Abstract
Background
In late December, 2019, patients presenting with viral pneumonia due to an unidentified microbial agent were reported in Wuhan, China. A novel coronavirus was subsequently identified as the causative pathogen, provisionally named 2019 novel coronavirus (2019-nCoV). As of Jan 26, 2020, more than 2000 cases of 2019-nCoV infection have been confirmed, most of which involved people living in or visiting Wuhan, and human-to-human transmission has been confirmed.
Methods
We did next-generation sequencing of samples from bronchoalveolar lavage fluid and cultured isolates from nine inpatients, eight of whom had visited the Huanan seafood market in Wuhan. Complete and partial 2019-nCoV genome sequences were obtained from these individuals. Viral contigs were connected using Sanger sequencing to obtain the full-length genomes, with the terminal regions determined by rapid amplification of cDNA ends. Phylogenetic analysis of these 2019-nCoV genomes and those of other coronaviruses was used to determine the evolutionary history of the virus and help infer its likely origin. Homology modelling was done to explore the likely receptor-binding properties of the virus.
Findings
The ten genome sequences of 2019-nCoV obtained from the nine patients were extremely similar, exhibiting more than 99·98% sequence identity. Notably, 2019-nCoV was closely related (with 88% identity) to two bat-derived severe acute respiratory syndrome (SARS)-like coronaviruses, bat-SL-CoVZC45 and bat-SL-CoVZXC21, collected in 2018 in Zhoushan, eastern China, but were more distant from SARS-CoV (about 79%) and MERS-CoV (about 50%). Phylogenetic analysis revealed that 2019-nCoV fell within the subgenus Sarbecovirus of the genus Betacoronavirus, with a relatively long branch length to its closest relatives bat-SL-CoVZC45 and bat-SL-CoVZXC21, and was genetically distinct from SARS-CoV. Notably, homology modelling revealed that 2019-nCoV had a similar receptor-binding domain structure to that of SARS-CoV, despite amino acid variation at some key residues.
Interpretation
2019-nCoV is sufficiently divergent from SARS-CoV to be considered a new human-infecting betacoronavirus. Although our phylogenetic analysis suggests that bats might be the original host of this virus, an animal sold at the seafood market in Wuhan might represent an intermediate host facilitating the emergence of the virus in humans. Importantly, structural analysis suggests that 2019-nCoV might be able to bind to the angiotensin-converting enzyme 2 receptor in humans. The future evolution, adaptation, and spread of this virus warrant urgent investigation.
Funding
National Key Research and Development Program of China, National Major Project for Control and Prevention of Infectious Disease in China, Chinese Academy of Sciences, Shandong First Medical University.
Introduction
Viruses of the family Coronaviridae possess a single-strand, positive-sense RNA genome ranging from 26 to 32 kilobases in length.1 Coronaviruses have been identified in several avian hosts,2, 3 as well as in various mammals, including camels, bats, masked palm civets, mice, dogs, and cats. Novel mammalian coronaviruses are now regularly identified.1 For example, an HKU2-related coronavirus of bat origin was responsible for a fatal acute diarrhoea syndrome in pigs in 2018.4
Among the several coronaviruses that are pathogenic to humans, most are associated with mild clinical symptoms,1 with two notable exceptions: severe acute respiratory syndrome (SARS) coronavirus (SARS-CoV), a novel betacoronavirus that emerged in Guangdong, southern China, in November, 2002,5 and resulted in more than 8000 human infections and 774 deaths in 37 countries during 2002–03;6 and Middle East respiratory syndrome (MERS) coronavirus (MERS-CoV), which was first detected in Saudi Arabia in 20127 and was responsible for 2494 laboratory-confirmed cases of infection and 858 fatalities since September, 2012, including 38 deaths following a single introduction into South Korea.8, 9
Research in context.
Evidence before this study
The causal agent of an outbreak of severe pneumonia in Wuhan, China, is a novel coronavirus, provisionally named 2019 novel coronavirus (2019-nCoV). The first cases were reported in December, 2019.
Added value of this study
We have described the genomic characteristics of 2019-nCoV and similarities and differences to other coronaviruses, including the virus that caused the severe acute respiratory syndrome epidemic of 2002–03. Genome sequences of 2019-nCoV sampled from nine patients who were among the early cases of this severe infection are almost genetically identical, which suggests very recent emergence of this virus in humans and that the outbreak was detected relatively rapidly. 2019-nCoV is most closely related to other betacoronaviruses of bat origin, indicating that these animals are the likely reservoir hosts for this emerging viral pathogen.
Implications of all the available evidence
By documenting the presence of 2019-nCoV in a sample of patients, our study extends previous evidence that this virus has led to the novel pneumonia that has caused severe disease in Wuhan and other geographical localities. Currently available data suggest that 2019-nCoV infected the human population from a bat reservoir, although it remains unclear if a currently unknown animal species acted as an intermediate host between bats and humans.
In late December, 2019, several patients with viral pneumonia were found to be epidemiologically associated with the Huanan seafood market in Wuhan, in the Hubei province of China, where a number of non-aquatic animals such as birds and rabbits were also on sale before the outbreak. A novel, human-infecting coronavirus,10, 11 provisionally named 2019 novel coronavirus (2019-nCoV), was identified with use of next-generation sequencing. As of Jan 28, 2020, China has reported more than 5900 confirmed and more than 9000 suspected cases of 2019-nCoV infection across 33 Chinese provinces or municipalities, with 106 fatalities. In addition, 2019-nCoV has now been reported in Thailand, Japan, South Korea, Malaysia, Singapore, and the USA. Infections in medical workers and family clusters were also reported and human-to-human transmission has been confirmed.12 Most of the infected patients had a high fever and some had dyspnoea, with chest radiographs revealing invasive lesions in both lungs.12, 13
We report the epidemiological data of nine inpatients, from at least three hospitals in Wuhan, who were diagnosed with viral pneumonia of unidentified cause. Using next-generation sequencing of bronchoalveolar lavage fluid samples and cultured isolates from these patients, 2019-nCoV was found. We describe the genomic characterisation of ten genomes of this novel virus, providing important information on the origins and cell receptor binding of the virus.
Methods
Patients and samples
Nine patients with viral pneumonia and negative for common respiratory pathogens, who presented to at least three hospitals in Wuhan, were included in this study. Eight of the patients had visited the Huanan seafood market before the onset of illness, and one patient (WH04) did not visit the market but stayed in a hotel near the market between Dec 23 and Dec 27, 2019 (table ). Five of the patients (WH19001, WH19002, WH19004, WH19008, and YS8011) had samples collected by the Chinese Center for Disease Control and Prevention (CDC) which were tested for 18 viruses and four bacteria using the RespiFinderSmart22 Kit (PathoFinder, Maastricht, Netherlands) on the LightCycler 480 Real-Time PCR system (Roche, Rotkreuz, Switzerland). Presence of SARS-CoV and MERS-CoV was tested using a previously reported method.14 All five CDC samples were negative for all common respiratory pathogens screened for. Four of the patients (WH01, WH02, WH03, and WH04) had samples collected by BGI (Beijing, China), and were tested for five viruses and one bacterium using the RespiPathogen 6 Kit (Jiangsu Macro & Micro Test, Nantong, China) on the Applied Biosystems ABI 7500 Real-Time PCR system (ThermoFisher Scientific, Foster City, CA, USA). All four samples were negative for the targeted respiratory pathogens.
Table.
Information about samples taken from nine patients infected with 2019-nCoV
|
Patient information |
Sample information |
Genome sequence obtained | |||||
|---|---|---|---|---|---|---|---|
| Exposure to Huanan seafood market | Date of symptom onset | Admission date | Sample type | Collection date | Ct value | ||
| Samples WH19001 and WH19005 | Yes | Dec 23, 2019 | Dec 29, 2019 | BALF and cultured virus | Dec 30, 2019 | 30·23 | Complete |
| Sample WH19002 | Yes | Dec 22, 2019 | NA | BALF | Dec 30, 2019 | 30·50 | Partial (27 130 nucleotides) |
| Sample WH19004 | Yes | NA | NA | BALF | Jan 1, 2020 | 32·14 | Complete |
| Sample WH19008 | Yes | NA | Dec 29, 2019 | BALF | Dec 30, 2019 | 26·35 | Complete |
| Sample YS8011 | Yes | NA | NA | Throat swab | Jan 7, 2020 | 22·85 | Complete |
| Sample WH01 | Yes | NA | NA | BALF | Dec 26, 2019 | 32·60 | Complete |
| Sample WH02 | Yes | NA | NA | BALF | Dec 31, 2019 | 34·23 | Partial (19 503 nucleotides) |
| Sample WH03 | Yes | Dec 26, 2019 | NA | BALF | Jan 1, 2020 | 25·38 | Complete |
| Sample WH04 | No* | Dec 27, 2019 | NA | BALF | Jan 5, 2020 | 25·23 | Complete |
Ct=threshold cycle. BALF=bronchoalveolar lavage fluid. NA=not available. 2019-nCoV=2019 novel coronavirus.
Patient stayed in a hotel near Huanan seafood market from Dec 23 to Dec 27, 2019, and reported fever on Dec 27, 2019.
Virus isolation
Special-pathogen-free human airway epithelial (HAE) cells were used for virus isolation. Briefly, bronchoalveolar lavage fluids or throat swabs from the patients were inoculated into the HAE cells through the apical surfaces. HAE cells were maintained in an air–liquid interface incubated at 37°C. The cells were monitored daily for cytopathic effects by light microscopy and the cell supernatants were collected for use in quantitative RT-PCR assays. After three passages, apical samples were collected for sequencing.
BGI sequencing strategy
All collected samples were sent to BGI for sequencing. 140 μL bronchoalveolar lavage fluid samples (WH01 to WH04) were reserved for RNA extraction using the QIAamp Viral RNA Mini Kit (52904; Qiagen, Heiden, Germany), according to the manufacturer's recommendations. A probe-captured technique was used to remove human nucleic acid. The remaining RNA was reverse-transcribed into cDNA, followed by the second-strand synthesis. Using the synthetic double-stranded DNA, a DNA library was constructed through DNA-fragmentation, end-repair, adaptor-ligation, and PCR amplification. The constructed library was qualified with an Invitrogen Qubit 2.0 Fluorometer (ThermoFisher, Foster City, CA, USA), and the qualified double-stranded DNA library was transformed into a single-stranded circular DNA library through DNA-denaturation and circularisation. DNA nanoballs were generated from single-stranded circular DNA by rolling circle amplification, then qualified with Qubit 2.0 and loaded onto the flow cell and sequenced with PE100 on the DNBSEQ-T7 platform (MGI, Shenzhen, China).
After removing adapter, low-quality, and low-complexity reads, high-quality genome sequencing data were generated. Sequence reads were first filtered against the human reference genome (hg19) using Burrows-Wheeler Alignment.15 The remaining data were then aligned to the local nucleotide database (using Burrows-Wheeler Alignment) and non-redundant protein database (using RapSearch),16 downloaded from the US National Center for Biotechnology Information website, which contain only coronaviruses that have been published. Finally, the mapped reads were assembled with SPAdes17 to obtain a high-quality coronavirus genome sequence.
Primers were designed with use of OLIGO Primer Analysis Software version 6.44 on the basis of the assembled partial genome, and were verified by Primer-Blast (for more details on primer sequencs used please contact the corresponding author). PCR was set up as follows: 4·5 μL of 10X buffer, 4 μL of dNTP mix (2·5 μmol/L), 1 μL of each primer (10 μmol/L), and 0·75 units of HS Ex Taq (Takara Biomedical Technology, Beijing, China), in a total volume of 30 μL. The cDNAs reverse transcribed from clinical samples were used as templates, and random primers were used. The following program was run on the thermocycler: 95°C for 5 min; 40 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min as determined by product size; 72°C for 7 min; and a 4°C hold. Finally, the PCR products were separated by agarose gel electrophoresis, and products of the expected size were sequenced from both ends on the Applied Biosystems 3730 DNA Analyzer platform (Applied Biosystems, Life Technologies, Foster City, CA, USA; for more details on expected size please contact the corresponding author).
Chinese CDC sequencing strategy
The whole-genome sequences of 2019-nCoV from six samples (WH19001, WH19005, WH19002, WH19004, WH19008, and YS8011) were generated by a combination Sanger, Illumina, and Oxford nanopore sequencing. First, viral RNAs were extracted directly from clinical samples with the QIAamp Viral RNA Mini Kit, and then used to synthesise cDNA with the SuperScript III Reverse Transcriptase (ThermoFisher, Waltham, MA, USA) and N6 random primers, followed by second-strand synthesis with DNA Polymerase I, Large (Klenow) Fragment (ThermoFisher). Viral cDNA libraries were prepared with use of the Nextera XT Library Prep Kit (Illumina, San Diego, CA, USA), then purified with Agencourt AMPure XP beads (Beckman Coulter, Brea, CA, USA), followed by quantification with an Invitrogen Qubit 2.0 Fluorometer. The resulting DNA libraries were sequenced on either the MiSeq or iSeq platforms (Illumina) using a 300-cycle reagent kit. About 1·2–5 GB of data were obtained for each sample.
The raw fastQ files for each virus sample were filtered using previously described criteria,18 then subjected to de novo assembly with the CLCBio software version 11.0.1. Mapped assemblies were also done using the bat-derived SARS-like coronavirus isolate bat-SL-CoVZC45 (accession number MG772933.1) as a reference. Variant calling, genome alignments, and sequence illustrations were generated with CLCBio software, and the assembled genome sequences were confirmed by Sanger sequencing.
Rapid amplification of cDNA ends (RACE) was done to obtain the sequences of the 5′ and 3′ termini, using the Invitrogen 5′ RACE System and 3′ RACE System (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's instructions. Gene-specific primers (appendix p 1) for 5′ and 3′ RACE PCR amplification were designed to obtain a fragment of approximately 400–500 bp for the two regions. Purified PCR products were cloned into the pMD18-T Simple Vector (TaKaRa, Takara Biotechnology, Dalian, China) and chemically competent Escherichia coli (DH5α cells; TaKaRa), according to the manufacturer's instructions. PCR products were sequenced with use of M13 forward and reverse primers.
Virus genome analysis and annotation
Reference virus genomes were obtained from GenBank using Blastn with 2019-nCoV as a query. The open reading frames of the verified genome sequences were predicted using Geneious (version 11.1.5) and annotated using the Conserved Domain Database.19 Pairwise sequence identities were also calculated using Geneious. Potential genetic recombination was investigated using SimPlot software (version 3.5.1)20 and phylogenetic analysis.
Phylogenetic analysis
Sequence alignment of 2019-nCoV with reference sequences was done with Mafft software (version 7.450).21 Phylogenetic analyses of the complete genome and major coding regions were done with RAxML software (version 8.2.9)22 with 1000 bootstrap replicates, employing the general time reversible nucleotide substitution model.
Development of molecular diagnostics for 2019-nCoV
On the basis of the genome sequences obtained, a real-time PCR detection assay was developed. PCR primers and probes were designed using Applied Biosystems Primer Express Software (ThermoFisher Scientific, Foster City, CA, USA) on the basis of our sequenced virus genomes. The specific primers and probe set (labelled with the reporter 6-carboxyfluorescein [FAM] and the quencher Black Hole Quencher 1 [BHQ1]) for orf1a were as follows: forward primer 5′-AGAAGATTGGTTAGATGATGATAGT-3′; reverse primer 5′-TTCCATCTCTAATTGAGGTTGAACC-3′; and probe 5′-FAM-TCCTCACTGCCGTCTTGTTGACCA-BHQ1-3′. The human GAPDH gene was used as an internal control (forward primer 5′-TCAAGAAGGTGGTGAAGCAGG-3′; reverse primer 5′-CAGCGTCAAAGGTGGAGGAGT-3′; probe 5′-VIC-CCTCAAGGGCATCCTGGGCTACACT-BHQ1-3′). Primers and probes were synthesised by BGI (Beijing, China). RT-PCR was done with an Applied Biosystems 7300 Real-Time PCR System (ThermoScientific), with 30 μL reaction volumes consisting of 14 μL of diluted RNA, 15 μL of 2X Taqman One-Step RT-PCR Master Mix Reagents (4309169; Applied Biosystems, ThermoFisher), 0·5 μL of 40X MultiScribe and RNase inhibitor mixture, 0·75 μL forward primer (10 μmol/L), 0·75 μL reverse primer (10 μmol/L), and 0·375 μL probe (10 μmol/L). Thermal cycling parameters were 30 min at 42°C, followed by 10 min at 95°C, and a subsequent 40 cycles of amplification (95°C for 15 s and 58°C for 45 s). Fluorescence was recorded during the 58°C phase.
Role of the funding source
The funder of the study had no role in data collection, data analysis, data interpretation, or writing of report. GFG and WS had access to all the data in the study, and GFG, WS, WT, WC, and GW were responsible for the decision to submit for publication.
Results
From the nine patients' samples analysed, eight complete and two partial genome sequences of 2019-nCoV were obtained. These data have been deposited in the China National Microbiological Data Center (accession number NMDC10013002 and genome accession numbers NMDC60013002-01 to NMDC60013002-10) and the data from BGI have been deposited in the China National GeneBank (accession numbers CNA0007332–35).
Based on these genomes, we developed a real-time PCR assay and tested the original clinical samples from the BGI (WH01, WH02, WH03, and WH04) again to determine their threshold cycle (Ct) values (table). The remaining samples were tested by a different real-time PCR assay developed by the Chinese CDC, with Ct values ranging from 22·85 to 32·41 (table). These results confirmed the presence of 2019-nCoV in the patients.
Bronchoalveolar lavage fluid samples or cultured viruses of nine patients were used for next-generation sequencing. After removing host (human) reads, de novo assembly was done and the contigs obtained used as queries to search the non-redundant protein database. Some contigs identified in all the samples were closely related to the bat SARS-like betacoronavirus bat-SL-CoVZC45 betacoronavirus.23 Bat-SL-CoVZC45 was then used as the reference genome and reads from each pool were mapped to it, generating consensus sequences corresponding to all the pools. These consensus sequences were then used as new reference genomes. Eight complete genomes and two partial genomes (from samples WH19002 and WH02; table) were obtained. The de novo assembly of the clean reads from all the pools did not identify any other long contigs that corresponded to other viruses at high abundance.
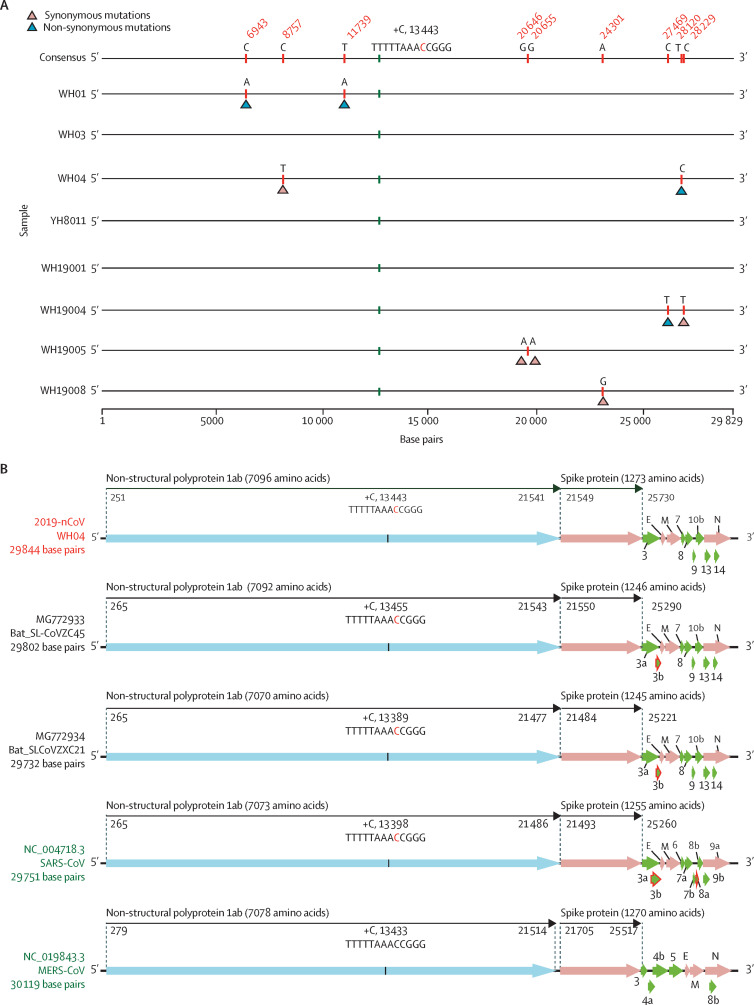
The eight complete genomes were nearly identical across the whole genome, with sequence identity above 99·98%, indicative of a very recent emergence into the human population (figure 1A ). The largest nucleotide difference was four mutations. Notably, the sequence identity between the two virus genomes from the same patient (WH19001, from bronchoalveolar lavage fluid, and WH19005, from cell culture) was more than 99·99%, with 100% identity at the amino acid level. In addition, the partial genomes from samples WH02 and WH19002 also had nearly 100% identity to the complete genomes across the aligned gene regions.
Figure 1.
Sequence comparison and genomic organisation of 2019-nCoV
(A) Sequence alignment of eight full-length genomes of 2019-nCoV, 29 829 base pairs in length, with a few nucleotides truncated at both ends of the genome. (B) Coding regions of 2019-nCoV, bat-SL-CoVZC45, bat-SL-CoVZXC21, SARS-CoV, and MERS-CoV. Only open reading frames of more than 100 nucleotides are shown. 2019-nCoV=2019 novel coronavirus. SARS-CoV=severe acute respiratory syndrome coronavirus. MERS-CoV=Middle East respiratory syndrome coronavirus.
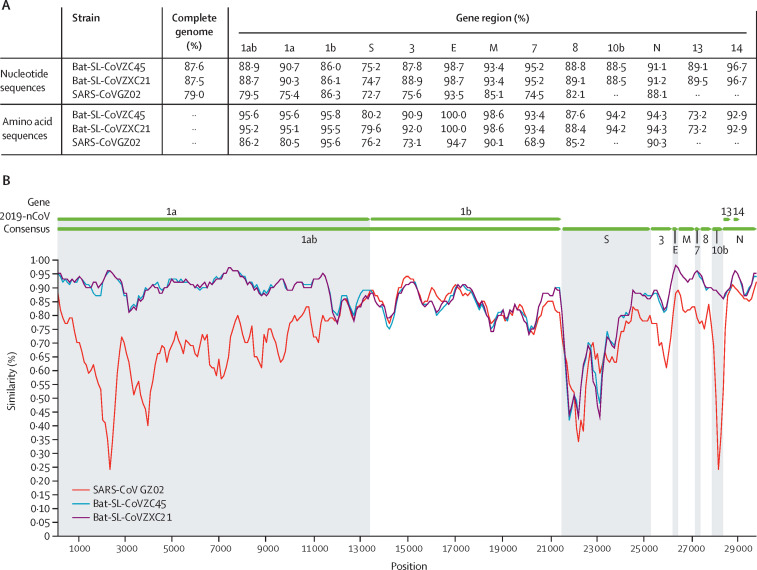
A Blastn search of the complete genomes of 2019-nCoV revealed that the most closely related viruses available on GenBank were bat-SL-CoVZC45 (sequence identity 87·99%; query coverage 99%) and another SARS-like betacoronavirus of bat origin, bat-SL-CoVZXC21 (accession number MG772934;23 87·23%; query coverage 98%). In five gene regions (E, M, 7, N, and 14), the sequence identities were greater than 90%, with the highest being 98·7% in the E gene (figure 2A ). The S gene of 2019-nCoV exhibited the lowest sequence identity with bat-SL-CoVZC45 and bat-SL-CoVZXC21, at only around 75%. In addition, the sequence identity in 1b (about 86%) was lower than that in 1a (about 90%; figure 2A). Most of the encoded proteins exhibited high sequence identity between 2019-nCoV and the related bat-derived coronaviruses (figure 2a). The notable exception was the spike protein, with only around 80% sequence identity, and protein 13, with 73·2% sequence identity. Notably, the 2019-nCoV strains were less genetically similar to SARS-CoV (about 79%) and MERS-CoV (about 50%). The similarity between 2019-nCoV and related viruses was visualised using SimPlot software, with the 2019-nCoV consensus sequence employed as the query (figure 2B).
Figure 2.
Sequence identity between the consensus of 2019-nCoV and representative betacoronavirus genomes
(A) Sequence identities for 2019-nCoV compared with SARS-CoV GZ02 (accession number AY390556) and the bat SARS-like coronaviruses bat-SL-CoVZC45 (MG772933) and bat-SL-CoVZXC21 (MG772934). (B) Similarity between 2019-nCoV and related viruses. 2019-nCoV=2019 novel coronavirus. SARS-CoV=severe acute respiratory syndrome coronavirus.
Comparison of the predicted coding regions of 2019-nCoV showed that they possessed a similar genomic organisation to bat-SL-CoVZC45, bat-SL-CoVZXC21, and SARS-CoV (figure 1B). At least 12 coding regions were predicted, including 1ab, S, 3, E, M, 7, 8, 9, 10b, N, 13, and 14 (figure 1B). The lengths of most of the proteins encoded by 2019-nCoV, bat-SL-CoVZC45, and bat-SL-CoVZXC21 were similar, with only a few minor insertions or deletions. A notable difference was a longer spike protein encoded by 2019-nCoV compared with the bat SARS-like coronaviruses, SARS-CoV, and MERS-CoV (figure 1B).
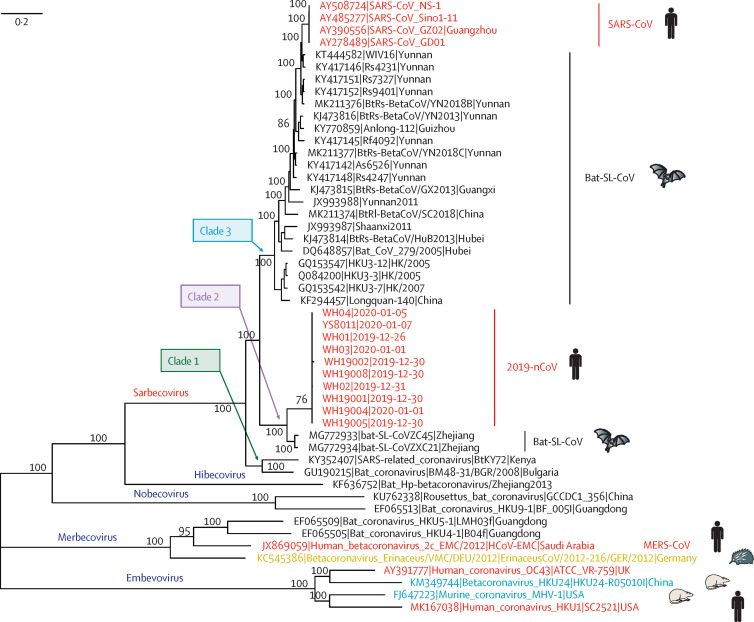
Phylogenetic analysis of 2019-nCoV and its closely related reference genomes, as well as representative betacoronaviruses, revealed that the five subgenera formed five well supported branches (figure 3 ). The subgenus Sarbecovirus could be classified into three well supported clades: two SARS-CoV-related strains from Rhinolophus sp from Bulgaria (accession number GU190215) and Kenya (KY352407) formed clade 1; the ten 2019-nCoV from Wuhan and the two bat-derived SARS-like strains from Zhoushan in eastern China (bat-SL-CoVZC45 and bat-SL-CoVZXC21) formed clade 2, which was notable for the long branch separating the human and bat viruses; and SARS-CoV strains from humans and many genetically similar SARS-like coronaviruses from bats collected from southwestern China formed clade 3, with bat-derived coronaviruses also falling in the basal positions (figure 3). In addition, 2019-nCoV was distinct from SARS-CoV in a phylogeny of the complete RNA-dependent RNA polymerase (RdRp) gene (appendix p 2). This evidence indicates that 2019-nCoV is a novel betacoronavirus from the subgenus Sarbecovirus.
Figure 3.
Phylogenetic analysis of full-length genomes of 2019-nCoV and representative viruses of the genus Betacoronavirus
2019-nCoV=2019 novel coronavirus. MERS-CoV=Middle East respiratory syndrome coronavirus. SARS-CoV=severe acute respiratory syndrome coronavirus.
As the sequence similarity plot revealed changes in genetic distances among viruses across the 2019-nCoV genome, we did additional phylogenetic analyses of the major encoding regions of representative members of the subgenus Sarbecovirus. Consistent with the genome phylogeny, 2019-nCoV, bat-SL-CoVZC45, and bat-SL-CoVZXC21 clustered together in trees of the 1a and spike genes (appendix p 3). By contrast, 2019-nCoV did not cluster with bat-SL-CoVZC45 and bat-SL-CoVZXC21 in the 1b tree, but instead formed a distinct clade with SARS-CoV, bat-SL-CoVZC45, and bat-SL-CoVZXC21 (appendix p 3), indicative of potential recombination events in 1b, although these probably occurred in the bat coronaviruses rather than 2019-nCoV. Phylogenetic analysis of the 2019-nCoV genome excluding 1b revealed similar evolutionary relationships as the full-length viral genome (appendix p 3).
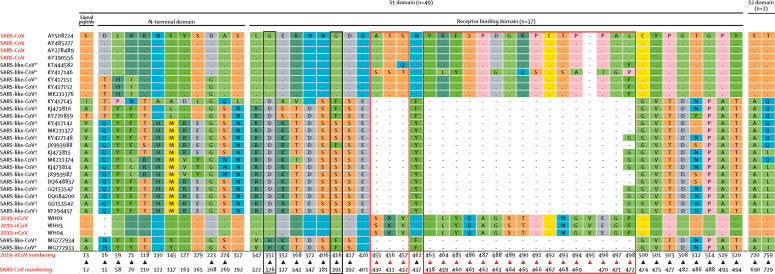
The envelope spike (S) protein mediates receptor binding and membrane fusion24 and is crucial for determining host tropism and transmission capacity.25, 26 Generally, the spike protein of coronaviruses is functionally divided into the S1 domain (especially positions 318–510 of SARS-CoV), responsible for receptor binding, and the S2 domain, responsible for cell membrane fusion.27 The 2019-nCoV S2 protein showed around 93% sequence identity with bat-SL-CoVZC45 and bat-SL-CoVZXC21—much higher than that of the S1 domain, which had only around 68% identity with these bat-derived viruses. Both the N-terminal domain and the C-terminal domain of the S1 domain can bind to host receptors.28 We inspected amino acid variation in the spike protein among the Sarbecovirus coronaviruses (figure 4 ). Although 2019-nCoV and SARS-CoV fell within different clades (figure 3), they still possessed around 50 conserved amino acids in S1, whereas most of the bat-derived viruses displayed mutational differences (figure 4). Most of these positions in the C-terminal domain (figure 4). In addition, a number of deletion events, including positions 455–457, 463–464, and 485–497, were found in the bat-derived strains (figure 4).
Figure 4.
Specific amino acid variations among the spike proteins of the subgenus sarbecovirus
Viruses are ordered by the tree topology (as shown in figure 3) from top to bottom. One-letter codes are used for amino acids. CoV=coronavirus. 2019-nCoV=2019 novel coronavirus. SARS=severe acute respiratory syndrome. *Bat-derived SARS-like viruses that can grow in human cell lines or in mice. †Bat-derived SARS-like viruses without experimental data available.
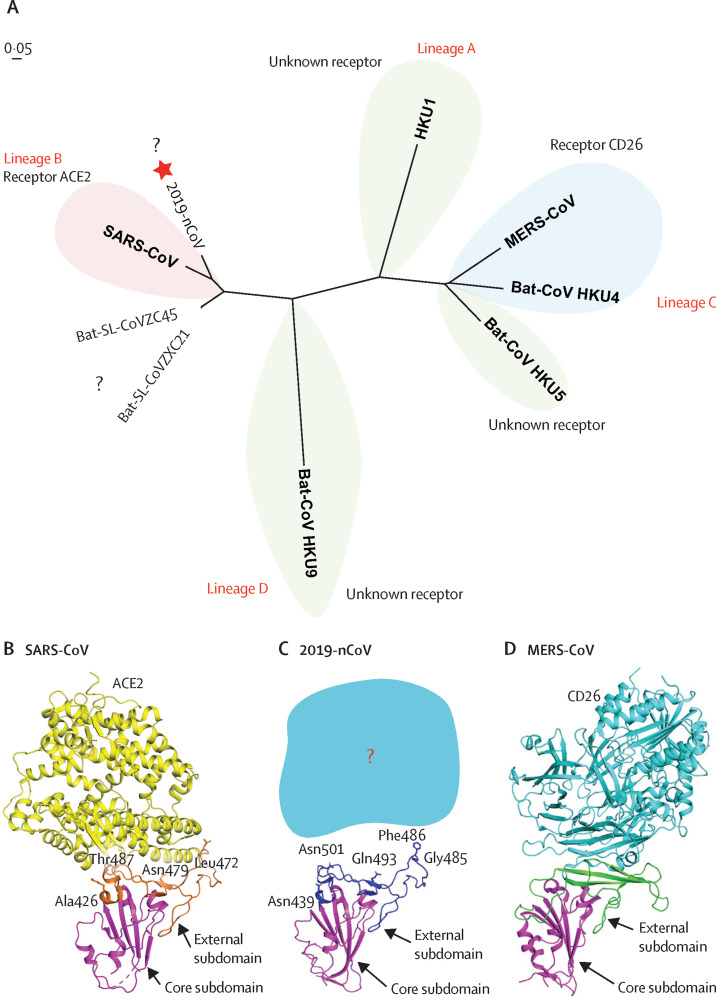
The receptor-binding domain of betacoronaviruses, which directly engages the receptor, is commonly located in the C-terminal domain of S1, as in SARS-CoV29 for lineage B, and MERS-CoV30, 31 and BatCoV HKU4,32 for lineage C (figure 5 ). Through phylogenetic analysis of the receptor-binding domain of four different lineages of betacoronaviruses (appendix p 4), we found that, although 2019-nCoV was closer to bat-SL-CoVZC45 and bat-SL-CoVZXC21 at the whole-genome level, the receptor-binding domain of 2019-nCoV fell within lineage B and was closer to that of SARS-CoV (figure 5A). The three-dimensional structure of 2019-nCoV receptor-binding domain was modelled using the Swiss-Model program33 with the SARS-CoV receptor-binding domain structure (Protein Data Bank ID 2DD8)34 as a template. This analysis suggested that, like other betacoronaviruses, the receptor-binding domain was composed of a core and an external subdomain (figure 5B–D). Notably, the external subdomain of the 2019-nCoV receptor-binding domain was more similar to that of SARS-CoV. This result suggests that 2019-nCoV might also use angiotensin-converting enzyme 2 (ACE2) as a cell receptor. However, we also observed that several key residues responsible for the binding of the SARS-CoV receptor-binding domain to the ACE2 receptor were variable in the 2019-nCoV receptor-binding domain (including Asn439, Asn501, Gln493, Gly485 and Phe486; 2019-nCoV numbering).
Figure 5.
Phylogenetic analysis and homology modelling of the receptor-binding domain of the 2019-nCoV, SARS-CoV, and MERS-CoV
(A) Phylogenetic analysis of the receptor-binding domain from various betacoronaviruses. The star highlights 2019-nCoV and the question marks means that the receptor used by the viruses remains unknown. Structural comparison of the receptor-binding domain of SARS-CoV (B), 2019-nCoV (C), and MERS-CoV (D) binding to their own receptors. Core subdomains are magenta, and the external subdomains of SARS-CoV, 2019-nCoV, and MERS CoV are orange, dark blue, and green, respectively. Variable residues between SARS-CoV and 2019-nCoV in the receptor-binding site are highlighted as sticks. CoV=coronavirus. 2019-nCoV=2019 novel coronavirus. SARS-CoV=severe acute respiratory syndrome coronavirus. MERS=Middle East respiratory syndrome coronavirus.
Discussion
From genomic surveillance of clinical samples from patients with viral pneumonia in Wuhan, China, a novel coronavirus (termed 2019-nCoV) has been identified.10, 11 Our phylogenetic analysis of 2019-nCoV, sequenced from nine patients' samples, showed that the virus belongs to the subgenus Sarbecovirus. 2019-nCoV was more similar to two bat-derived coronavirus strains, bat-SL-CoVZC45 and bat-SL-CoVZXC21, than to known human-infecting coronaviruses, including the virus that caused the SARS outbreak of 2003.
Epidemiologically, eight of the nine patients in our study had a history of exposure to the Huanan seafood market in Wuhan, suggesting that they might have been in close contact with the infection source at the market. However, one patient had never visited the market, although he had stayed in a hotel near the market before the onset of their illness. This finding suggests either possible droplet transmission or that the patient was infected by a currently unknown source. Evidence of clusters of infected family members and medical workers has now confirmed the presence of human-to-human transmission.12 Clearly, this infection is a major public health concern, particularly as this outbreak coincides with the peak of the Chinese Spring Festival travel rush, during which hundreds of millions of people will travel through China.
As a typical RNA virus, the average evolutionary rate for coronaviruses is roughly 10−4 nucleotide substitutions per site per year,1 with mutations arising during every replication cycle. It is, therefore, striking that the sequences of 2019-nCoV from different patients described here were almost identical, with greater than 99·9% sequence identity. This finding suggests that 2019-nCoV originated from one source within a very short period and was detected relatively rapidly. However, as the virus transmits to more individuals, constant surveillance of mutations arising is needed.
Phylogenetic analysis showed that bat-derived coronaviruses fell within all five subgenera of the genus Betacoronavirus. Moreover, bat-derived coronaviruses fell in basal positions in the subgenus Sarbecovirus, with 2019-nCoV most closely related to bat-SL-CoVZC45 and bat-SL-CoVZXC21, which were also sampled from bats.23 These data are consistent with a bat reservoir for coronaviruses in general and for 2019-nCoV in particular. However, despite the importance of bats, several facts suggest that another animal is acting as an intermediate host between bats and humans. First, the outbreak was first reported in late December, 2019, when most bat species in Wuhan are hibernating. Second, no bats were sold or found at the Huanan seafood market, whereas various non-aquatic animals (including mammals) were available for purchase. Third, the sequence identity between 2019-nCoV and its close relatives bat-SL-CoVZC45 and bat-SL-CoVZXC21 was less than 90%, which is reflected in the relatively long branch between them. Hence, bat-SL-CoVZC45 and bat-SL-CoVZXC21 are not direct ancestors of 2019-nCoV. Fourth, in both SARS-CoV and MERS-CoV, bats acted as the natural reservoir, with another animal (masked palm civet for SARS-CoV35 and dromedary camels for MERS-CoV)36 acting as an intermediate host, with humans as terminal hosts. Therefore, on the basis of current data, it seems likely that the 2019-nCoV causing the Wuhan outbreak might also be initially hosted by bats, and might have been transmitted to humans via currently unknown wild animal(s) sold at the Huanan seafood market.
Previous studies have uncovered several receptors that different coronaviruses bind to, such as ACE2 for SARS-CoV29 and CD26 for MERS-CoV.30 Our molecular modelling showed structural similarity between the receptor-binding domains of SARS-CoV and 2019-nCoV. Therefore, we suggest that 2019-nCoV might use ACE2 as the receptor, despite the presence of amino acid mutations in the 2019-nCoV receptor-binding domain. Although a previous study using HeLa cells expressing ACE2 proteins showed that 2019-nCoV could employ the ACE2 receptor,37 whether these mutations affect ACE2 binding or change receptor tropism requires further study.
Recombination has been seen frequently in coronaviruses.1 As expected, we detected recombination in the Sarbecoviruses analysed here. Our results suggest that recombination events are complex and are more likely occurring in bat coronaviruses than in 2019-nCoV. Hence, despite its occurrence, recombination is probably not the reason for emergence of this virus, although this inference might change if more closely related animal viruses are identified.
In conclusion, we have described the genomic structure of a seventh human coronavirus that can cause severe pneumonia and have shed light on its origin and receptor-binding properties. More generally, the disease outbreak linked to 2019-nCoV again highlights the hidden virus reservoir in wild animals and their potential to occasionally spill over into human populations.
Data sharing
Data are available on various websites and have been made publicly available (more information can be found in the first paragraph of the Results section).
Acknowledgments
Acknowledgments
This work was supported by the National Key Research and Development Programme of China (2016YFD0500301, 2020YFC0840800, 2020YFC0840900), the National Major Project for Control and Prevention of Infectious Disease in China (2017ZX10104001, 2018ZX10101002, 2018ZX10101004, and 2018ZX10732-401), the State Key Research Development Program of China (2019YFC1200501), the Strategic Priority Research Programme of the Chinese Academy of Sciences (XDB29010102), and the Academic Promotion Programme of Shandong First Medical University (2019QL006 and 2019PT008). WS was supported by the Taishan Scholars Programme of Shandong Province (ts201511056).
Contributors
GFG, WT, WS, WC, WX, and GW designed the study. RL, XZ, PN, HW, WW, BH, NZ, XM, WZ, LZ, JC, YM, JW, YL, JY, ZX, JM, WJL, and DW did the experiments. BY, FZ, and ZH provided samples. WS, WC, WT, JL, HS, YB, LW, TH, and HZ analysed data. WS, WT, and JL wrote the report. ECH and GFG revised the report.
Declaration of interests
We declare no competing interests.
Supplementary Material
References
- 1.Su S, Wong G, Shi W, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- 3.Ismail MM, Tang AY, Saif YM. Pathogenicity of turkey coronavirus in turkeys and chickens. Avian Dis. 2003;47:515–522. doi: 10.1637/5917. [DOI] [PubMed] [Google Scholar]
- 4.Zhou P, Fan H, Lan T, et al. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556:255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peiris JS, Guan Y, Yuen KY. Severe acute respiratory syndrome. Nat Med. 2004;10(suppl 12):S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan-Yeung M, Xu RH. SARS: epidemiology. Respirology. 2003;8(suppl):S9–S14. doi: 10.1046/j.1440-1843.2003.00518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 8.Lee J, Chowell G, Jung E. A dynamic compartmental model for the Middle East respiratory syndrome outbreak in the Republic of Korea: a retrospective analysis on control interventions and superspreading events. J Theor Biol. 2016;408:118–126. doi: 10.1016/j.jtbi.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JY, Kim YJ, Chung EH, et al. The clinical and virological features of the first imported case causing MERS-CoV outbreak in South Korea, 2015. BMC Infect Dis. 2017;17:498. doi: 10.1186/s12879-017-2576-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan W, Zhao X, Ma X, et al. A novel coronavirus genome identified in a cluster of pneumonia cases—Wuhan, China 2019–2020. China CDC Weekly. 2020;2:61–62. [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001017. published online Jan 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan JFW, Yuan S, Kok KH, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020 doi: 10.1016/S0140-6736(20)30154-9. published online Jan 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 doi: 10.1016/S0140-6736(20)30183-5. published online Jan 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niu P, Shen J, Zhu N, Lu R, Tan W. Two-tube multiplex real-time reverse transcription PCR to detect six human coronaviruses. Virol Sin. 2016;31:85–88. doi: 10.1007/s12250-015-3653-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao Y, Tang H, Ye Y. RAPSearch2: a fast and memory-efficient protein similarity search tool for next-generation sequencing data. Bioinformatics. 2012;28:125–126. doi: 10.1093/bioinformatics/btr595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nurk S, Bankevich A, Antipov D, et al. In: Research in computational molecular biology (RECOMB 2013): lecture notes in computer science. Deng M, Jiang R, Sun F, Zhang X, editors. vol 7821. Springer; Berlin: 2013. Assembling genomes and mini-metagenomes from highly chimeric reads; pp. 158–170. [Google Scholar]
- 18.Pan M, Gao R, Lv Q, et al. Human infection with a novel, highly pathogenic avian influenza A (H5N6) virus: virological and clinical findings. J Infect. 2016;72:52–59. doi: 10.1016/j.jinf.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 19.Marchler-Bauer A, Bo Y, Han L, et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017;45:D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lole KS, Bollinger RC, Paranjape RS, et al. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakamura T, Yamada KD, Tomii K, Katoh K. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics. 2018;34:2490–2492. doi: 10.1093/bioinformatics/bty121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu D, Zhu C, Ai L, et al. Genomic characterization and infectivity of a novel SARS-like coronavirus in Chinese bats. Emerg Microbes Infect. 2018;7:154. doi: 10.1038/s41426-018-0155-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu G, Wang Q, Gao GF. Bat-to-human: spike features determining ‘host jump’ of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015;23:468–478. doi: 10.1016/j.tim.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Q, Wong G, Lu G, Yan J, Gao GF. MERS-CoV spike protein: targets for vaccines and therapeutics. Antiviral Res. 2016;133:165–177. doi: 10.1016/j.antiviral.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He Y, Zhou Y, Liu S, et al. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine. Biochem Biophys Res Commun. 2004;324:773–781. doi: 10.1016/j.bbrc.2004.09.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li F. Evidence for a common evolutionary origin of coronavirus spike protein receptor-binding subunits. J Virol. 2012;86:2856–2858. doi: 10.1128/JVI.06882-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 30.Lu G, Hu Y, Wang Q, et al. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang N, Shi X, Jiang L, et al. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013;23:986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Q, Qi J, Yuan Y, et al. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe. 2014;16:328–337. doi: 10.1016/j.chom.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waterhouse A, Bertoni M, Bienert S, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prabakaran P, Gan J, Feng Y, et al. Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody. J Biol Chem. 2006;281:15829–15836. doi: 10.1074/jbc.M600697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guan Y, Zheng BJ, He YQ, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 36.Alagaili AN, Briese T, Mishra N, et al. Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. mBio. 2014;5:e00884–e00914. doi: 10.1128/mBio.00884-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou P, Yang X-L, Wang X-G, et al. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. bioRxiv. 2020 doi: 10.1101/2020.01.22.914952. published online Jan 23. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available on various websites and have been made publicly available (more information can be found in the first paragraph of the Results section).