Abstract
Xanthomonas campestris pv. musacearum (Xcm) is the causal agent of banana xanthomonas wilt, a major threat to banana production in eastern and central Africa. The pathogen is present in very high levels within infected plants and can be transmitted by a broad range of mechanisms; therefore early specific detection is vital for effective disease management. In this study, a polyclonal antibody (pAb) was developed and deployed in a lateral flow device (LFD) format to allow rapid in‐field detection of Xcm. Published Xcm PCR assays were also independently assessed: only two assays gave specific amplification of Xcm, whilst others cross‐reacted with non‐target Xanthomonas species. Pure cultures of Xcm were used to immunize a rabbit, the IgG antibodies purified from the serum and the resulting polyclonal antibodies tested using ELISA and LFD. Testing against a wide range of bacterial species showed the pAb detected all strains of Xcm, representing isolates from seven countries and the known genetic diversity of Xcm. The pAb also detected the closely related Xanthomonas axonopodis pv. vasculorum (Xav), primarily a sugarcane pathogen. Detection was successful in both naturally and experimentally infected banana plants, and the LFD limit of detection was 105 cells mL−1. Whilst the pAb is not fully specific for Xcm, Xav has never been found in banana. Therefore the LFD can be used as a first‐line screening tool to detect Xcm in the field. Testing by LFD requires no equipment, can be performed by non‐scientists and is cost‐effective. Therefore this LFD provides a vital tool to aid in the management and control of Xcm.
Keywords: diagnostics, ELISA, LFD, polyclonal antibody
Introduction
Xanthomonas campestris pv. musacearum (Xcm) is a Gram‐negative member of the gamma‐Proteobacteria that is the causal agent of banana xanthomonas wilt (BXW), considered to be one of the greatest threats to banana productivity in the Great Lakes region of eastern Africa (Tripathi et al., 2009). It was first found in Ethiopia around 40 years ago on enset (Yirgou & Bradbury, 1978) and is now widespread in east and central Africa. Since the early 2000s, the disease has been reported in Uganda, the Democratic Republic of the Congo, Rwanda, Tanzania, Kenya and Burundi (Adikini et al., 2011).
Xcm causes symptoms of foliar wilting and yellowing, leading to uneven fruit ripening and death of the plant. Symptom development is rapid, typically <1 month after inoculation, and leads to high levels of the pathogen within the plant typified by pockets of bacterial ooze when the plants are cut (Tripathi et al., 2009; Adikini et al., 2011). The economic impact of Xcm is high due to total loss of yield and rapid death of the mother plant, preventing the creation of more plants by division of suckers, a primary mode of cultivation (Tripathi et al., 2007). The disease affects almost all banana cultivars with only a small number able to avoid insect‐borne infection due to the differing morphological structure of the male buds (Adikini et al., 2013). Therefore recent approaches to control have included genetic engineering to create transgenic bananas with resistance to Xcm (Tripathi et al., 2010; Namukwaya et al., 2012) and the development of in vitro screening methods for rapid determination of cultivar resistance/susceptibility to Xcm infection (Tripathi et al., 2008).
Xcm can be transmitted in numerous ways including insect vectors, the use of contaminated farm equipment, transmission within the mat to suckers, soilborne infection and planting of infected symptomless plantlets (Tripathi et al., 2008; Adikini et al., 2013). A range of management strategies has been developed to limit Xcm spread. These have been demonstrated to be highly effective when employed properly, causing reduction of disease incidence to <10% per year in regions of Uganda. However, the approaches, including restricting movement, tool sterilization, removal of male buds and burying infected plants, are often poorly taken up by farmers due to the high financial and labour costs (Tushemereirwe et al., 2006).
The Xanthomonas genus is large, composed of many species and pathovars that cause disease in several hundred plant species. The genus is composed of two groups, with Xcm sitting within Group 2 along with many other important plant pathogens (Rodriguez‐R et al., 2012). The taxonomy of species and pathovars is very complex and still evolving, meaning confusion can exist over the identity of isolates within culture collections and that species identification can be problematic for diagnostic laboratories (Parkinson et al., 2009). Xcm was initially described as Xanthomonas musacearum and was reclassified Xcm in 1978 (Young et al., 1978). It has recently been proposed that Xcm should be reclassified as X. vasicola pv. musacearum (Xvm) due to its close relatedness to strains of X. vasicola. Indeed, partial gyrB sequences have shown 100% nucleotide sequence identity between Xcm and some X. vasicola isolates, and FAME and rep‐PCR methods group Xcm with X. vasicola (Aritua et al., 2008; Tripathi et al., 2009).
Recent whole‐genome comparisons of Xcm and X. vasicola pv. vasculorum (Xvv) are also corroborative, finding significantly greater nucleotide identity with each other than with any other sequenced xanthomonad. Importantly, all Xcm isolates form a clade closely related to but distinct from Xvv (Studholme et al., 2010; Wasukira et al., 2012). Studied Xcm isolates have demonstrated very little genetic diversity, with comparisons of whole genomes showing at least 99·99% nucleotide identity (Wasukira et al., 2012). Despite this, genome‐wide analyses looking for single nucleotide polymorphism (SNP) variation have proposed two major sub‐lineages within Xcm, each corresponding to a distinct geographic region. Whilst this is based on only 86 SNPs and a relatively small number of isolates, it splits sub‐lineage I containing isolates from Ethiopia, the Democratic Republic of the Congo and Rwanda from sub‐lineage II comprising isolates from Uganda, Tanzania, Burundi and Kenya (Wasukira et al., 2012). Numerous published data sets support the reclassification of Xcm as Xvm; however, further pathogenicity testing is required before this can be formally recognized as a pathovar designation (Aritua et al., 2008).
A primary driver for Xcm‐specific detection tools is to allow discrimination from numerous other diseases of banana that can cause similar wilt type symptoms, including moko, bugtok and blood disease caused by bacteria and the fungal disease fusarium wilt (Adikini et al., 2011). Typically, Xcm detection is based upon visual symptom expression and culture‐based morphological assessment. Recently, semiselective media for Xcm have been developed (Mwangi et al., 2007; Tripathi et al., 2007) but this approach has been found to be lacking as it is time‐consuming, requires confirmatory testing and often misses early stages of infection (Adikini et al., 2011). Biochemical tests can be used in some cases to resolve species and pathovars; however, approaches such as metabolomic profiling (e.g. Biolog) are time‐consuming, whilst fatty acid methyl ester (FAME) analysis is costly (Aritua et al., 2008; Adikini et al., 2011). As an alternative, molecular approaches have been investigated to allow the specific detection of Xcm including conventional PCR (Lewis Ivey et al., 2010; Adikini et al., 2011; Adriko et al., 2012) or rep‐PCR (Versalovic et al., 1994). Since the recent discovery of sub‐lineages within Xcm, Wasukira et al. (2012) have developed a PCR‐RFLP assay to allow discrimination of the lineages. Whilst these approaches can provide specific detection, they require well equipped laboratories, highly trained staff and are not readily amenable to field applications.
Lateral flow devices (LFDs) are an established technology for the rapid detection of plant pathogens, and are one of the primary techniques used for ‘point‐of‐care’ or ‘in‐field’ pathogen detection (Boonham et al., 2008). LFDs provide an identification in minutes, have no technological requirements and are simple to use, so they are an important identification tool despite recent molecular detection advances (Lane et al., 2007; De Boer & López, 2012). The requirement of such diagnostic approaches for Xcm is essential to facilitate elimination of residual sources of infection and improve disease management (Ocimati et al., 2013). This is critical for diseases such as BXW where, by the time symptoms are visible, the pathogen has spread systemically and therefore poses a high risk of further transmission (Adikini et al., 2013), and where latent infection can occur with long incubation periods (Ocimati et al., 2013). Therefore this study set out to develop an antibody‐based diagnostic kit for Xcm that could be deployed in a low‐resource setting, providing a cost‐effective diagnosis that can be performed by untrained personnel. A further advantage of this approach for diseases of this kind is that testing can occur simultaneously with control. Divorcing sampling from testing by sending samples to the laboratory can potentially lead to delays and loss of urgency on the part of growers. To enable this, polyclonal antibodies were generated to Xcm and formatted into a lateral flow device for in‐field detection.
Materials and methods
Bacterial cultures used and their maintenance
A range of Xcm isolates, closely related Xanthomonas species including Xvv, Xanthomonas axonopodis pv. vasculorum (Xav) and Xanthomonas vasicola pv. holcicola (Xvh), more distantly related Xanthomonas species and other bacterial species known to cause disease on banana were selected from the National Collection of Plant Pathogenic Bacteria (NCPPB), held at Fera, York, UK (Table 1). This included 32 isolates representing eight pathovars of Xanthomonas, and Enterobacter sp., Serratia marcescens, Pseudomonas marginalis pv. marginalis and Ralstonia solanacearum (all associated with diseases of banana or known endophytes of banana). Melissococcus plutonius was sourced from the National Bee Unit (Fera, York, UK).
Table 1.
Bacterial isolates used in the study, their plant host and country of origin and results generated by ELISA, LFD and PCR testing
| NCPPB number | Species | Country of origin | Host species | ELISAa | LFDb | PCRc | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GspDm | BXW | Xcm12 | Xcm35 | Xcm36 | Xcm38 | Xcm44 | Xcm47 | Xcm48 | ||||||
| 4389 | Xanthomonas campestris pv. musacearum | Rwanda | Musa sp. | + | + | + | + | + | + | + | + | + | + | + |
| 4390 | X. campestris pv. musacearum | Rwanda | Musa sp. | + | + | + | + | + | + | + | + | + | + | + |
| 4391 | X. campestris pv. musacearum | Rwanda | Musa sp. | + | + | + | + | + | + | + | + | + | + | + |
| 4378 | X. campestris pv. musacearum | Uganda | Musa sp. | + | + | + | + | + | + | + | + | + | + | + |
| 4379 | X. campestris pv. musacearum | Uganda | Musa sp. | + | + | + | + | + | + | + | + | + | + | + |
| 4380 | X. campestris pv. musacearum | Uganda | Musa sp. | + | + | + | + | + | + | + | + | + | + | + |
| 4381 | X. campestris pv. musacearum | Uganda | Musa sp. | + | + | + | + | + | + | + | + | + | + | + |
| 4383 | X. campestris pv. musacearum | Uganda | Musa sp. | + | + | + | + | + | + | + | + | + | + | + |
| 4434 | X. campestris pv. musacearum | Kenya | Musa sp. | + | + | + | + | + | + | + | + | + | + | + |
| 4433 | X. campestris pv. musacearum | Burundi | Musa sp. | + | + | + | + | + | + | + | + | + | + | + |
| 2005 | X. campestris pv. musacearum | Ethiopia | Ensete ventricosum | + | + | + | + | + | + | + | + | + | + | + |
| 2251 | X. campestris pv. musacearum | Ethiopia | Musa sp. | + | + | + | + | + | + | + | + | + | + | + |
| 4387 | X. campestris pv. musacearum | D.R.Congo | Musa sp. | + | + | + | + | + | + | + | + | + | + | + |
| 4388 | X. campestris pv. musacearum | D.R.Congo | Musa sp. | + | + | + | + | + | + | + | + | + | + | + |
| 4392 | X. campestris pv. musacearum | Tanzania | Musa sp. | + | + | + | + | + | + | + | + | + | + | + |
| 4393 | X. campestris pv. musacearum | Tanzania | Musa sp. | + | + | + | + | + | + | + | + | + | + | + |
| 4394 | X. campestris pv. musacearum | Tanzania | Musa sp. | + | + | + | + | + | + | + | + | + | + | + |
| 206 | Xanthomonas vasicola pv. vasculorum | South Africa | Zea mays | − | − | − | + | + | − | + | + | + | + | + |
| 702 | X. vasicola pv. vasculorum | Zimbabwe | Saccharum officinarum | − | − | − | + | + | − | + | + | + | + | + |
| 890 | X. vasicola pv. vasculorum | South Africa | S. officinarum | − | − | − | + | + | − | + | + | + | + | + |
| 989 | X. vasicola pv. holicola | USA | Holcus sp. | − | − | − | + | + | − | + | + | + | + | + |
| 1060 | X. vasicola pv. holicola | Ethiopia | Sorghum vulgare | − | − | − | + | + | − | + | + | + | + | + |
| 2417 | X. vasicola pv. holicola | New Zealand | S. vulgare | − | − | − | + | + | − | + | + | + | + | + |
| 186 | Xanthomonas axonopodis pv. vasculorum | Mauritius | Thysanolaena maxima | + | + | − | − | − | − | − | + | + | − | − |
| 796 | X. axonopodis pv. vasculorum | Mauritius | S. officinarum | + | + | − | − | − | − | − | Weak + | + | − | − |
| 899 | X. axonopodis pv. vasculorum | Reunion | S. officinarum | + | + | − | − | − | − | − | NS | + | − | − |
| 1630 | Xanthomonas arboricola pv. celebensis | New Zealand | Musa sp. | − | − | − | − | − | − | − | NS | − | − | − |
| 529 | X. campestris pv. campestris | UK | Brassica oleracea var. capitata | − | − | − | − | − | − | − | Weak + | − | − | − |
| 2985 | X. campestris pv. perlagonii | New Zealand | Pelargonium peltatum | − | − | − | − | Weak + | − | − | Weak + | Weak + | − | Weak + |
| 4031 | X. campestris pv. perlogonii | UK | Pelargonium × hortorum | − | − | − | − | − | − | − | NS | − | − | − |
| 422 | X. campestris pv. vesicatoria | New Zealand | Lycopersicon esculentum | − | − | − | − | − | − | − | Weak + | − | − | − |
| 701 | X. campestris pv. vesicatoria | Zimbabwe | L. esculentum | − | − | − | − | − | − | − | Weak + | − | − | − |
| 1131 | Xanthomonas sp. | Eastern Samoa | Musa paradisiaca | − | − | − | − | − | − | − | Weak + | − | − | − |
| 1132 | Xanthomonas sp. | Western Samoa | Musa canksii var. samoensis | − | − | − | − | − | − | − | Weak + | − | − | − |
| 2198 | Ralstonia solanacearum | Trinidad | Musa sp. | − | − | − | − | − | − | − | NS | − | − | − |
| 2315 | R. solanacearum | Peru | Musa sp. | − | − | − | − | − | − | − | − | − | − | − |
| 3205 | R. solanacearum | Guyana | Musa sp. (plantain) | − | − | − | − | − | − | − | − | − | − | − |
| 3214 | R. solanacearum | Colombia | Plantago sp. | − | − | − | − | − | − | − | − | − | − | − |
| 4168 | Enterobacter sp. | Unknown | Unknown | − | − | − | − | − | − | − | NS | − | − | − |
| 2641 | Serratia marcescens biotype A4a | USA | Medicago sativa | − | − | − | − | − | − | − | − | − | − | − |
| 1232 | Pseudomonas marginalis pv. marginalis | Uganda | Musa sp. | − | − | − | − | − | − | − | − | − | − | − |
| n/a | Melissococcus plutonius | UK | n/a | − | − | − | − | − | − | − | − | − | − | − |
| n/a | Healthy Musa cv. Tropicana | n/a | Musa sp. | − | − | − | − | − | − | − | NS | − | − | − |
| n/a | Xcm‐infected Musa (field sample) | Uganda | Musa sp. | + | + | + | + | + | + | + | + | + | + | + |
ELISA results using purified pAb at 1:32 000 dilution, average of triplicate wells; +, positive with OD > 1·7; −, negative with OD < 0·3.
LFD results: +, test and control line positive; −, only control line positive.
PCR results: +, positive; −, negative; Weak +, weak positive; NS, non‐specific amplification.
All Xanthomonas strains were grown on yeast dextrose chalk (YDC) agar incubated at 28°C for 48 h. Ralstonia solanacearum was grown on SMSA agar, supplemented immediately prior to use with 0·1 g L−1 polymixin B sulphate, 0·1 g L−1 bacitracin, 0·05 g L−1 2,3,5‐triphenyltetrazolium chloride, 0·005 g L−1 crystal violet, 0·005 g L−1 chloramphenicol and 0·0005 g L−1 penicillin G, and incubated at 25°C for 48 h. Melisococcus plutonius was grown anaerobically on M110 media (2·5 g L−1 peptone, 10 g L−1 glucose, 2 g L−1 soluble starch, 2·5 g L−1 yeast extract, 5 g L−1 neopeptone, 2 g L−1 trypticase, 4·33 g L−1 K2HPO4 and 3·42 g L−1 KH2PO4, 15 g L−1 agar technical no. 1, pH 7·2, supplemented immediately prior to use with 1 g L−1 cysteine.HCl) incubated at 37°C for approximately 2 weeks. Pseudomonas marginalis pv. marginalis, Enterobacter sp. and S. marcescens were grown on King's B media incubated at 25°C for 48 h, 37°C for 24 h and 37°C for 48 h, respectively. ACDP (Advisory Committee on Dangerous Pathogens) hazard group 2 species (Enterobacter sp. and S. marcescens) were formalin‐fixed prior to use by the addition of 2·5 μL mL−1 37% formalin.
Inoculation of banana plants and field infected samples
Young (20–25 cm in height) Musa cv. Tropicana and cv. Cavendish Dwarf plants were artificially inoculated with Xcm. Xcm was grown on YDC agar as above, colonies resuspended in sterile water and the concentration adjusted to 1 × 109 cells mL−1 by spectrophotometry. Inoculation was performed by injection of 200 μL inoculum into the pseudostem of the plant approximately 2·5 cm above soil level. Control plants were either uninoculated or inoculated with sterile water. Post‐inoculation the plants were grown in a glasshouse maintained at 28°C day, 20°C night with 16 h supplemented lighting. After inoculation, 100 μL of the inoculum was spread onto the YDC agar and incubated as above to confirm viability. Symptom development was monitored and leaf material harvested and stored at −20°C for later use. Sections of pseudostem and leaves from 16 field‐grown Musa plants showing typical symptoms of Xcm infection were collected in Uganda and shipped to the UK prior to storage at 4°C.
Production of polyclonal antibody and LFD development
An 8‐month‐old female New Zealand cross rabbit was used for the production of polyclonal antibodies, and prior to immunization a blood sample was collected. The immunogen was composed of a mix of six Xcm strains all collected from Musa sp. (NCPPB4378 from Uganda, NCPPB4387 from the Democratic Republic of the Congo, NCPPB4389 from Rwanda, NCPPB4392 from Tanzania, NCPPB4433 from Burundi and NCPPB4434 from Kenya) encompassing the geographic variability of Xcm. To prepare the immunogen, each isolate was first grown on YDC agar before being transferred to nutrient broth (Oxoid) and grown at 28°C at 200 rpm for 24 h. Cells were pelleted by centrifugation, resuspended in phosphate‐buffered saline, concentrations adjusted by spectrophotometry and equally pooled to form the immunogen. The immunogen was mixed equally with Freund's complete adjuvant to give a final concentration of 1·25 × 108 cells mL−1 and the rabbit immunized subcutaneously with 1 mL. Three further immunizations of the immunogen mixed equally with incomplete Freund's adjuvant to a final concentration of 1·25 × 108 cells mL−1 were administered subcutaneously at 4‐week intervals. Four weeks after the final immunization a blood sample was taken and the serum analysed for immune response by ELISA (see below). Further blood samples were taken at 4‐week intervals, the serum separated from whole blood by centrifugation and stored at –30°C prior to use. The IgG antibodies were purified from post‐immunization serum by HiTrap Protein G column (GE Healthcare) following the manufacturer's protocol. The protein concentration of the purified IgG antibody was determined by extinction coefficient calculation at 280 nm. Purified IgG antibodies were provided to Forsite Diagnostics Ltd (York, UK) for the production of a lateral flow device (Danks & Barker, 2000).
ELISA testing
PTA‐ELISA was performed as follows: 96‐well ELISA plates were coated with 100 μL of bacteria (Table 1) at a concentration of 107 cells mL−1 in coating buffer (0·015 m Na2CO3, 0·046 m NaHCO3, pH 9·6), or 100 μL of plant material extract (2 g plant material in 20 mL coating buffer, ground using a hand homogenizer (BIOREBA)) and incubated overnight at 2–8°C. Plates were washed three times with phosphate‐buffered saline + Tween‐20 (PBS‐T; 1·4 m NaCl, 0·015 m KH2PO4, 0·08 m Na2HPO4, 0·03 m KCl, pH 7·4, 0·05% Tween‐20) and blocked with 200 μL PBS‐T containing 1% bovine serum albumin (BSA) for 1 h at 33°C. Plates were washed as above and 100 μL polyclonal antibody applied, either pre‐immunization serum, post‐immunization serum or purified antibody or serum from a rabbit immunized with Cucumber vein yellowing virus (as a negative control) at various dilutions in PBS‐T containing 0·2% BSA and incubated for 1 h at 33°C. Plates were washed as above and 100 μL goat anti‐rabbit alkaline phosphatase was applied, diluted 1:4000 in PBS‐T containing 0·2% BSA, and incubated for 1 h at 33°C. Plates were washed as above and 100 μL of 1 mg mL−1 ρ‐nitrophenyl phosphate in substrate buffer (10% diethanolamine, 0·002 m MgCl2, pH 9·8) was added. The plates were kept in the dark at room temperature for 30 min and the absorbance measured at 405 nm on a plate reader (Thermomax Microplate reader) using SOFT max pro software.
To determine the sensitivity of the ELISA, bacterial cells were serially diluted from 108 cells mL−1 through eight log‐dilutions and 100 μL applied per well. Wells of M. plutonius bacterial culture and M. plutonius monoclonal antibody were included in each experiment as a procedural positive control. Healthy Musa cultivars Tropicana and Cavendish Dwarf, Musa cv. Tropicana experimentally inoculated with Xcm NCPPB4434 and 16 field plants with symptoms (presumed Xcm‐infected) were tested by ELISA, with sections of either leaf or pseudostem prepared as described above.
LFD testing
When testing LFDs, samples were prepared in buffer A provided by Forsite Diagnostics Ltd and in all cases 100 μL was applied to each LFD. Each LFD was allowed to run for 10 min before assessing the results. A positive result was indicated by both a test (T) line and control (C) line, a negative result but successfully run LFD was indicated by a C line alone, and a failed LFD run indicated by no lines. Controls of buffer A and healthy Musa were included in all experiments.
Bacterial cultures of target and non‐target species (Table 1) were diluted to 108 cells mL−1 in buffer A. To test plant material, 5 cm sections of leaf or pseudostem were cut into approximately 1 cm sections and placed in an extraction bottle (Forsite Diagnostics Ltd). This was either allowed to stand for 5 min to allow the sap/bacteria to ooze into the buffer, or homogenized by shaking manually for 1 min. To determine the sensitivity of the LFDs, Xcm NCPPB4378 was serially diluted from 108 cells mL−1 through six log‐dilutions in buffer A. To test sensitivity of the LFD in a plant exudate background, cultured bacteria were diluted in healthy Musa leaf sap exuded into buffer A. Healthy Musa cultivars Tropicana and Cavendish Dwarf, healthy sugarcane, Musa cv. Tropicana experimentally inoculated with Xcm NCPPB4434 and the 16 field plants with symptoms (presumed Xcm‐infected) were tested by LFD, with sections of either leaf or pseudostem prepared as described above.
Evaluation of PCR assays
PCR was performed using published protocols designed for the detection of Xcm (Table 2). In all cases, 2× ReddyMix PCR Master Mix with 1·5 mm MgCl2 (Thermo Scientific) was used with the cycling conditions as originally described. PCR was performed in a GeneAmp 9700 thermocycler using 1 μL DNA as template (concentration as extracted). Assays used were: the BXW assay of Lewis Ivey et al. (2010) using 500 nm each of primers BXW‐1 and BXW‐3; the GspD assay of Adriko et al. (2012) using 400 nm each of primers Gsp DM‐F2 and Gsp DM‐R3; and assays Xcm12, Xcm35, Xcm36, Xcm38, Xcm44, Xcm47 and Xcm48 of Adikini et al. (2011) using 400 nm of each forward and reverse primer. All primers were synthesized by Eurofins‐MWG‐Operon. PCR products (10 μL) were separated by gel electrophoresis in 1·5% agarose gel in 1× Tris‐borate‐EDTA buffer, stained with ethidium bromide and visualized using a UV transilluminator.
Table 2.
Details of the PCR assays evaluated in the study
| Primer | Sequence (5′–3′) | Target | Amplicon size (bp) | References |
|---|---|---|---|---|
| BXW‐1 | GTCGTTGGCACCATGCTCA | hrpB operon | 214 | Lewis Ivey et al. (2010) |
| BXW‐3 | TCCGACCGATACGGCT | |||
| GspDm‐F2 | GCGGTTACAACACCGTTCAAT | GspD gene | 265 | Adriko et al. (2012) |
| GspDm‐R3 | AGGTGGAGTTGATCGGAATG | |||
| Xcm 12‐F | GCCGGCGTGCGCAACTATCTG | ATP‐binding hypothetical protein | 360 | Adikini et al. (2011) |
| Xcm 12‐R | GCCATCCGCAAACAATCGCAACCT | |||
| Xcm35‐F | GAGCGCGAGGAAACGGGGAAGT | Non‐coding region | 480 | Adikini et al. (2011) |
| Xcm35‐R | TTGTGTTCGCCCAACCCTCTCAGT | |||
| Xcm36‐F | GCTTCGGCGGAGGCGTGCTAAT | Hypothetical protein | 420 | Adikini et al. (2011) |
| Xcm36‐R | TCGGCCGGGCGAGAACTTGAA | |||
| Xcm38‐F | CCGCCGGTCGCAATGTGGGTAAT | Polymorphic membrane protein | 650 | Adikini et al. (2011) |
| Xcm38‐R | CAGCGGCGCCGGTGTATTGAGTG | |||
| Xcm44‐F | AATAGCCCGGGTGATTGTCC | Hypothetical protein | 350 | Adikini et al. (2011) |
| Xcm44‐R | AGCCGGCAGCTACGATGAG | |||
| Xcm47‐F | GCTGCGTAATGGGCGAGATGATGC | Putative membrane protein | 370 | Adikini et al. (2011) |
| Xcm47‐R | GCTGCCGCCGGTTTGGTTTGT | |||
| Xcm48‐F | CCCGCGATCACTTCCAACAAACAC | Non‐coding region | 450 | Adikini et al. (2011) |
| Xcm48‐R | GCTCAATCGCCGGAGGGAGAATC |
Results
Specificity of the ELISA and LFD
To assess the specificity of the pAb, ELISA was conducted testing Xcm, a wide range of Xanthomonas species, and other bacterial species known to cause disease in banana. All 17 Xcm strains, representing isolates from seven countries (Burundi, the Democratic Republic of the Congo, Ethiopia, Kenya, Rwanda, Tanzania and Uganda) encompassing both sub‐lineages I and II, and therefore representing the known genetic diversity of Xcm, could be readily detected. However, the pAb was also found to detect all strains of Xav tested. All the other Xanthomonas species and pathovars tested were negative, as were other bacterial species (Table 1). Control samples of sap, leaf and pseudostem from healthy Musa cv. Tropicana and Cavendish Dwarf were tested by ELISA and found to give negative results in all cases.
The specificity of the LFD was found to be identical to that of the ELISA, detecting all Xcm strains tested along with Xav strains, but not any other species tested (Table 1). Healthy Musa cv. Tropicana and Cavendish Dwarf sap, leaf and pseudostem and healthy sugarcane leaf were tested by LFD and gave negative results in all cases. The LFD gave unambiguous, easy to interpret results with clear differentiation between positive and negative samples, in both pure culture and infected plant material (Fig. 1).
Figure 1.

Detection by the banana xanthomonas wilt lateral flow device. From left to right; Pure bacterial culture at 108 cells mL−1 of Xanthomonas campestris pv. musacearum (Xcm) NCPPB4378, Xanthomonas axonopodis pv. vasculorum NCPPB796, Xanthomonas campestris pv. campestris NCPPB529, plant material of healthy Musa cv. Tropicana, healthy Saccharum and Musa cv. Tropicana experimentally infected with Xcm NCPPB4434.
Sensitivity of the ELISA and LFD
The sensitivity of the ELISA with the purified pAb was determined using eight Xcm isolates. When testing bacteria from pure culture, the limit of detection ranged from 106 to 102 cells mL−1. However, when testing in a sap exudate background, to mimic actual samples, the limit of detection was reduced to 108 cells mL−1 for seven of the isolates. One isolate (NCPPB4434), which was only detectable at 106 cells mL−1 in pure culture, could not be detected in sap exudate.
The sensitivity of the LFD was first determined using pure culture of Xcm NCPPB4378 in buffer where the limit of detection was 105 cells mL−1. To determine the effect upon the limit of detection in a plant background, Xcm NCPPB4433 was serially diluted in leaf exudate and whole‐leaf extract where the limit of detection was decreased to 106 cells mL−1. Six further Xcm isolates were tested in a sap exudate background and the limit of detection ranged from 106 to 103 cells mL−1, most typically being 105 cells mL−1. Lower dilutions yielded fainter but still clearly visible test lines (Fig. 2) that became easier to view upon drying of the membrane. To determine if a hook effect (Amarasiri Fernando & Wilson, 1992) may have a deleterious effect on detection by the LFD at high bacterial titres, Xcm NCPPB4433 was tested at 1010 and 109 cells mL−1 in buffer A and buffer A with sap exudate, all of which could be detected.
Figure 2.
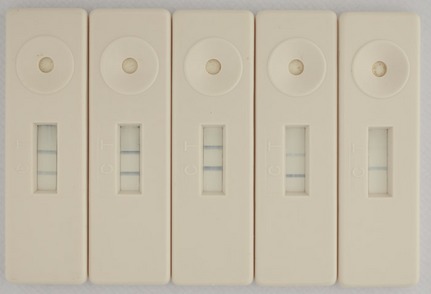
Sensitivity of the lateral flow device, tested with Xanthomonas campestris pv. musacearum NCPPB2251. Bacterial culture serially diluted in banana sap/buffer A from 108 cells mL−1 (left) to 104 cells mL−1 (right).
Testing of naturally and experimentally infected plant samples by ELISA and LFD
Leaf samples showing moderate wilt symptoms from Musa experimentally infected with Xcm NCPPB4434 could be detected by both ELISA and the LFD. Sixteen of the Musa samples from Uganda with symptoms, suspected of Xcm infection, were tested. Despite the fact that these samples were noticeably degraded to the extent that they were starting to rot following shipping to the UK, when tested by ELISA, nine samples tested positive and two samples tested negative. Five of the samples gave positive results in ELISA control wells lacking primary antibody. It is speculated that this may be due to the degraded nature of the samples, with a component of the sample causing non‐specific binding of the secondary antibody. However, when tested by LFD all 16 samples were positive.
PCR evaluation
All previously published PCR assays were tested against a wide range of bacteria, to independently evaluate their performance characteristics with regard to specificity. The evaluation included the Xcm‐specific assays of Adikini et al. (2011) and Adriko et al. (2012) and the Xcm, Xav and Xvh assay of Lewis Ivey et al. (2010).
All primer sets were able to detect all strains of Xcm tested, representing isolates from seven different countries (Burundi, the Democratic Republic of the Congo, Ethiopia, Kenya, Rwanda, Tanzania and Uganda) and both sub‐lineages I and II of Xcm (Table 1). The assays could also detect successfully either pure bacterial culture or naturally infected plant material.
The GspD assay of Adriko et al. (2012) was found to provide specific amplification of only Xcm and no other species. In the present study, the assay Xcm35 of Adikini et al. (2011) was found to be specific and only detected Xcm. However, the remaining six assays cross‐reacted with other species, with all detecting a minimum of Xvv and Xvh. The least specific assay, Xcm38, gave positive results with all Xanthomonas species tested as well as Ralstonia solanacearum and Enterobacter sp.
The Lewis Ivey et al. (2010) assay detected all strains of Xcm and Xvh; however, it did not amplify any strains of Xav, but did amplify all strains of Xvv tested. A possibility for this anomaly is the complexity arising from the challenges with Xanthomonas species and pathovar taxonomy, and the proposed but not formally accepted renaming of Xav as Xvv. In this study, the species names are as listed in NCPPB; however, some isolates were also studied by Lewis Ivey et al. (2010) and Adriko et al. (2012) and have contrasting species designations. For example, NCPPB702 is described as Xvv in this study but Xav by Lewis Ivey et al. (2010). Interestingly, Lewis Ivey found that only some Xav strains were PCR positive, for example finding Xav NCPPB702 to be PCR positive and Xav NCPPB796 to be PCR negative. These two isolates were included in the present study, where NCPPB702 was classified as Xvv and was PCR positive, and NCPPB796 was classified as Xav and was PCR negative. Therefore it seems highly likely this apparent difference in PCR assay specificity is due to the species names assigned to Xav and Xvv isolates. Using the NCPPB species names the assay specificity was shown to be specific for all Xvv isolates (PCR positive) and all Xav isolates were PCR negative and not detected.
Unclassified Xanthomonas isolates
The unidentified isolates NCPPB1131 and NCPPB1132, both Xanthomonas species isolated from Eastern and Western Samoa on Musa species, were negative with all methods, with the exception of the very non‐specific PCR assay Xcm38. Interestingly, based upon whole genome sequencing, Studholme et al. (2011) concluded that NCPPB4393 was in fact not an Xcm isolate as initially described but was X. sacchari. However, in this study NCPPB4393 has tested positive with all of the Xcm‐specific detection methods (ELISA, LFD and PCR assays) indicating that it is indeed an Xcm isolate.
Discussion
BXW is a devastating disease; however, it has been demonstrated that effective management can cause substantial decreases in disease prevalence. Crucial to a successful management strategy is the ability to specifically detect the pathogen and with sufficient sensitivity to identify latent infections. The antibody developed in this study has been formatted for use in both ELISA and LFD, therefore providing an alternate laboratory test if used in ELISA, and allowing in‐field testing with the LFD. The substantial reduction of the ELISA limit of detection when testing in a plant background compared to pure bacterial culture is probably due to the much larger number of proteins competing for binding sites and therefore reducing the sensitivity, an effect considerably reduced when testing by LFD. Five samples were found to fail under ELISA testing due to degradation of samples during shipping, and two samples were negative, presumably due to levels of infection below the limit of detection, whereas all of these were positive when tested by LFD. This is particularly relevant when testing samples from remote locations, when time to send samples to the laboratory is prolonged, and demonstrates the practical advantages of the LFD where detection is possible after other techniques have failed.
The pAb was found to be specific to Xcm, detecting all strains representing the geographic spread of Xcm; however, it also cross‐reacted with Xav. Whilst it would be preferable for the pAb to be fully specific to Xcm, Xav, the causal agent of a gumming disease of sugarcane, is found in grass species and maize. There are currently no instances of Xav being found in banana. Moreover, it has been demonstrated that Xav cannot cause disease in banana via artificial inoculation. Therefore the antibody generated in this study represents an important tool, with sufficient specificity that when testing banana will robustly and sensitively detect Xcm. Both the ELISA and LFD have been demonstrated to detect Xcm in infected plant material and can accordingly be used as an initial screening tool to rule out wilt caused by other bacterial or fungal diseases with similar symptoms. The LFD has a broad detection range and has been demonstrated to readily detect infected plant material.
A number of molecular detection tools have been developed to allow laboratory‐based detection of Xcm. Evaluation of these has led to conflicting reports regarding assay performance appearing within the literature, with Adriko et al. (2012) reporting that the Adikini et al. (2011) assays are not specific for Xcm, detecting between two and nine non‐banana Xanthomonas strains. In the present independent assessment, only two primer pairs (GspD of Adriko et al. (2012) and Xcm35 of Adikini et al. (2011)) gave specific amplification of Xcm, with other published PCR assays cross‐reacting with other Xanthomonas species. In particular, the Xcm assay panel of Adikini et al. (2011) was found to show significant cross‐reactivity, with six of the seven assays found to be non‐specific.
The Lewis Ivey et al. (2010) assay was designed to amplify all Xcm strains and was published as also detecting some strains of Xav and Xvh, a finding confirmed by Adriko et al. (2012). The assay performance in the authors' hands is in contrast to this; however, this appears to be due to the non‐formalized renaming of Xav as Xvv. Therefore the assay was found to specifically detect Xcm, Xvh and Xvv. Two unidentified Xanthomonas species isolates (NCPPB1131 and NCPPB1132) were found to be negative with all Xcm‐specific detection techniques; therefore this supports the finding of Studholme et al. (2011) that they fall within the X. sacchari clade.
Whilst this study shows that some of the published PCR assays can provide specific detection of Xcm, these techniques are still contingent upon a well‐equipped laboratory with highly trained staff, and unlike LFDs, results are not instantaneous, with samples requiring transportation to the laboratory followed by testing which may take between many hours to days. Therefore the development of simple, rapid detection strategies which can be performed in the field, such as LFDs, are invaluable. LFDs are suited to this setting, being cost‐effective, portable, easy to use and providing almost instantaneous results. Crucially, they can then allow prompt action when infected plants are found, vital in the management of diseases such as Xcm that use multiple modes of transmission, or alternatively used to test planting material at borders to prevent introduction of Xcm into countries or regions where it is not yet present. Based on the results of this study, the LFD can be deployed for in‐field detection of Xcm. If further confirmatory testing is required, two Xcm‐specific PCR assays (GspD of Adriko et al. (2012) and Xcm35 of Adikini et al. (2011)) have been identified from the literature, which can subsequently be used in a laboratory setting. Large‐scale field testing of the LFD is currently underway, and it is hoped that this new detection approach, the first published primary literature report of an LFD for Xcm, will become a vital asset to the detection and management of BXW.
Acknowledgements
This work was funded by Bioversity International who receiving funding from the McKnight Foundation, with additional support from ASARECA Funds provided to IITA.
References
- Adikini S, Tripathi L, Beed F, Tusiime G, Magembe EM, Kim DJ, 2011. Development of a specific molecular tool for detecting Xanthomonas campestris pv. musacearum . Plant Pathology 60, 443–52. [Google Scholar]
- Adikini S, Beed F, Tusiime G et al, 2013. Spread of Xanthomonas campestris pv. musacearum in banana plants: implications for management of banana Xanthomonas wilt disease. Canadian Journal of Plant Pathology 35, 458–68. [Google Scholar]
- Adriko J, Aritua V, Mortensen CN, Tushemereirwe WK, Kubiriba J, Lund OS, 2012. Multiplex PCR for specific and robust detection of Xanthomonas campestris pv. musacearum in pure culture and infected plant material. Plant Pathology 61, 489–97. [Google Scholar]
- Amarasiri Fernando S, Wilson GS, 1992. Studies of the ‘hook’ effect in the one‐step sandwich immunoassay. Journal of Immunological Methods 151, 47–66. [DOI] [PubMed] [Google Scholar]
- Aritua V, Parkinson N, Thwaites R et al, 2008. Characterization of the Xanthomonas sp. causing wilt of enset and banana and its proposed reclassification as a strain of X. vasicola . Plant Pathology 57, 170–7. [Google Scholar]
- Boonham N, Glover R, Tomlinson J, Mumford R, 2008. Exploiting generic platform technologies for the detection and identification of plant pathogens. European Journal of Plant Pathology 121, 355–63. [Google Scholar]
- Danks C, Barker I, 2000. On‐site detection of plant pathogens using lateral‐flow devices. EPPO Bulletin 30, 421–6. [Google Scholar]
- De Boer SH, López MM, 2012. New grower‐friendly methods for plant pathogen monitoring. Annual Review of Phytopathology 50, 197–218. [DOI] [PubMed] [Google Scholar]
- Lane CR, Hobden E, Walker L et al, 2007. Evaluation of a rapid diagnostic field test kit for identification of Phytophthora species, including P. ramorum and P. kernoviae at the point of inspection. Plant Pathology 56, 828–35. [Google Scholar]
- Lewis Ivey ML, Tusiime G, Miller SA, 2010. A polymerase chain reaction assay for the detection of Xanthomonas campestris pv. musacearum in banana. Plant Disease 94, 109–14. [DOI] [PubMed] [Google Scholar]
- Mwangi M, Mwebaze M, Bandyopadhyay R et al, 2007. Development of a semiselective medium for isolating Xanthomonas campestris pv. musacearum from insect vectors, infected plant material and soil. Plant Pathology 56, 383–90. [Google Scholar]
- Namukwaya B, Tripathi L, Tripathi JN, Arinaitwe G, Mukasa SB, Tushemereirwe WK, 2012. Transgenic banana expressing Pflp gene confers enhanced resistance to Xanthomonas wilt disease. Transgenic Research 21, 855–65. [DOI] [PubMed] [Google Scholar]
- Ocimati W, Ssekiwoko F, Karamura E, Tinzaara W, Eden‐Green S, Blomme G, 2013. Systemicity of Xanthomonas campestris pv. musacearum and time to disease expression after inflorescence infection in East African highland and Pisang Awak bananas in Uganda. Plant Pathology 62, 777–85. [Google Scholar]
- Parkinson N, Cowie C, Heeney J, Stead D, 2009. Phylogenetic structure of Xanthomonas determined by comparison of gyrB sequences. International Journal of Systematic and Evolutionary Microbiology 59, 264–74. [DOI] [PubMed] [Google Scholar]
- Rodriguez‐R LM, Grajales A, Arrieta‐Ortiz ML, Salazar C, Restrepo S, Bernal A, 2012. Genomes‐based phylogeny of the genus Xanthomonas . BMC Microbiology 12, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studholme DJ, Kemen E, MacLean D et al, 2010. Genome‐wide sequencing data reveals virulence factors implicated in banana Xanthomonas wilt. FEMS Microbiology Letters 310, 182–92. [DOI] [PubMed] [Google Scholar]
- Studholme DJ, Wasukira A, Paszkiewicz K et al, 2011. Draft genome sequences of Xanthomonas sacchari and two banana‐associated Xanthomonads reveal insights into the Xanthomonas Group 1 Clade. Genes 2, 1050–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi L, Tripathi JN, Tushemereirwe WK, Bandyopadhyay R, 2007. Development of a semi‐selective medium for isolation of Xanthomonas campestris pv. musacearum from banana plants. European Journal of Plant Pathology 117, 177–86. [Google Scholar]
- Tripathi L, Odipio J, Tripathi JN, Tusiime G, 2008. A rapid technique for screening banana cultivars for resistance to Xanthomonas wilt. European Journal of Plant Pathology 121, 9–19. [Google Scholar]
- Tripathi L, Mwangi M, Abele S, Aritua V, Tushemereirwe WK, Bandyopadhyay R, 2009. Xanthomonas wilt: a threat to banana production in East and Central Africa. Plant Disease 93, 440–51. [DOI] [PubMed] [Google Scholar]
- Tripathi L, Mwaka H, Tripathi JN, Tushemereirwe WK, 2010. Expression of sweet pepper Hrap gene in banana enhances resistance to Xanthomonas campestris pv. musacearum . Molecular Plant Pathology 11, 721–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tushemereirwe WK, Okaasai O, Kubiriba J et al, 2006. Status of banana bacterial wilt in Uganda. African Crop Science Journal 14, 73–82. [Google Scholar]
- Versalovic J, Schneider M, de Bruijn FJ, Lupski JR, 1994. Genomic fingerprinting of bacteria using repetitive sequence‐based polymerase chain reaction. Methods in Molecular and Cellular Biology 5, 25–40. [Google Scholar]
- Wasukira A, Tayebwa J, Thwaites R et al, 2012. Genome‐wide sequencing reveals two major sub‐lineages in the genetically monomorphic pathogen Xanthomonas campestris pathovar musacearum . Genes 3, 361–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yirgou D, Bradbury JF, 1978. Bacterial wilt of enset (Ensete ventricosum) incited by Xanthomonas musacearum sp. n. Phytopathology 58, 111–2. [Google Scholar]
- Young JM, Dye DW, Bradbury JF, Panagopoulos CG, Robbs CF, 1978. A proposed nomenclature and classification for plant pathogenic bacteria. New Zealand Journal of Agricultural Research 21, 153–77. [Google Scholar]


