Abstract
Background
The recent global outbreak of coronavirus disease (COVID-19) is affecting many countries worldwide. Iran is one of the top 10 most affected countries. Search engines provide useful data from populations, and these data might be useful to analyze epidemics. Utilizing data mining methods on electronic resources’ data might provide a better insight into the COVID-19 outbreak to manage the health crisis in each country and worldwide.
Objective
This study aimed to predict the incidence of COVID-19 in Iran.
Methods
Data were obtained from the Google Trends website. Linear regression and long short-term memory (LSTM) models were used to estimate the number of positive COVID-19 cases. All models were evaluated using 10-fold cross-validation, and root mean square error (RMSE) was used as the performance metric.
Results
The linear regression model predicted the incidence with an RMSE of 7.562 (SD 6.492). The most effective factors besides previous day incidence included the search frequency of handwashing, hand sanitizer, and antiseptic topics. The RMSE of the LSTM model was 27.187 (SD 20.705).
Conclusions
Data mining algorithms can be employed to predict trends of outbreaks. This prediction might support policymakers and health care managers to plan and allocate health care resources accordingly.
Keywords: coronavirus, COVID-19, prediction, incidence, Google Trends, linear regression, LSTM, pandemic, outbreak, public health
Introduction
Recently, a respiratory disease originating from coronavirus occurred in Wuhan City of China. Since the first positive case of this virus was in 2019, this coronavirus was named coronavirus disease (COVID-19) by the World Health Organization (WHO) [1]. Some hypotheses attribute the origin of this virus to seafood and bats [2].
COVID-19 has spread globally and has affected most countries; it was defined as a pandemic by the WHO in March 2020 [3]. As of March 21, 2020, COVID-19 has affected 186 countries and territories around the world, with more than 280,000 confirmed cases and 11,842 deaths [4]. Iran is one of the top 10 countries affected by this virus [4].
As COVID-19 is spreading rapidly worldwide, prediction models can help in health resource management and planning for prevention purposes. Google search data is one information resource that contains useful information to predict and estimate epidemics [5]. Data mining algorithms and techniques are well-known tools for predictive model development and data analysis. They can implicitly extract useful information from raw data [6-8]. The extracted knowledge can be used in different areas such as the health care industry. Recently, a large amount of data was generated in health care, including those on patients, diseases, and diagnoses.
The tasks in data mining fall into two categories: (1) descriptive tasks that deal with the general properties of the data and (2) predictive tasks, wherein the goal is to build models that can estimate the mapping from inputs to outputs by using a sample of data called training data. The trained models can be deployed to make predictions of the outputs for unseen inputs. These techniques are more flexible and efficient for exploratory analysis than the traditional statistical analysis [9].
In this study, data mining models were used to build predictive models from Google search data to predict the incidence of COVID-19 in Iran.
Methods
Dataset
The daily new cases of coronavirus (daily incidence) from February 15, 2020, to March 18, 2020, in Iran were obtained from the Worldometer website [10].
Google Trends [11] was searched for concepts related to COVID-19, from February 10, 2020, to March 18, 2020. The related concepts were suggested by one of the authors. A dataset consisting of 10 input features including the previous day’s search trends, cases of the previous day, and a target value (new cases of that day) was created. The total number of entries was calculated for the 37 days. The list of features is shown in Table 1. The terms in square brackets were searched in the corresponding Persian language words. The “pd” postfix in the features’ name indicates that the features are related to the previous day.
Table 1.
Features used for predicting new COVID-19 cases.
| Feature name | Description |
| [Corona]_pd | The interest of “Corona” search term in Persian for the previous day in Iran |
| COVID-19_pda | The interest of “COVID-19” search term for the previous day in Iran |
| Coronavirus_pd | The interest of “Coronavirus” topic for the previous day in Iran |
| [Antiseptic selling]_pd | The interest of “Antiseptic selling” search term in Persian for the previous day in Iran |
| [Antiseptic buying]_pd | The interest of “Antiseptic buying” search term in Persian for the previous day in Iran |
| [Hand washing]_pd | The interest of “Handwashing” search term in Persian for the previous day in Iran |
| Hand sanitizer_pd | The interest of “Hand sanitizer” topic for the previous day in Iran |
| Ethanol_pd | The interest of “Ethanol” topic for the previous day in Iran |
| Antiseptic_pd | The interest of “Antiseptic” topic for the previous day in Iran |
| Cases_pd | COVID-19 Incidence of the previous day in Iran |
| New cases | COVID-19 Incidence of prediction day in Iran (Label) |
aCOVID-19: coronavirus disease
Google Trends does not provide absolute search numbers but instead, provides a measure entitled interest over time, which is described as “A value of 100 is the peak popularity for the term. A value of 50 means that the term is half as popular. A score of 0 means there was not enough data for this term” [11]; for consistency, the values of the daily new cases were transformed to the range between 0 to 100.
Modeling and Evaluation
Linear Regression
One of the data mining techniques used for prediction tasks is linear regression. In a problem with one predictor, this technique tries to find the best line to fit. That line could relate the predictor and prediction values. The extended version of this one-predictor regression is called multiple linear regression and is used for multiple-predictor problems [12]. We used this type of linear regression in this study.
Long Short-Term Memory
Long short-term memory (LSTM) is an artificial recurrent neural network that is an effective model for the prediction of time series where data are sequential [9]. By storing the past in hidden states, they can predict the outputs more accurately. In this study, the aim was to estimate the number of positive COVID-19 cases through time; as this is a well-suited task for the LSTM model, we used this model in our study.
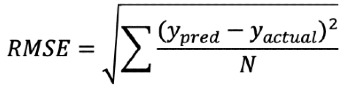
The linear regression model and a 3-layer LSTM model (Figure 1) are employed to predict the daily new cases. RapidMiner Studio 9.3.001 (RapidMiner GmbH) and Python 3.7.3 (Python Software Foundation) were used for modeling and evaluation. Tensorflow (Google Brain Team) and Keras (François Cholle) were used as frameworks for training LSTM models. In addition, 10-fold cross-validation was used to evaluate the performance of the models, and the root mean square error (RMSE) metric was chosen for performance evaluation:
Figure 1.
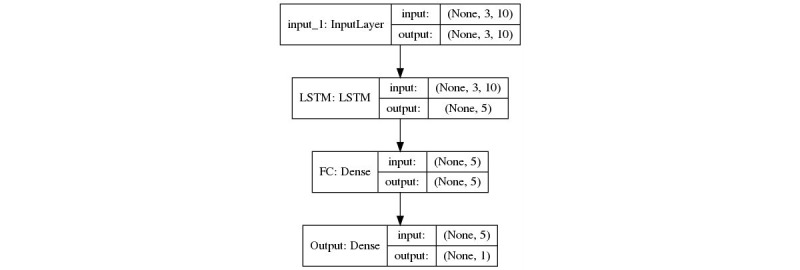
Proposed LSTM network architecture. LSTM: long short-term memory.
Results
The features’ effect in the linear regression model is shown in Table 2. The RMSE for the linear regression model was 7.562 (SD 6.492). The LSTM RMSE was 27.187 (SD 20.705). The training and validation loss of the LSTM model is shown in Figure 2. The predictions made by these models are shown in Figure 3.
Table 2.
Features’ effect on new daily cases in the linear regression model.
| Feature | Coefficient (SE) |
t value | P value |
| [Corona]_pd | –1.58 (0.77) | –2.05 | .05 |
| COVID-19_pda | 0.27 (0.12) | 2.28 | .03 |
| Coronavirus_pd | 1.55 (0.69) | 2.26 | .03 |
| [Antiseptic selling] _pd | –0.09 (0.11) | –0.78 | .44 |
| [Antiseptic buying] _pd | 0.32 (0.14) | 2.33 | .03 |
| [Hand washing] _pd | 0.44 (0.15) | 3.01 | .006 |
| Hand sanitizer_pd | –2.01 (0.50) | –4.00 | <.001 |
| Antiseptic | 1.52 (0.54) | 2.80 | .009 |
| New cases_pd | 1.03 (0.17) | 6.05 | <.001 |
aCOVID-19: coronavirus disease
Figure 2.
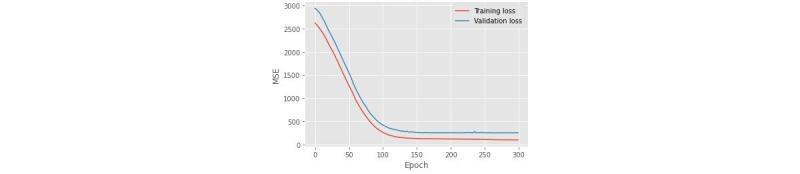
Training and validation loss of the long short-term memory model. MSE: mean squared error.
Figure 3.
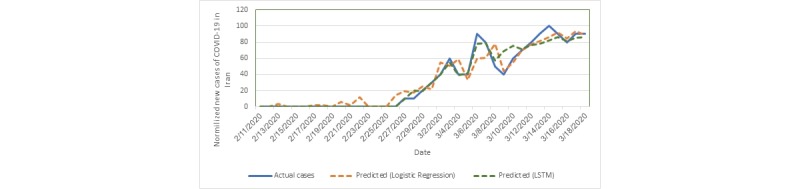
Actual and predicted new cases of COVID-19. LSTM: long short-term memory; COVID-19: coronavirus disease.
Discussion
In this study, we proposed the use of prediction models for COVID-19 incidence in Iran using Google Trends data. Although the predictions are not very precise, they can be helpful for a base idea to build accurate models from more aggregation of data.
The features’ effect of the linear regression model shows that besides new cases in the previous day, hand sanitizer, handwashing, and antiseptic topics were the focus of the population. It could be inferred that people’s worries were increasing, and they were seeking prevention solutions. The lags in the prediction might have originated from other countries’ incidence. In other words, the population could be more sensitive and engaged in their health care after hearing news about the epidemic in other countries. This model could be used for other types of interventions such as assessing individuals’ awareness and engagement. Health authorities might use these data for measuring the information’s broadcast effect on the population and obtaining feedback from the search statistics.
The LSTM model showed a fluctuated performance in the folds while training loss was low. This indicates overfitting in the LSTM model because of the limited amount of training data. However, a low training error shows that the LSTM model can extract the pattern in the data. Therefore, we believe that by increasing the amount of training data, the LSTM model can outperform other models for this task. In addition, owing to the presence of just a few samples in each test fold (4 instances) and the subsequent high variation in RMSE, evaluation of the LSTM model was repeated with 3-fold cross-validation and the obtained RMSE was 13.45 (SD 7.90).
Past work on influenza and Zika virus predictions, for example, in the study by Santillana et al [13] in 2015, proposed a machine learning method for predicting influenza in the United States. In their study, the authors used data from Google searches along with Twitter data, hospital visit records, and a surveillance system. They provided multiple estimates to have an unbiased and more accurate prediction. They also showed that social media contains important information for effectively predicting disease incidence.
In 2017, McGough et al [14] also proposed a Zika virus prediction system by using Zika-related Google searches, Twitter microblogs, and a digital surveillance system. They also showed that the internet-based sources were useful to predict weekly Zika cases. In another related study in 2016, Majumder et al [15] used HealthMap surveillance data and Google Trends to predict cases of Zika virus in Colombia [15] and showed that digital surveillance data could be useful for the prediction of Zika cases. Further, in 2017, Teng et al [16] proposed a prediction model for Zika virus using search data from Google Trends and built the model using an autoregressive integrated moving average. They found a strong correlation between Zika-related searches and Zika cases. For the incidence prediction of influenza, socioenvironmental factors were considered when developing an epidemiological model named Susceptible Exposed to Infectious Recovered (SEIR) [17]. The model supports decision makers to factor the mass media and climate factors into the classical epidemic models. Another study emphasized the importance of environmental factors for the development of an influenza prediction model [18]. The findings of these studies along with our study show that internet resources could be helpful in pandemic forecasting.
The easy-to-obtain Google search data is a more dynamic and available source in comparison with traditional data sources. It could be a representation of the population’s thoughts, concerns, conditions, and needs in multiple periods. The major strength of this study is use of these data to predict the epidemiology of COVID-19 for the first time in the country.
In contrast, one major limitation of this study is the limited access to the Google Search data. Since Google Trends just provides data based on “interest” measure, more accurate and informative models could be built if the absolute search frequency is available for the researchers. It is worth mentioning that we used some of the keywords related to COVID-19 for extracting Google search frequencies; the selected keywords may have been incomplete and further research could aim to identify the most relevant set of keywords. In addition, future research should combine other data sources such as social media information, people’s contacts with the special call center for COVID-19, mass media, environmental and climate factors, and screening registries. Furthermore, in the broader context, such predictions could be made for other countries and even globally.
In conclusion, the data mining models could help policymakers and health managers to plan health care resources and control the prevention of an epidemic outbreak. The availability of high-quality and timely data in the early stages of the outbreak collaboration of the researchers to analyze the data could have positive effects on health care resource planning.
Abbreviations
- COVID-19
coronavirus disease
- LSTM
long short-term memory
- RMSE
root mean square error
- SEIR
Susceptible Exposed to Infectious Recovered
- WHO
World Health Organization
Footnotes
Conflicts of Interest: None declared.
References
- 1.Guo Y, Cao Q, Hong Z, Tan Y, Chen S, Jin H, Tan K, Wang D, Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil Med Res. 2020 Mar 13;7(1):11. doi: 10.1186/s40779-020-00240-0. https://mmrjournal.biomedcentral.com/articles/10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giovanetti M, Benvenuto Domenico, Angeletti Silvia, Ciccozzi Massimo. The first two cases of 2019-nCoV in Italy: Where they come from? J Med Virol. 2020 May;92(5):518–521. doi: 10.1002/jmv.25699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet. 2020 Feb;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prathap L, Jagadeesan V, Suganthirababu P, Ganesan D. Online Journal of Health and Allied Sciences. 2017. [2020-04-07]. Association of quantitative and qualitative dermatoglyphic variable and DNA polymorphism in female breast cancer population https://www.ojhas.org/issue62/2017-2-2.pdf.
- 5.Yang S, Santillana M, Kou SC. Accurate estimation of influenza epidemics using Google search data via ARGO. Proc Natl Acad Sci U S A. 2015 Nov 24;112(47):14473–8. doi: 10.1073/pnas.1515373112. http://www.pnas.org/cgi/pmidlookup?view=long&pmid=26553980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piatetsky-Shapiro G. ACM SIGKDD Explorations Newsletter. 2000. Jan 01, [2020-04-07]. Knowledge Discovery in Databases: An Overview https://dl.acm.org/doi/10.1145/846183.846197.
- 7.Rushing J, Ramachandran R, Nair U, Graves S, Welch R, Lin H. ADaM: a data mining toolkit for scientists and engineers. Computers & Geosciences. 2005 Jun;31(5):607–618. doi: 10.1016/j.cageo.2004.11.009. [DOI] [Google Scholar]
- 8.Dangare CS, Apte SS. Improved Study of Heart Disease Prediction System using Data Mining Classification Techniques. IJCA. 2012 Jun 30;47(10):44–48. doi: 10.5120/7228-0076. [DOI] [Google Scholar]
- 9.Sherstinsky A. Fundamentals of Recurrent Neural Network (RNN) and Long Short-Term Memory (LSTM) network. Physica D: Nonlinear Phenomena. 2020 Mar;404:132306. doi: 10.1016/j.physd.2019.132306. [DOI] [Google Scholar]
- 10.Haggag A, Ibrahim Khalil A, Elzahed E, Abdelhamid M. Study of Fingerprints Pattern in Breast Cancer Patients Insharkia Governorate, A Case –Control Retrospective Clinical Study. Zagazig University Medical Journal. 2018 Jan 01;24(1):66–71. doi: 10.21608/zumj.2018.13006. [DOI] [Google Scholar]
- 11.Tahergorabi Z, Moodi M, Mesbahzadeh B. Breast Cancer: A preventable disease. Journal of Birjand University of Medical. 2014;21(2):126–141. [Google Scholar]
- 12.Bregman J, Mackenthun KM. Environmental Impact Statements. Chelsea: MI Lewis Publication; 2006. [Google Scholar]
- 13.Santillana M, Nguyen AT, Dredze M, Paul MJ, Nsoesie EO, Brownstein JS. Combining Search, Social Media, and Traditional Data Sources to Improve Influenza Surveillance. PLoS Comput Biol. 2015 Oct 29;11(10):e1004513. doi: 10.1371/journal.pcbi.1004513. http://dx.plos.org/10.1371/journal.pcbi.1004513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGough SF, Brownstein JS, Hawkins JB, Santillana M. Forecasting Zika Incidence in the 2016 Latin America Outbreak Combining Traditional Disease Surveillance with Search, Social Media, and News Report Data. PLoS Negl Trop Dis. 2017 Jan 13;11(1):e0005295. doi: 10.1371/journal.pntd.0005295. http://dx.plos.org/10.1371/journal.pntd.0005295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Majumder MS, Santillana M, Mekaru SR, McGinnis DP, Khan K, Brownstein JS. Utilizing Nontraditional Data Sources for Near Real-Time Estimation of Transmission Dynamics During the 2015-2016 Colombian Zika Virus Disease Outbreak. JMIR Public Health Surveill. 2016 Jun 01;2(1):e30. doi: 10.2196/publichealth.5814. https://publichealth.jmir.org/2016/1/e30/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teng Y, Bi D, Xie G, Jin Y, Huang Y, Lin B, An X, Feng D, Tong Y. Dynamic Forecasting of Zika Epidemics Using Google Trends. PLoS One. 2017 Jan 6;12(1):e0165085. doi: 10.1371/journal.pone.0165085. http://dx.plos.org/10.1371/journal.pone.0165085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niakan Kalhori S, Ghazisaeedi M, Azizi R, Naserpour A. Studying the influence of mass media and environmental factors on influenza virus transmission in the US Midwest. Public Health. 2019 May;170:17–22. doi: 10.1016/j.puhe.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 18.Naserpor A, Niakan Kalhori SR, Ghazisaeedi M, Azizi R, Hosseini Ravandi M, Sharafie S. Modification of the Conventional Influenza Epidemic Models Using Environmental Parameters in Iran. Healthc Inform Res. 2019 Jan;25(1):27–32. doi: 10.4258/hir.2019.25.1.27. https://www.e-hir.org/DOIx.php?id=10.4258/hir.2019.25.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]


