1: transmissible gastroenteritis virus (TGEV), NC_002306; human coronavirus 229E (HCoV-229E), NC_002645; porcine epidemic diarrhea virus (PEDV), NC_003436. Group 2: mouse hepatitis virus A59 (MHV-A59), NC_001846; bovine coronavirus (BCoV-Lun) AF391542. Group 3: infectious bronchitis virus (IBV), strains Beaudette (NC_001451) and LX4 (AY223860). Torovirus: equine torovirus (EToV), X52374. A multiple protein alignment of these sequences was generated with the help of the ClustalX1.82 program65 and was adjusted manually. Two regions of poor conservation were removed from the alignment, which was converted subsequently into the nucleotide form. All columns containing gaps were removed. The resulting alignment contains the following SARS-CoV sequences fused: 13,623–13,859, 14,310–18,857 and 20,076–21,482. It included 5487 characters with 3207 of them being parsimony-informative. Using the PAUP program (version 4.0.0d55) and parsimony criterion, an exhaustive tree search of the 135,135 evaluated trees identified the best tree having a score of 10,927 and the second best tree having a score of 10,964; the worst tree had a score of 13,611. A total of 1000 bootstrap trials were conducted using the parsimony criterion and a branch-and-bound search to generate a bootstrap 50% majority-rule consensus tree. The frequency of occurrence of particular bifurcations in bootstraps is indicated at the nodes. Similar trees with similar high bootstrap support above 960 were obtained using the NJ method that was applied to distance matrices obtained for either nucleotide or amino acid alignments (not shown).
Supplementary data 1.
Supplementary data 1
In conclusion, SARS-CoV is distantly related to established group 2 coronaviruses, a relationship comparable to that observed in group 1 between porcine epidemic diarrhoea coronavirus (PEDV) and human coronavirus 229E (HCoV-229E) on the one hand, and transmissible gastroenteritis coronavirus (TGEV) and related viruses on the other hand (Figure 2). Accordingly, the lack of antigenic cross-reactivity observed between distant group-mates in group 124 may be observed between SARS-CoV and the established group 2 viruses. Thus, SARS-CoV may be the first identified representative of a larger cluster that could be called subgroup 2b, if the established group 2 coronaviruses would be referred to as subgroup 2a. The 2b cluster should include the immediate ancestor of SARS-CoV, which may circulate in the field. If close relatives of SARS-CoV were to be identified in animal hosts, the virus would represent the second example of a group 2 coronavirus that may have crossed the animal–human barrier. The first putative case is that of the bovine coronavirus (BCoV) and human coronavirus OC43 (HCoV-OC43),30 two viruses that are so closely related at the genetic level30., 31. that they can be considered to be the same virus species.
2.2. Two proteinases are predicted to cleave the SARS-CoV replicase polyproteins into 16 subunits, the largest of these having a unique domain organization
A detailed comparison of the SARS-CoV replicase with that of its closest known relatives in group 2, mouse hepatitis coronavirus (MHV) and BCoV (Figure 1), revealed a replicase proteolytic processing scheme and domain organization that, with some notable exceptions (see below), proved to be typical for group 2 viruses.11., 32. Using the conserved signatures of the cleavage sites recognized by coronavirus proteinases11., 12., 33., 34. and their flanking sequences, we predict the generation of 16 replicase subunits through proteolysis mediated by 3CLpro (11 cleavages) and PL2pro (three cleavages) (Figure 1 and Table 1).
The most conspicuous differences between known group 2 coronaviruses and SARS-CoV were identified in nsp3, the largest replicase subunit that is encoded by ORF1a (Table 1). Unlike all other coronaviruses, SARS-CoV does not have an ortholog of papain-like proteinase 1 (PL1pro; see close-up in Figure 1),13., 35. which was probably lost during evolution of this lineage. This observation implies that the three cleavages in the N-terminal half of pp1a must all be performed by the conserved PL2pro,36., 47. a downstream-located paralog of PL1pro. The ortholog of this proteinase appears to dominate over PL1pro in HCoV-229E,32 and is the only active PLpro in avian infectious bronchitis coronavirus (IBV).32., 37. Immediately upstream of PL2pro, we identified a 375 amino acid residue “orphan domain” in SARS-CoV (called SUD for SARS-CoV unique domain; Figure 1), which is not present in other coronaviruses. The corresponding ORF1a region differs profoundly among group 1 coronaviruses. In one of these viruses (TGEV), and in the group 3 IBV, this region contains just a few amino acid residues, essentially fusing PL2pro to the upstream X domain. In contrast, HCoV-229E and PEDV share a conserved domain in this position. Interestingly, nsp3 also was the main site of replicase differences between BCoV variants isolated from respiratory and intestinal samples from an animal that had died during an outbreak of fatal shipping pneumonia.20 Due to the plausible multifunctionality of nsp3, which may be involved in the control of subgenomic mRNA synthesis,13., 38. the gross internal rearrangements and point mutations in this protein may have pleiotropic effect(s) on SARS-CoV properties, including its pathogenic potential.
2.3. SARS-CoV produces eight subgenomic mRNAs to express the ORFs located in the 3′-proximal part of the genome
In a striking parallel with the unique features of nsp3, the 3′-proximal part of the SARS-CoV genome contains five ORFs (6, 7a, 7b, 8a and 8b) that are not present in established group 2 coronaviruses and for which no obvious homologs could be identified upon sequence comparison. Furthermore, SARS-CoV lacks counterparts for two genes inserted between replicase ORF1b and the S gene in subgroup 2a viruses (see the close-up in Figure 1).6., 7. All these ORFs (from 2 to 9b) are predicted to be expressed from sg mRNAs in SARS-CoV. In members of the genus Coronavirus and the related family Arteriviridae, all sg mRNAs are 3′-coterminal with the viral genome, and contain a common 5′ leader sequence that is identical with that of the genome.6., 7., 9., 39. The fusion of the leader to the coding part (or “body”) of each of the sg RNAs involves a discontinuous step in RNA synthesis, which is currently believed to occur during minus strand synthesis, thus producing composite subgenomic negative-stranded templates for sg mRNA synthesis (Figure 3(C)).19., 39., 40. Leader-to-body joining is guided by a base-pairing interaction involving conserved transcription-regulating sequences (TRSs; also previously termed “intergenic sequences (IGSs)” in coronaviruses), which are found at the 3′ end of the genomic leader (leader TRS) and at the 5′ end of each of the sg RNA bodies (body TRSs), often located exactly between two genes, but sometimes located within the coding sequence of an upstream gene (Figure 1, Figure 3).
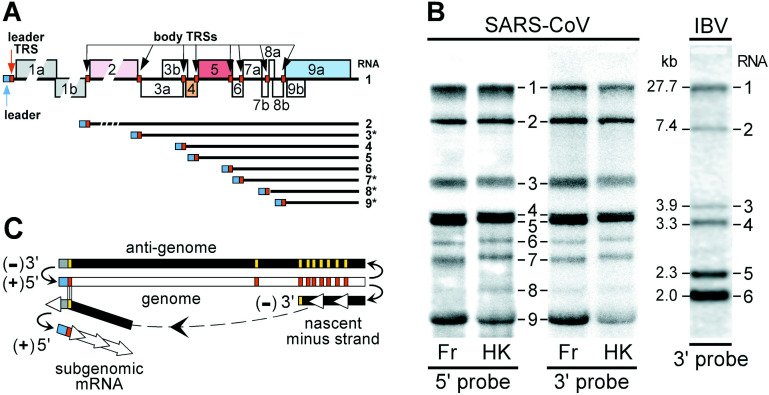
Figure 3.
SARS-CoV subgenomic mRNA synthesis. (A) Organization of ORFs in the 3′ end of the SARS-CoV genome with predicted leader and body TRSs indicated by small boxes. The subgenomic mRNAs resulting from the use of these TRSs for leader-to-body fusion are depicted below, with mRNAs predicted to be functionally bicistronic indicated with an asterisk (∗). (B) Hybridization analysis of intracellular viral RNA from Vero cells infected with SARS-CoV, Frankfurt-1 (Fr) and HKU-39849 (HK) isolates. See Materials and Methods for technical details. Oligonucleotides complementary to sequences from the SARS-CoV leader sequence and to a region in the genomic 3′ end both recognized a set of nine RNA species (the genome (RNA1) and eight subgenomic RNAs) confirming the presence of common 5′ and 3′ sequences. RNA from Vero cells infected with avian infectious bronchitis virus (IBV), which produces only five subgenomic mRNAs of known sizes41 was run in the same gel and used as a size marker. (C) Model for nidovirus subgenomic RNA synthesis by discontinuous extension of minus strands.19., 39. Whereas genome replication relies on continuous minus strand synthesis (antigenome), subgenomic minus strands would be produced by attenuation of nascent strand synthesis at a body TRS (red bar), followed by translocation of the nascent strand to the leader TRS in the genomic template. Following base-pairing between the body TRS complement at the 3′ end of the minus strand and the leader TRS, RNA synthesis would resume to complete the subgenomic minus strand that would then serve as template for the transcription of subgenomic mRNAs.
In the SARS-CoV genome we readily identified a potential leader TRS (5′-CUAAACGAACUUU-3′) that has a 6–11 nucleotides match with a number of sequences in the 3′ end of the genome, many of which are positioned immediately upstream of viral genes (Figure 3(A)). As recognized also by others,4., 5., 34. the sequence 5′-ACGAAC-3′ is absolutely conserved and can be considered the core of the SARS-CoV TRS. Based on the SARS-CoV sequence with the largest 5′-terminal segment (accession number AY2787415), the SARS-CoV leader sequence is (at least) 72 nucleotides long, similar to e.g. that of BCoV, with which it has a striking 20 out of 21 nucleotides match immediately upstream of the leader TRS (5′-GAUCUCUUGUAGAUCUGUUC-3′). On the basis of the location of putative body TRSs, the synthesis of nine mRNAs by SARS-CoV was expected: the genomic mRNA (RNA1) and eight subgenomic mRNAs with sizes of approximately 8.4, 4.6, 3.8, 3.5, 3.0, 2.6, 2.1 and 1.8 kb (including 5′ leader and 3′ poly(A)-tail). However, in the first published experimental analysis of the SARS-CoV-specific mRNAs generated in infected Vero cells, the synthesis of only five viral mRNAs could be confirmed.5
To investigate SARS-CoV RNA synthesis in more detail, Vero cells were infected with SARS-CoV isolates Frankfurt-13 and HKU-39849,1 and intracellular RNA was analyzed by hybridization with oligonucleotide probes complementary to a part of the 5′ leader sequence and a sequence just upstream of the 3′ poly(A) tail. The coronavirus IBV,41 which also replicates in Vero cells, was used as control and size marker. As illustrated in Figure 3(B), the genomic RNA and all eight predicted subgenomic transcripts were detected with both SARS-CoV probes, confirming the fact that these RNAs contain both common 5′-terminal and common 3′-terminal sequences. Remarkably, a slight mobility shift was observed for RNAs 7 and larger of the Frankfurt-1 isolate. The subsequent sequence analysis of this virus revealed that this was due to a 45 nt in-frame deletion in ORF7b,34 probably the first documented example of SARS-CoV genetic adaptation to cell culture conditions. The confirmation of leader-body fusion sites of the SARS-CoV subgenomic mRNAs will be published elsewhere.34 Remarkably, up to four of the eight SARS-CoV subgenomic mRNAs (3, 7, 8, and 9) may be functionally bicistronic (Table 2), as observed occasionally for other coronavirus subgenomic mRNAs.6., 7.
2.4. The replicase of coronaviruses includes a variety of putative RNA-processing enzymes
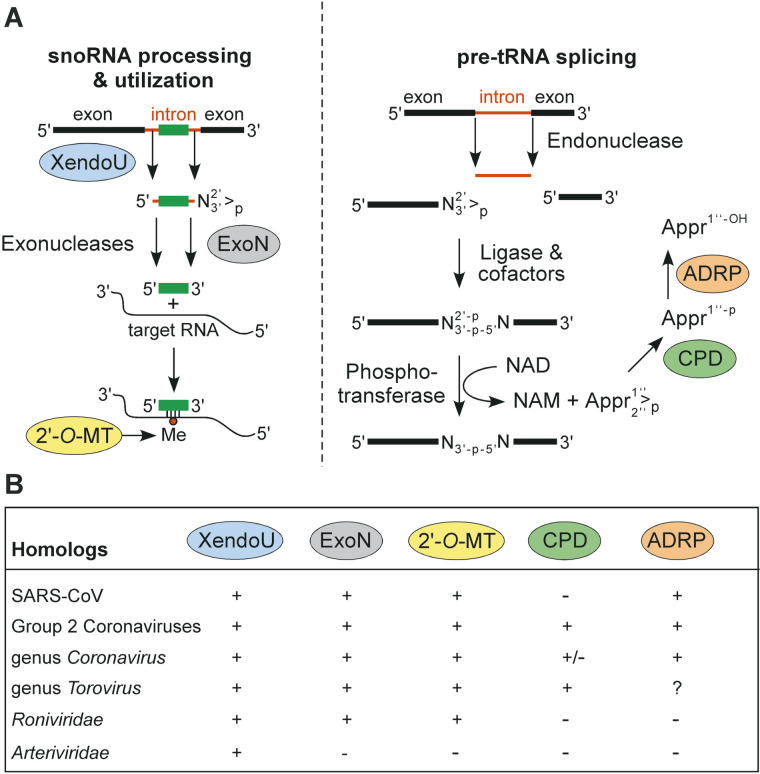
The production of a complex and diverse set of RNA molecules by nidoviruses (including SARS-CoV) is linked to an unparalleled complexity of their giant replicase, which contains a variety of (putative) enzymatic functions and a number of completely uncharacterized domains (Figure 1).18 We have initiated the characterization of coronavirus replicase by comparative genomics,12 and have regularly updated this analysis through recent years).18., 32. Our continuing analysis has now identified distant coronavirus homologs of not less than five cellular enzymes that are associated with RNA processing (Figure 4): poly(U)-specific endoribonuclease (XendoU42), a 3′-to-5′ exonuclease (ExoN) that belongs to the DEDD superfamily,43 S-adenosylmethionine-dependent ribose 2′-O-methyltransferase (2′-O-MT) of the RrmJ family,44 adenosine diphosphate-ribose 1″-phosphatase (ADRP45), and cyclic phosphodiesterase (CPD).45., 46. In the SARS-CoV proteome, conserved domains presumably associated with these activities were mapped (from the N to C terminus) to the X domain47 of nsp3 (ADRP), the N-terminal domain of nsp14 (ExoN), a “nidovirus-specific” replicase domain26., 48. in the C-terminal part of nsp15 (XendoU), and nsp16 (2′-O-MT). The CPD-related domain is not conserved in SARS-CoV, but was identified in the product of ORF249 of established group 2 coronaviruses, and in the very C-terminal domain of the torovirus ORF1a polyprotein,50 as well as in some double-stranded RNA rotaviruses.
Figure 4.
Sequence alignments of protein families that include cellular enzymes involved in RNA processing and their nidovirus homologs. Our in-depth comparative sequence analysis (see Materials and Methods) revealed a statistically significant relationship between functionally uncharacterized proteins (domains) of nidoviruses, including SARS-CoV, and five protein families that include enzymes involved in two nuclear RNA processing pathways: intron excision to produce mature tRNA58 and the production of intron-encoded box C/D small nucleolar RNA (snoRNA) from its host pre-mRNA (Figure 5).59 Shown are alignments for key regions of a few selected members of the following groups of enzymes: (A) XendoU family; (B) ExoN family; (C) 2′-O-MT family; (D) CPD family; and (E) ADRP family. These protein families may be known also under other names. Cellular homologs, not necessarily including proteins involved in the discussed RNA processing pathways, are listed in the top segment of each alignment and nidovirus proteins in the bottom segment. In the CPD family, along with group 2 coronavirus representatives, proteins of two rotaviruses (double-stranded RNA viruses), which were identified in this study, are listed. In both segments, residues are highlighted independently: black for absolutely conserved residues and different shades of grey to indicate different levels of conservation; amino acid similarity groups used were: (i) D, E, N, Q; (ii) S, T; (iii) K, R; (iv) F, W, Y; and (v) I, L, M, V. Positions occupied by identical or similar residues in all proteins under comparison are indicated with an asterisk (∗) and colon (:), respectively, in the inter-segment row. For the ExoN family, three motifs conserved in the DEDD superfamily and Zn-finger unique for the ExoN family are indicated. Database accession numbers for nidovirus genome sequences: SARS-CoV, Entrez Genomes accession number NC_004718 (AY274119); MHV-A59, NC_001846; BCoV-Lun, AF391542; HCoV-229E, NC_002645; IBV-B, NC_001451; PEDV, NC_003436; TGEV, NC_002306; equine torovirus (EToV), X52374; equine arteritis virus (EAV), X53459; porcine reproductive and respiratory syndrome virus (PRRSV), M96262; gill-associated virus (GAV), AF227196. Abbrevations and NCBI protein database ID number or SwissProt names of the remaining protein sequences are: (A) Npun 0562, hypothetical protein of Nostoc punctiforme, ZP_00106190; Poliv smB, pancreatic protein of Paralichthys olivaceus, BAA88246; Celeg Pp11, placental protein 11-like precursor of Caenorhabditis elegans, NP_492590); Xlaev endoU, endoU protein of Xenopus laevis, CAD45344; pp1b, ORF1b-encoded part of nidovirus replicase polyprotein 1ab. (B) Yeast PAN2, PAB-dependent poly(A)-specific ribonuclease subunit PAN2 of Saccharomyces cerevisiae, P53010; Mycge DPO3, DNA polymerase III polC-type, containing exonuclease domain, of Mycoplasma genitalium, P47277; Bacsu DING, probable ATP-dependent helicase dinG homolog, containing exonuclease domain, of Bacillus subtilis, P54394; Ecoli DP3E, DNA polymerase III, epsilon chain, containing exonuclease domain, of Escherichia coli, P03007 (PDB: 1J53 and 1J54); Ecoli RNT, exoribonuclease T of Escherichia coli, P30014. (C) Hsap AKA, A-kinase anchoring protein 18 gamma of Homo sapiens, AAF28106; Athal CPD1, putative CPD1 of Arabidopsis thaliana, CAA16750; Athal CPD2, putative CPD2 of Arabidopsis thaliana, CAA16751; yeast YG59, hypothetical 26.7 kDa protein of yeast, P53314; Ecoli LIGT, 2′-5′ RNA ligase of Escherichia coli, P37025; ns2, non-structural protein (ORF2-encoded) of the coronaviruses HCoV-O43 (AAA74377), BCoV-Quebec (P18517), and MHV-A59 (P19738); EToV pp1a, C-terminal fragment of EToV pp1a, S11237; HRoV VP3, VP3 of human rotavirus, BAA84964; ARoV VP3, VP3 of avian rotavirus PO-13, BAA24128. (D) Ecoli o177, putative polyprotein of Escherichia coli, AAC74129; Hsap Y1268a, KIAA1268 protein of Homo sapiens, BAA86582; Hsap H2A1.1, histone macroH2A1.1 of Homo sapiens, AAC33434; yeast YMX7, hypothetical 32.1 kDa protein of yeast, Q04299; yeast YBN2, hypothetical 19.9 kDa protein of yeast, P38218. (E) Yeast YBR1, putative ribosomal RNA methyltransferase (rRNA (uridine-2′-O-)-methyltransferase) of yeast, P38238; yeast SPB1, putative rRNA methyltransferase SPB1 of yeast, P25582; yeast YGN6, putative ribosomal RNA methyltransferase YGL136c (rRNA (uridine-2′-O-)-methyltransferase) of yeast, P53123; Ecoli FTSJ, cell division protein of Escherichia coli, NP_417646.
The conservation in the ExoN, 2′-O-MT and CPD-related domains of nidoviruses includes the catalytic and other active-site residues identified in the prototype cellular enzymes. Although the active-site residues of the ADRP and XendoU families are yet to be characterized, the most conserved amino acids of these families are found in their putative nidovirus homologs. Some of the nidovirus domains may contain unique and conserved additional domains. For instance, we noted that the nidovirus ExoN homologs contain an additional conserved domain resembling a mononuclear Zn-finger (Figure 4(B)) between the universally conserved blocks I and II, which include the catalytic residues (two Asp and one Glu).51 Another Zn-finger-like module has been inserted between blocks II and III in the ExoN homolog of roniviruses, a subset of nidoviruses (data not shown). Our combined observations indicate that the nidovirus homologs of these cellular RNA processing enzymes must be enzymatically active, although they may have evolved to act on specific (and unique) substrates or have additional unique components.
The newly predicted enzymes could be involved in the metabolism of virus and/or cellular RNAs. For instance, the 2′-O-MT activity could be used to produce the 5′-cap of viral mRNAs, as was demonstrated for a homologous flavivirus enzyme.52 Based on a parallel with some cellular DNA-processing homologs, like exonuclease I53 and the exonuclease domain of DNA polymerases,54 it is tempting to speculate on a link between the ExoN activity and RNA proofreading, repair, and/or recombination. The first two activities are not known in RNA viruses, and recombination commonly proceeds through the copy-choice mechanism with RdRp switching templates to produce chimeric nascent chains.55 However, due to the extreme sizes of their giant genomes, coronaviruses may differ from other RNA viruses and share an unprecedented similarity with DNA-based life-forms in the mechanisms of genome biosynthesis and maintenance. If confirmed, these unusual properties would explain the preliminary reports on the resistance of SARS-CoV to ribavirin, a drug that was shown to force other RNA viruses into “error catastrophe”.56 The experimental verification of these predictions will be an important step in increasing our understanding of the functional roles these putative enzymes play in the replicative cycle of SARS-CoV and related viruses. Extensive attempts to demonstrate the 2′-O-MT activity of several coronaviruses (which was also recently predicted by others57) in a 5′-RNA-capping reaction have not produced conclusive evidence so far (J.Z. and A.E.G., unpublished results). This development indicates that, as before with other distant nidovirus homologs (e.g. the helicase),15 the translation of bioinformatics predictions into a functional description is likely to be a laborious and time-consuming process, involving mainly the identification of virus-specific substrates and proper assay conditions.
In this respect, we have made an observation that both provides additional support for the provisional assignments made above and may help in the experimental verification of the predicted activities. When the five enzyme families listed (Figure 4) above were analyzed as a single dataset, it became apparent that representatives of these families cooperate in two nuclear intron RNA processing pathways. These pathways are functionally antagonistic: intron excision aimed at the synthesis of mature tRNA58 and the production of intron-encoded box C/D small nucleolar RNA (snoRNA) from its host pre-mRNA59 (Figure 5(A)). In the first pathway, XendoU initiates a cascade of poorly characterized endo- and exonuclease reactions that may involve ExoN, a homolog of the yeast Rrp6p exosome component,60 ultimately leading to the production of mature U16 and U86 snoRNAs. Subsequently, these snoRNAs may be utilized in diverse rRNA processing events involving nucleotide methylation by fibrillarin, a 2′-O-MT,61 and assisted by helicase(s).59 Strikingly, the homologs of three cellular enzymes from this pathway, encoded in the replicases of all nidoviruses except for arteriviruses, are genetically clustered in a single protein block (nsp14–nsp16) immediately downstream of the RNA-helicase (nsp13) (Figure 1, Figure 4). Because of the proximity of these four domains to each other, their expression must be tightly coordinated at the level of 3CLpro proteolysis and by the upstream ORF1a/ORF1b ribosomal frameshift signal.
Figure 5.
Nidoviruses encode homologs of cellular enzymes involved in RNA processing. (A) The cellular pathways for processing of pre-U16 snoRNA and pre-tRNA splicing are summarized, with relevant enzymatic activities indicated. For details, see the text. Homologs of the highlighted enzymes have been identified in nidoviruses (see also Figure 1 and the text). (B) Table summarizing the conservation of homologs of the cellular enzymes presumably involved in RNA processing in SARS-CoV and different nidovirus groups.
In the other pathway, which involves tRNA-processing, the utilization of a 2′-phosphate group of a splicing intermediate involves the conversion of adenosine diphosphate ribose 1″-2″ cyclic phosphate (Appr>p) by CPD62 into adenosine diphosphate ribose 1″-phosphate (Appr-1″-p), of which the phosphate group may be further processed by an ADRP.45 Both these activities may drive the production of mature tRNA. Although the nidovirus homologs of CPD and ADRP remain to be characterized, they are not under the control of the ORF1a/ORF1b ribosomal frameshift signal (Figure 1) and may thus, unlike the ORF1b-encoded enzymes, be produced in larger quantities.
The nidovirus homologs of the five RNA processing enzymes discussed above may interfere with these or similar cellular RNA processing pathways to reprogram the cell for the benefit of virus reproduction. It seems even more conceivable that they, alone or in concert with other enzymes like the RdRp or helicase, are involved directly in viral RNA synthesis, particularly in transcription, which, in an apparent parallel with snoRNA-driven processes,59 is guided by conserved oligonucleotide base-pairing interactions (Figure 3(C)). The viral enzymes, like their cellular counterparts, might be part of separate pathways or, alternatively, cooperate in a single pathway in which the XendoU, ExoN and 2′-O-MT homologs provide RNA specificity, and the CPD and ADRP homologs modulate the pace through processing of compound(s) containing 2′-phosphate groups. In this respect, we note that both the XendoU/ExoN/2′-O-MT and CPD/ADRP cellular pathways start with an endoribonuclease-mediated cleavage to produce molecule(s) with 2′-3′-cyclic phosphate termini (Figure 5), indicating the structural basis for possible cooperation of the coronavirus homologs of these enzymes in a single pathway. The expected functional hierarchy of the five putative nidovirus enzymes (Figure 5(A)) is supported by their corresponding evolutionary conservation, with the XendoU homolog being absolutely conserved and the CPD homolog being least conserved among nidoviruses (Figure 5(B)).
3. Concluding Remarks
The availability and comparative analysis of the SARS-CoV genome and proteome set the stage for the extensive biological characterization of this emerging pathogen and the development of anti-SARS-CoV strategies. Our conclusion that SARS-CoV is distantly related to group 2 coronaviruses (Figure 2) implies that viruses from this group, in particular the extensively studied mouse hepatitis virus and its derivatives lacking non-essential CPD-like and HE genes, may be the best available models for both in vitro and in vivo studies, in particular where the synthesis of viral macromolecules and the structure and function of the replication complex are involved. A detailed comparative characterization of the BCoV/HCoV-OC43 pair may provide invaluable insights into the processes of adaptation of a non-human coronavirus to a human host, which should be highly relevant to understanding the emergence of SARS-CoV. The SARS-CoV genome (Figure 1) lacks genes that are common in group 2 viruses, like PL1pro and CPD-like and HE genes, but encodes a number of unique protein sequences, underlining the ability of coronaviruses to the gross evolution. The comparative studies presented here have tentatively identified both known and novel viral enzymes (Figure 1, Figure 5), most of which may be involved in RNA processing and have homologs of which the tertiary structure has been solved (Figure 1). Intriguing parallels have been drawn between these putative viral enzymes and characterized, but distant cellular homologs that will guide the functional dissection of the replicases of SARS-CoV and related viruses and may put the mechanism of coronavirus RNA synthesis in a completely new perspective. The newly described putative enzymes of SARS-CoV double the list of potential targets for the design of antiviral strategies aimed at controlling this emerging virus infection.33., 34.
4. Materials and Methods
4.1. Analysis of intracellular SARS-CoV RNA
Vero cells were infected with SARS-CoV (Frankfurt 1 or HKU-39849) at an MOI of 0.01 or were mock infected. At the onset of cytopathogenic effect (approximately 40 hours post infection), intracellular RNA was isolated by cell lysis for ten minutes at room temperature with 5% (w/v) lithium dodecyl sulfate in LET buffer (10 mM Tris–HCl (pH 7.4), 100 mM LiCl, 1 mM EDTA), containing 20 μg/ml of proteinase K. After shearing of the cellular DNA using a syringe, lysates were incubated at 42 °C for 15 minutes, extracted with phenol (pH 4.0) and chloroform, and RNA was ethanol-precipitated. The RNAs were separated in denaturing 1% (w/v) agarose gels containing 2.2 M formaldehyde and Mops buffer (10 mM Mops (sodium salt) (pH 7), 5 mM sodium acetate, 1 mM EDTA). Dried gels were used for direct hybridization with 32P-labeled oligonucleotides SARSV001 (5′-CGAGGTTGGTTGGCTTTTCCTG-3′) and SARSV002 (5′-CACATGGGGATAGCACTAC-3′), which are complementary to sequences in the SARS-CoV leader sequence and the genomic 3′ end, respectively. After hybridization, gels were analyzed using a Personal FX Molecular Imager and Quantity One software (both from Bio-Rad).
4.2. Methods for bioinformatics
Genpeptides, Conserved domain (CD)63 and protein family (Pfam)64 databases were used in this study. Amino acid sequence alignments were generated using ClustalX1.8165 and Dialign266 programs assisted by Blosum position-specific matrices,67 and were processed for presentation using GeneDoc.68 Multiple sequence alignments were converted into hidden Markov model (HMM) profiles using HMMER2.01 software.69 Sequence databases were searched in default mode, unless stated otherwise, using the HMMER2.01 package.64., 69. and a family of Blast programs.70
The expectation values of similarity (E) of 0.05 or lower for Blast searches and 0.1 or lower for HMMER-mediated searches were considered to be statistically significant.71 Database searches with nidovirus proteins (Table 1, Table 2) and their alignments were conducted in an iterative mode until no new homologs were identified. Also, sequences that were identified below the threshold during the last iteration were used to initiate reciprocal searches that might have resulted in new significant matches. This approach worked for all protein families described here, except for the identification of the relationship between the nidovirus ExoN family and cellular DEDD superfamily, which is known to be extremely diverse.43 In this latter case, using the MAST program,72 we found a strong match (p=3 e−10) between the most conserved motif III of a DEDD protein and a conserved block of the ExoN family that facilitated the identification of the two other motifs in the nidovirus proteins having a non-typical intermotif spacing partially occupied by Zn-finger(s) (see the text and Figure 4). Furthermore, we observed an approximately 30 times selective increase of the global similarity between the ExoN family and DEDD proteins, after the coronavirus sequences were modified artificially by removing putative Zn-fingers that are not present in the DEDD proteins. In the HMMER-mediated searches of >106 sequences using this Zn-finger-deficient ExoN family as a query, numerous DEDD proteins were retrieved immediately after the nidovirus proteins, starting with E=0.81. The relatively poor statistics of these hits were due to the failure by HMMER to align all three motifs.
Cluster phylogenetic trees were reconstructed using the neighbour-joining algorithm described by Saitou & Nei73 with the Kimura correction,74 and were evaluated with 1000 bootstrap trials, as implemented in the ClustalX1.81 program. Parsimonious trees were generated using exhaustive search and evaluated with bootstrap branch-and-bound search using a UNIX version of the PAUP∗ 4.0.0d55 program that is included in the GCG-Wisconsin Package programs. The resulting trees were visualized using the TreeView program.75
Acknowledgements
We acknowledge the work of many colleagues in the nidovirus field whom we were unable to cite due to space limitations. We thank Dr H. F. Rabenau and Dr H. W. Doerr (Johann Wolfgang Goethe Universität Frankfurt am Main, Germany), and Dr G. van Doornum and Dr A.D. Osterhaus (Erasmus Universiteit Rotterdam, The Netherlands) for providing samples of tissue culture grown SARS-CoV. We are grateful to Karol Miaskiewicz, Annie Sharman and Vladimir Nikolayev for software administration, and to the Advanced Biomedical Computer Center (Frederick, MD) for access to the computer resources. We thank Dave Cavanagh, Dongwan Yoo, David Brian, Luis Enjuanes, Gijsbert van Willigen, Fred Wassenaar, Henk van der Meij, Ria Dubbeldeman, Corrie Verbree, Erwin van den Born, Olga Slobodskaya, and Yvonne van der Meer for reagents, technical assistance, critical reading of the manuscript, and/or helpful discussions.
Edited by J. Karn
References
- 1.Peiris J.S.M., Lai S.T., Poon L.L.M., Guan Y., Yam L.Y.C., Lim W. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 3.Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 4.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 5.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 6.Lai M.M.C., Holmes K.V. Coronaviruses. In: Knipe D.M., Howley P.M., editors. Fields Virology. Lippincott; Philadelphia, PA: 2001. pp. 1163–1185. [Google Scholar]
- 7.Siddell S.G. The Coronaviridae. In: Fraenkel-Conrat H., Wagner R.R., editors. The Viruses. Plenum Press; New York: 1995. [Google Scholar]
- 8.Enjuanes L., Spaan W.J.M., Snijder E.J., Cavanagh D. Order Nidovirales. In: van Regenmortel M.H.V., Fauquet C.M., Bishop D.H.L., Carstens E.B., Estes M.K., Lemon S.M., editors. Virus Taxonomy. Academic Press; New York: 2000. pp. 827–834. [Google Scholar]
- 9.Snijder E.J., Meulenberg J.J.M. Arteriviruses. In: Knipe D.M., Howley P.M., editors. Fields Virology. Lippincott; Philadelphia, PA: 2001. pp. 1205–1220. [Google Scholar]
- 10.Brierley I., Digard P., Inglis S.C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 12.Gorbalenya A.E., Koonin E.V., Donchenko A.P., Blinov V.M. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucl. Acids Res. 1989;17:4847–4861. doi: 10.1093/nar/17.12.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herold J., Siddell S.G., Gorbalenya A.E. A human RNA viral cysteine proteinase that depends upon a unique Zn2+-binding finger connecting the two domains of a papain-like fold. J. Biol. Chem. 1999;274:14918–14925. doi: 10.1074/jbc.274.21.14918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Dinten L.C., van Tol H., Gorbalenya A.E., Snijder E.J. The predicted metal-binding region of the arterivirus helicase protein is involved in subgenomic mRNA synthesis, genome replication, and virion biogenesis. J. Virol. 2000;74:5213–5223. doi: 10.1128/jvi.74.11.5213-5223.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seybert A., Hegyi A., Siddell S.G., Ziebuhr J. The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5′-to-3′ polarity. RNA. 2000;6:1056–1068. doi: 10.1017/s1355838200000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Meer Y., Snijder E.J., Dobbe J.C., Schleich S., Denison M.R., Spaan W.J.M., Krijnse Locker J. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J. Virol. 1999;73:7641–7657. doi: 10.1128/jvi.73.9.7641-7657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gosert R., Kanjanahaluethai A., Egger D., Bienz K., Baker S.C. RNA replication of Mouse Hepatitis Virus takes place at double-membrane vesicles. J. Virol. 2002;76:3697–3708. doi: 10.1128/JVI.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorbalenya A.E. Big nidovirus genome. When count and order of domains matter. Advan. Expt. Biol. Med. 2001;494:1–17. [PubMed] [Google Scholar]
- 19.Sawicki S.G., Sawicki D.L. Coronaviruses use discontinuous extension for synthesis of subgenome-length negative strands. Advan. Expt. Biol. Med. 1995;380:499–506. doi: 10.1007/978-1-4615-1899-0_79. [DOI] [PubMed] [Google Scholar]
- 20.Chouljenko V.N., Lin X.Q., Storz J., Kousoulas K.G., Gorbalenya A.E. Comparison of genomic and predicted amino acid sequences of respiratory and enteric bovine coronaviruses isolated from the same animal with fatal shipping pneumonia. J. Gen. Virol. 2001;82:2927–2933. doi: 10.1099/0022-1317-82-12-2927. [DOI] [PubMed] [Google Scholar]
- 21.Hegyi A., Friebe A., Gorbalenya A.E., Ziebuhr J. Mutational analysis of the active centre of coronavirus 3C-like proteases. J. Gen. Virol. 2002;83:581–593. doi: 10.1099/0022-1317-83-3-581. [DOI] [PubMed] [Google Scholar]
- 22.Stephensen C.B., Casebolt D.B., Gangopadhyay N.N. Phylogenetic analysis of a highly conserved region of the polymerase gene from 11 coronaviruses and development of a consensus polymerase chain reaction assay. Virus Res. 1999;60:181–189. doi: 10.1016/S0168-1702(99)00017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzales, J. M., Gomez-Puertas, P., Cavanagh, D., Gorbalenya, A.E. & Enjuanes, L. (2003). A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch. Virol. In the press. [DOI] [PMC free article] [PubMed]
- 24.Sanchez C.M., Jimenez G., Laviada M.D., Correa I., Sune C., Bullido M. Antigenic homology among coronaviruses related to transmissible gastroenteritis virus. Virology. 1990;174:410–417. doi: 10.1016/0042-6822(90)90094-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snijder E.J., Horzinek M.C. Toroviruses: replication, evolution and comparison with other members of the coronavirus-like superfamily. J. Gen. Virol. 1993;74:2305–2316. doi: 10.1099/0022-1317-74-11-2305. [DOI] [PubMed] [Google Scholar]
- 26.Snijder E.J., den Boon J.A., Bredenbeek P.J., Horzinek M.C., Rijnbrand R., Spaan W.J.M. The carboxyl-terminal part of the putative Berne virus polymerase is expressed by ribosomal frameshifting and contains sequence motifs which indicate that toro- and coronaviruses are evolutionarily related. Nucl. Acids Res. 1990;18:4535–4542. doi: 10.1093/nar/18.15.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snijder E.J., den Boon J.A., Spaan W.J.M., Weiss M., Horzinek M.C. Primary structure and post-translational processing of the Berne virus peplomer protein. Virology. 1990;178:355–363. doi: 10.1016/0042-6822(90)90332-L. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.den Boon J.A., Snijder E.J., Krijnse Locker J., Horzinek M.C., Rottier P.J.M. Another triple-spanning envelope protein among intracellularly budding RNA viruses: the torovirus E protein. Virology. 1991;182:655–663. doi: 10.1016/0042-6822(91)90606-C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Haan C.A., Masters P.S., Shen X., Weiss S., Rottier P.J. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology. 2002;296:177–189. doi: 10.1006/viro.2002.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X.M., Herbst W., Kousoulas K.G., Storz J. Biological and genetic characterization of a hemagglutinating coronavirus isolated from a diarrhoeic child. J. Med. Virol. 1994;44:152–161. doi: 10.1002/jmv.1890440207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasseville A.M., Boutin M., Gelinas A.M., Dea S. Sequence of the 3′-terminal end (8.1 kb) of the genome of porcine haemagglutinating encephalomyelitis virus: comparison with other haemagglutinating coronaviruses. J. Gen. Virol. 2002;83:2411–2416. doi: 10.1099/0022-1317-83-10-2411. [DOI] [PubMed] [Google Scholar]
- 32.Ziebuhr J., Thiel V., Gorbalenya A.E. The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involves two paralogous papain-like proteases that cleave the same peptide bond. J. Biol. Chem. 2001;276:33220–33232. doi: 10.1074/jbc.M104097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 34.Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 35.Baker S.C., Yokomori K., Dong S., Carlisle R., Gorbalenya A.E., Koonin E.V., Lai M.M. Identification of the catalytic sites of a papain-like cysteine proteinase of murine coronavirus. J. Virol. 1993;67:6056–6063. doi: 10.1128/jvi.67.10.6056-6063.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanjanahaluethai A., Baker S.C. Identification of mouse hepatitis virus papain-like proteinase 2 activity. J. Virol. 2000;74:7911–7921. doi: 10.1128/jvi.74.17.7911-7921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim K.P., Ng L.F., Liu D.X. Identification of a novel cleavage activity of the first papain-like proteinase domain encoded by open reading frame 1a of the coronavirus Avian infectious bronchitis virus and characterization of the cleavage products. J. Virol. 2000;74:1674–1685. doi: 10.1128/jvi.74.4.1674-1685.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tijms M.A., van Dinten L.C., Gorbalenya A.E., Snijder E.J. A zinc finger-containing papain-like protease couples subgenomic mRNA synthesis to genome translation in a positive-stranded RNA virus. Proc. Natl Acad. Sci. USA. 2001;98:1889–1894. doi: 10.1073/pnas.041390398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pasternak A.O., van den Born E., Spaan W.J.M., Snijder E.J. Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis. EMBO J. 2000;20:7220–7228. doi: 10.1093/emboj/20.24.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sethna P.B., Hung S.L., Brian D.A. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc. Natl Acad. Sci. USA. 1989;86:5626–5630. doi: 10.1073/pnas.86.14.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boursnell M.E., Brown T.D.K., Foulds I.J., Green P.F., Tomley F.M., Binns M.M. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J. Gen. Virol. 1987;68:57–77. doi: 10.1099/0022-1317-68-1-57. [DOI] [PubMed] [Google Scholar]
- 42.Laneve P., Altieri F., Fiori M.E., Scaloni A., Bozzoni I., Caffarelli E. Purification, cloning, and characterization of XendoU, a novel endoribonuclease involved in processing of intron-encoded small nucleolar RNAs in Xenopus laevis. J. Biol. Chem. 2003;278:13026–13032. doi: 10.1074/jbc.M211937200. [DOI] [PubMed] [Google Scholar]
- 43.Zuo Y., Deutscher M.P. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucl. Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bugl H., Fauman E.B., Staker B.L., Zheng F., Kushner S.R., Saper M.A. RNA methylation under heat shock control. Mol. Cell. 2000;6:349–360. doi: 10.1016/s1097-2765(00)00035-6. [DOI] [PubMed] [Google Scholar]
- 45.Martzen M.R., McCraith S.M., Spinelli S.L., Torres F.M., Fields S., Grayhack E.J., Phizicky E.M. A biochemical genomics approach for identifying genes by the activity of their products. Science. 1999;286:1153–1155. doi: 10.1126/science.286.5442.1153. [DOI] [PubMed] [Google Scholar]
- 46.Nasr F., Filipowicz W. Characterization of the Saccharomyces cerevisiae cyclic nucleotide phosphodiesterase involved in the metabolism of ADP-ribose 1″,2″-cyclic phosphate. Nucl. Acids Res. 2000;28:1676–1683. doi: 10.1093/nar/28.8.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gorbalenya A.E., Koonin E.V., Lai M.M. Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha- and coronaviruses. FEBS Letters. 1991;288:201–205. doi: 10.1016/0014-5793(91)81034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.den Boon J.A., Snijder E.J., Chirnside E.D., de Vries A.A.F., Horzinek M.C., Spaan W.J.M. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J. Virol. 1991;65:2910–2920. doi: 10.1128/jvi.65.6.2910-2920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bredenbeek P.J., Noten A.F., Horzinek M.C., Spaan W.J. Identification and stability of a 30-kDa nonstructural protein encoded by mRNA 2 of mouse hepatitis virus in infected cells. Virology. 1990;175:303–306. doi: 10.1016/0042-6822(90)90212-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Snijder E.J., den Boon J.A., Horzinek M.C., Spaan W.J.M. Comparison of the genome organization of toro- and coronaviruses: evidence for two nonhomologous RNA recombination events during Berne virus evolution. Virology. 1991;180:448–452. doi: 10.1016/0042-6822(91)90056-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bernad A., Blanco L., Lazaro J.M., Martin G., Salas M. A conserved 3′–5′ exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell. 1989;59:219–228. doi: 10.1016/0092-8674(89)90883-0. [DOI] [PubMed] [Google Scholar]
- 52.Egloff M.P., Benarroch D., Selisko B., Romette J.L., Canard B. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 2002;21:2757–2768. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Breyer W.A., Matthews B.W. Structure of Escherichia coli exonuclease I suggests how processivity is achieved. Nature Struct. Biol. 2000;7:1125–1128. doi: 10.1038/81978. [DOI] [PubMed] [Google Scholar]
- 54.Brautigam C.A., Sun S., Piccirilli J.A., Steitz T.A. Structures of normal single-stranded DNA and deoxyribo-3′-S-phosphorothiolates bound to the 3′-5′ exonucleolytic active site of DNA polymerase I from Escherichia coli. Biochemistry. 1999;38:696–704. doi: 10.1021/bi981537g. [DOI] [PubMed] [Google Scholar]
- 55.Nagy P.D., Simon A.E. New insights into the mechanisms of RNA recombination. Virology. 1997;235:1–9. doi: 10.1006/viro.1997.8681. [DOI] [PubMed] [Google Scholar]
- 56.Crotty S., Maag D., Arnold J.J., Zhong W., Lau J.Y., Hong Z. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nature Med. 2000;6:1375–1379. doi: 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- 57.Feder M., Pas J., Wyrwicz L.S., Bujnicki J.M. Molecular phylogenetics of the RrmJ/fibrillarin superfamily of ribose 2′-O-methyltransferases. Gene. 2003;302:129–138. doi: 10.1016/s0378-1119(02)01097-1. [DOI] [PubMed] [Google Scholar]
- 58.Abelson J., Trotta C.R., Li H. tRNA splicing. J. Biol. Chem. 1998;273:12685–12688. doi: 10.1074/jbc.273.21.12685. [DOI] [PubMed] [Google Scholar]
- 59.Filipowicz W., Pogacic V. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 2002;14:319–327. doi: 10.1016/s0955-0674(02)00334-4. [DOI] [PubMed] [Google Scholar]
- 60.Allmang C., Kufel J., Chanfreau G., Mitchell P., Petfalski E., Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang H., Boisvert D., Kim K.K., Kim R., Kim S.H. Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 Å resolution. EMBO J. 2000;19:317–323. doi: 10.1093/emboj/19.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Culver G.M., Consaul S.A., Tycowski K.T., Filipowicz W., Phizicky E.M. tRNA splicing in yeast and wheat germ. A cyclic phosphodiesterase implicated in the metabolism of ADP-ribose 1″,2″-cyclic phosphate. J. Biol. Chem. 1994;269:24928–24934. [PubMed] [Google Scholar]
- 63.Marchler-Bauer A., Panchenko A.R., Shoemaker B.A., Thiessen P.A., Geer L.Y., Bryant S.H. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucl. Acids Res. 2002;30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bateman A., Birney E., Durbin R., Eddy S.R., Finn R.D., Sonnhammer E.L. Pfam 3.1: 1313 multiple alignments and profile HMMs match the majority of proteins. Nucl. Acids Res. 1999;27:260–262. doi: 10.1093/nar/27.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl. Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morgenstern B. DIALIGN 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics. 1999;15:211–218. doi: 10.1093/bioinformatics/15.3.211. [DOI] [PubMed] [Google Scholar]
- 67.Henikoff S., Henikoff J.G. Position-based sequence weights. J. Mol. Biol. 1994;243:574–578. doi: 10.1016/0022-2836(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 68.Nicholas K.B., Nicholas N.H.B.J., Deerfield D.W. GeneDoc: analysis and visualization of genetic variation. EMBNET News. 1997;4:1–4. [Google Scholar]
- 69.Eddy S.R. Hidden Markov models. Curr. Opin. Struct. Biol. 1996;6:361–365. doi: 10.1016/s0959-440x(96)80056-x. [DOI] [PubMed] [Google Scholar]
- 70.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl. Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ponting C.P., Schultz J., Copley R.R., Andrade M.A., Bork P. Evolution of domain families. Advan. Protein Chem. 2000;54:185–244. doi: 10.1016/s0065-3233(00)54007-8. [DOI] [PubMed] [Google Scholar]
- 72.Bailey T.L., Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- 73.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 74.Kimura M. The Neutral Theory of Molecular Evolution Cambridge University Press. Cambridge; NY: 1983. [Google Scholar]
- 75.Page R.D. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 76.Hofmann M.A., Chang R.Y., Ku S., Brian D.A. Leader–mRNA junction sequences are unique for each subgenomic mRNA species in the bovine coronavirus and remain so throughout persistent infection. Virology. 1993;196:163–171. doi: 10.1006/viro.1993.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Senanayake S.D., Brian D.A. Bovine coronavirus I protein synthesis follows ribosomal scanning on the bicistronic N mRNA. Virus Res. 1997;48:101–105. doi: 10.1016/S0168-1702(96)01423-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu D.X., Inglis S.C. Internal entry of ribosomes on a tricistronic mRNA encoded by infectious bronchitis virus. J. Virol. 1992;66:6143–6154. doi: 10.1128/jvi.66.10.6143-6154.1992. (Erratum in J. Virol.66, 6840) [DOI] [PMC free article] [PubMed] [Google Scholar]
Further reading
Supplementary Material comprising one Figure is available on Science Direct