Abstract
Opioid addiction is recognized as a chronic relapsing brain disease resulting from repeated exposure to opioid drugs. Cellular and molecular mechanisms underlying the ability of organism to return back to the physiological norm after cessation of drug supply are not fully understood. The aim of this work was to extend our previous studies of morphine-induced alteration of rat forebrain cortex protein composition to the hippocampus. Rats were exposed to morphine for 10 days and sacrificed 24 h (groups +M10 and −M10) or 20 days after the last dose of morphine (groups +M10/−M20 and −M10/−M20). The six altered proteins (≥2-fold) were identified in group (+M10) when compared with group (−M10) by two-dimensional fluorescence difference gel electrophoresis (2D-DIGE). The number of differentially expressed proteins was increased to thirteen after 20 days of the drug withdrawal. Noticeably, the altered level of α-synuclein, β-synuclein, α-enolase and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was also determined in both (±M10) and (±M10/−M20) samples of hippocampus. Immunoblot analysis of 2D gels by specific antibodies oriented against α/β-synucleins and GAPDH confirmed the data obtained by 2D-DIGE analysis. Label-free quantification identified nineteen differentially expressed proteins in group (+M10) when compared with group (−M10). After 20 days of morphine withdrawal (±M10/−M20), the number of altered proteins was increased to twenty. We conclude that the morphine-induced alteration of protein composition in rat hippocampus after cessation of drug supply proceeds in a different manner when compared with the forebrain cortex. In forebrain cortex, the total number of altered proteins was decreased after 20 days without morphine, whilst in hippocampus, it was increased.
Introduction
Opioid dependence and withdrawal syndrome are the leading problems associated with licit and illicit opioid use. In spite of the large literature on opioid addiction, there is only little available information about the durability and reversibility of potentially adverse effects of opioid treatment and withdrawal. From a clinical point of view, drug withdrawal is the leading pathophysiological state driving opioid dependence and addictive behaviors [1,2]. Here, we focused our attention on morphine as the prototypical opioid agonist with which all others are compared.
Our previous results, in accordance with the data of Sim et al. [3], Sim-Selley et al. [4] and Maher et al. [5], indicated a desensitization of (2-D-alanine2-4-methylphenylalanine-5-glycineol)-enkephalin (DAMGO)- and (2-D-alanine-5-D-leucine)-enkephalin (DADLE)-stimulated G-protein responses in plasma membranes (PM) isolated from forebrain cortex (FBC) of rats exposed to morphine for 10 days [6]. Behavioral tests proved that these rats developed a tolerance to this drug as there was no significant difference between control (−M10) and morphine-treated (+M10) rats in the sensitivity to heat stimulation (hot-plate test), which was determined as a delay in hind paw withdrawal. Tolerance to morphine in (+M10) rats was also evidenced by hind paw withdrawal test. Precipitation of morphine withdrawal state by naloxone resulted in a rapid and dramatic opiate abstinence syndrome. There were no detectable signs of abstinence syndrome (such as body shakes, teeth clatter, vacuous chewing, ptosis, irritability to touch, diarrhea), in the (−M10) animals.
In opioid-dependent subjects, the body has adapted to perpetually high opioid tone by making homeostatic adjustments in anti-opioid systems [7]. In an effort to clarify biochemical mechanisms of development of opiate tolerance and dependence, our previous results showed that in rat FBC, such homeostatic adjustments were accompanied by a reversible and specific up-regulation of adenylyl cyclases I and II (ACI and ACII). Importantly, the up-regulation of ACI and ACII disappeared after 20 days of morphine withdrawal (±M10/−M20) [8,9].
Proteomic analyses of the consequences of a 10-day morphine treatment and subsequent 20-day drug withdrawal on FBC, which were based on CBB-staining of 2D gels and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS/MS), showed that the number of altered proteins was decreased from 28 (determined in ±M10 rats) to 14 (determined in ±M10/−M20 rats). When using label-free quantification (LFQ), the number of altered proteins was decreased from 113 to 19 [10]. Thus, we brought a straightforward evidence for the ability of the organism to oppose the drastic, morphine-induced change of the target tissue protein composition with the aim to return to the physiological norm after a complete removal of the drug.
The chronic administration of opioid drugs was reported to modulate synaptic transmission and plasticity of hippocampus and inhibit adult neurogenesis [11]. Chronic opioid administration also resulted in obtuseness of spatial memory and increase in the expression of proteins functionally associated with apoptosis and neurotoxicity [12]. In contrast to chronic opioid treatment, opioid withdrawal was associated with enhanced hippocampal plasticity [13]. Interestingly in the context with studies of prolonged morphine effect on the brain, the chronic antidepressant treatment was found to increase neurogenesis in adult rat hippocampus [14].
As neurodegeneration was frequently described as a pathological state accompanying addiction to opioid drugs and hippocampus is regarded as one of the key brain areas [15,16], together with striatum [17] and cerebellum [18], which exhibit neurogenesis in adult brain, the aim of this work was to extend our previous proteomic studies of morphine-induced alteration of rat forebrain cortex protein composition to the hippocampus.
The two-dimensional fluorescence difference gel electrophoresis (2D-DIGE) and MALDI-TOF MS/MS was used for comparison of protein profiling in hippocampal samples prepared from (+M10), (−M10), (+M10/−M20) and (−M10/−M20) groups of rats, [19,20]. Results obtained by 2D-DIGE were verified by immunoblot analysis of 2D gels and a gel-free, label free proteomic analysis, MaxLFQ [21].
Materials and methods
Chemicals
Acrylamide and bis-acrylamide were from SERVA (Heidelberg, Germany). Immobiline DryStrips, Pharmalyte buffer (broad pH range 3–10), Immobiline DryStrip cover fluid and Amersham™ CyDye DIGE Fluors (minimal dyes) for 2D-DIGE were purchased from GE Healthcare (Piscataway Township, NJ). Complete protease inhibitor cocktail was from Roche Diagnostic (Mannheim, Germany). All others chemicals were of the highest purity available and purchased from Sigma-Aldrich (St. Louis, USA).
Antibodies
The α- and β-synucleins were identified by mouse monoclonal antibody purchased from Santa Cruz, Inc. (Dallas, USA): α/β-synuclein (F-11, sc-514908, 1:500 dilution). glyceraldehyde-3P-dehydrogenase (GAPDH)- and actin-oriented antibodies were also from Santa Cruz, Inc. (Dallas, USA): GAPDH (FL-335, sc-25778, 1:5000 dilution), actin (I-19, sc-1616, 1:2500 dilution). Beta-actin polyclonal antibody was purchased from Bioss Antibodies: β-actin (bs-0061R, 1:5000 dilution). Sheep anti-mouse IgG-HRP (NA931V, 1:10000 dilution) was from GE Healthcare UK and goat anti-rabbit IgG-HRP (sc-2004, 1:10000 dilution) was from Santa Cruz, Inc. (Dallas, USA).
Morphine treatment of experimental animals
Young adult morphine-naive male Wistar rats (220–250 g) were exposed to increasing doses of morphine (10–40 mg/kg) dissolved in 0.9% NaCl for 10 consecutive days as described before [8,10,22]. Food (Altromin standard diet, Germany) and drinking water was provided ad libitum. The morphine-treated rats were sacrificed 24 h (group +M10) or 20 days (group +M10/−M20) after the last dose of morphine. Control animals (−M10, −M10/−M20) received 0.9% NaCl for 10 days and were sacrificed in parallel with morphine-treated rats.
Rats were killed by decapitation under ether narcosis, hippocampi dissected, separated from other parts of brain, washed from remaining blood, snap frozen in liquid nitrogen and stored at −80 °C until use. The experiments were approved by Animal Care and Use Committee of the Institute of Physiology of the Czech Academy of Sciences to be in agreement with Animal Protection Law of the Czech Republic as well as the European Communities Council Directive (86/609/EEC).
Preparation of post-nuclear supernatant fraction (PNS) from rat hippocampus
The hippocampal tissue pieces were minced, diluted in 250 mM sucrose, 20 mM Tris-HCl, 3 mM MgCl2, 1 mM EDTA (pH 7.6) containing protease inhibitor cocktail and fresh phenylmethylsulfonyl fluoride (1 mM final concentration), homogenized for 5 min (2 g w. w. per 10 ml) and centrifuged for 5 min (1200 × g). The 1200 × g supernatant represented the PNS fraction.
2D-DIGE
Samples of PNS containing 1 mg protein were precipitated with ice-cold acetone overnight at– 20 °C. After centrifugation at 9000 × g for 20 min at 4 °C, the supernatant was removed and the pellet was extracted with ice-cold 6% trichloroacetic acid (TCA) for 1.5 h on ice. After centrifugation at 9000 × g for 10 min at 4 °C, the supernatant was discarded and the pellet washed with 600 μl of ice-cold 96% ethanol for 1 h at room temperature. The mixture was centrifuged at 9000 × g for 10 min at 4 °C and the remaining pellet was solubilized with 600 μl lysis buffer containing 30 mM Tris, 7 M urea, 2 M thiourea, 4% (w/v) 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS), pH was adjusted to 8.5 with 50 mM NaOH. Each 50 μg of protein sample (−M10, +M10 and −M10/−M20, +M10/−M20) was labeled with 600 pmol of cyanine CyDye DIGE Fluor minimal dyes Cy3, Cy5 or Cy2 according to Jágr et al. [20].
In the first set of experiments, samples from (+M10) group were labeled with Cy3 and samples from control group (−M10) were labeled with Cy5. In the second set of experiments, samples prepared from rats sacrificed 20 days after morphine withdrawal (+M10/−M20) were labeled with Cy3; samples from the control group (−M10/−M20) were labeled with Cy5. Cy2 was always used to label a mixture of equal amounts of protein taken from all samples of PNS. Cy2-labeled protein mix thus represented an internal standard.
Samples were briefly vortexed and incubated on ice for 30 min in the dark. Staining was terminated by the addition of 10 mM lysine for 10 min on ice in the dark. Dye-labeled PNS samples were combined in such a way that each mixture was comprised of protein samples from (+M10, −M10) or (+M10/−M20, −M10/−M20) groups plus the aliquot of the internal standard (1:1:1, v/v/v). Finally, the mixture of labeled samples was mixed with an equal amount of sample buffer (8 M urea, 130 mM dithiothreitol (DTT), 4% (w/v) CHAPS, 2% (v/v) BioLyte 3–10 buffer (Bio-Rad).
All the samples (250 μl) were then transferred into a groove of the Immobiline DryStrip Reswelling Tray (GE Healthcare). Immobiline DryStrips (linear pH gradient 3–11 NL, 13 cm) were placed into the protein samples and rehydrated overnight. The exposure of protein labeled with CyDyes to all light sources was kept to a minimum. Isoelectric focusing was performed using the Multiphor II system (GE Healthcare) at 14 °C in the following manner: 150 V for 5 h, 500 V for 1 h, 3500 V for 12 h and 500 V for 3 h. The focused strips were stored at– 20 °C or immediately used.
The strips were rinsed thoroughly with ultrapure water, dried quickly on filter paper and equilibrated in 5 ml of equilibration buffer (50 mM Tris-HCl pH 6.8, 6 M urea, 0.1 mM EDTA, 2% SDS, 30% glycerol and 0.01% bromophenol blue) containing 1% DTT for 10 min in order to reduce disulfide bridges and other oxidized groups. Subsequently, the strips were alkylated in equilibration buffer containing 2.5% iodoacetamide for 10 min. Molecular weight markers were loaded onto a piece of filter paper and placed close to the alkaline side of the strip. The strip and molecular marker were covered with 0.5% agarose. Gels were run vertically at a constant current of 20 mA for 20 min and then at 90 mA for 4 h till the bromophenol blue dye reached the end of the gel. The apparatus was cooled to 15 °C using the Hoefer SE 600 unit (GE Healthcare). All the 2D-DIGE analyses were performed three times. After electrophoresis, the gels were washed with ultrapure water for 2×15 min before scanning.
2D image analysis
Three different gel images were obtained from one gel at the appropriate wavelength. They are Cy2 (blue 488 nm laser and 520 nm band pass emission filter), Cy3 (green 532 nm laser and 580 nm band pass emission filter) and Cy5 (red 633 nm laser and 670 nm band pass emission filter) by using a Pharos FX™ scanner (Bio-Rad) at a resolution of 50 μm.
The analysis of DIGE gels was done using PDQuest™ software (Bio-Rad), version 7.3.1. and 8.0.1. Student´s t-test was used to calculate significant differences in relative abundances of protein spot features in the (−M10) vs. (+M10) or (−M10/−M20) vs. (+M10/−M20) PNS samples. Protein levels showing significant quantitative differences at least 2-fold (p≤0.05) were selected for mass spectrometric analysis. P-values were calculated via GraphPadPrism4 software.
Colloidal Coomassie staining
For MS analysis, the gels were stained by colloidal Coomassie brilliant blue G-250 (CBB) to enable the visual detection according to Fountoulakis et al. [23]. The fresh 2D gels were immediately fixed in 50% methanol/7% acetic acid for 1 h and incubated with CBB (17% ammonium sulfate, 34% methanol, 3% orthophosphoric acid and 0.1% CBB) overnight with gentle agitation. After staining, the gels were washed several times in ultrapure sterile water and stored in 1% acetic acid at 4 °C.
MALDI-TOF MS/MS
Selected spots with significantly changed expression (≥2-fold) were cut out from 2D gels and processed as described previously [10,22]. Briefly, chopped 1x1x1 mm pieces were covered with 100 μl of 50 mM ammonium bicarbonate (ABC) buffer in 50% acetonitrile (ACN) (buffer A) with 50 mM DTT. After sonication, the supernatant was removed and each gel spot was mixed with 100 μl of buffer A with 50 mM iodoacetamide (IAA). After sonication, the supernatant was discarded and replaced with 100 μl of buffer A with 50 mM DTT. After sonication, the supernatant was discarded, and the samples were sonicated for 5 min in 100 μl of HPLC/MS-grade water. The water was then discarded, and the samples were again sonicated for 5 min in 100 μl of ACN. The ACN was removed, 5 ng of trypsin in 10 μl of 50 mM ABC was added to each sample, and the samples were incubated at 37 °C overnight. Trifluoroacetic acid (TFA) and ACN were added to reach final concentration of 1% TFA and 30% ACN. After sonication, 0.5 μl aliquot of trypsin digest was transferred onto MALDI target and let to dry. Subsequently, 0.5 μl drop of alpha-cyano-4-hydroxycinnamic acid solution (10 mg/ml in 50% ACN) was transferred onto MALDI target and let to dry again. Samples were measured using a 4800 Plus MALDI TOF/TOF analyzer (Applied Biosystems/MDS Sciex) equipped with a Nd:YAG laser (355 nm, firing rate 200 Hz).
The data were analyzed using in house running Mascot server 2.2.07 and matched against comprehensive UniProt database of protein sequences (27929 sequences; 14725510 residues). Database search criteria were as follows: enzyme = trypsin; taxonomy = Rattus norvegicus. Cystein carbamidomethylation was set as fixed modification, methionine oxidation and deamidation as variable modifications, respectively. Peptide mass tolerance was set to ±100 ppm, and fragment mass tolerance to ±0.4 Da with a maximum of two missed cleavages. Only hits that were scored as significant (MASCOT score ≥57, p<0.05) were accepted. All MALDI-TOF MS/MS analyses were performed in duplicates.
2D immunoblots
Samples of PNS containing 200–2000 μg of protein were prepared for isoelectric focusing as described above (2D-DIGE). Solubilization was performed with 250 μl IEF sample buffer containing 7 M urea, 2 M thiourea, 4% CHAPS, 1% DTT, 1% ampholines pH 3–10 and 0.01% bromophenol blue for 3 h at room temperature. Equilibration steps and SDS-PAGE was performed as described above (2D-DIGE).
After SDS-PAGE, the proteins were transferred to nitrocellulose (GE Healthcare, 1300 mA, 2.5 h) by using TE62 Standard Transfer Tank (Hoefer). All other steps were performed as described before [24]. The significance of the difference between (−M10) vs. (+M10) or (−M10/−M20) vs. (+M10/−M20) samples of PNS was analyzed by Student´s t-test and GraphPadPrism4. Results represent the average ± SEM.
Label-free quantification (MaxLFQ)
The 200 μg of hippocampal PNS samples (+M10), (−M10), (+M10/−M20) and (−M10/−M20) were processed as described previously [10]. The 2 μg of the peptide mixture were loaded on trap column PepMap 300 C 18 column, 5 mm x 300 μm ID, 5 μm particles, 300 Å pore size (163589, Thermo Scientific). Individual peptides were separated by high-performance liquid chromatography (HPLC) using EASY-Spray column, 50 cm x 75 μm ID, PepMap C18, 2 μm particles, 100 Å pore size (ES 803, Thermo Scientific). The separation of peptides was achieved via a linear gradient for 2 hours between mobile phase A (2% acetonitrile, 0.1% formic acid) and B (80% acetonitrile, 0.1% formic acid). Separation was started by running the system with 2% mobile phase B, followed by gradient elution to 40% B.
All data were analyzed and quantified with MaxQuant software. The false discovery rate (FDR) was set to 1% for both proteins and peptides and a minimum length of seven amino acids was specified. The Andromeda search engine was used for the MS/MS spectra search against the UniProt Rattus norvegicus database of protein sequences. Enzyme specificity was set as C-terminal to Arg and Lys, also allowing cleavage at proline bonds and a maximum of two missed cleavages. Dithiomethylation of cysteine was selected as fixed modification and N-terminal protein acetylation and methionine oxidation as variable modifications. The “match between runs” feature of MaxQuant was used to transfer identifications to other LC-MS/MS runs based on their masses and retention time (maximum deviation 0.7 min) and this was also used in quantification experiments. Quantifications were performed with the label-free algorithms according to Cox et al. [21]. Binary logarithms of intensity ratios were then median calculated for each group and the difference between control and sample was determined. Only at least 1.7-fold significant differences calculated for at least 2 measured values from 3 replicates were taken into consideration.
Protein determination
Lowry method was used for determination of protein concentration in all hippocampal samples of PNS (+M10, −M10, +M10/−M20 and −M10/−M20) using bovine serum albumin (Sigma, Fraction V) as a standard. Data were calculated by fitting the calibration curve as a quadratic equation.
Results
Comparative proteomic analysis of rat hippocampus from animals exposed to morphine for 10 days (+M10) and from control animals (−M10) by 2D-DIGE and MALDI-TOF MS/MS
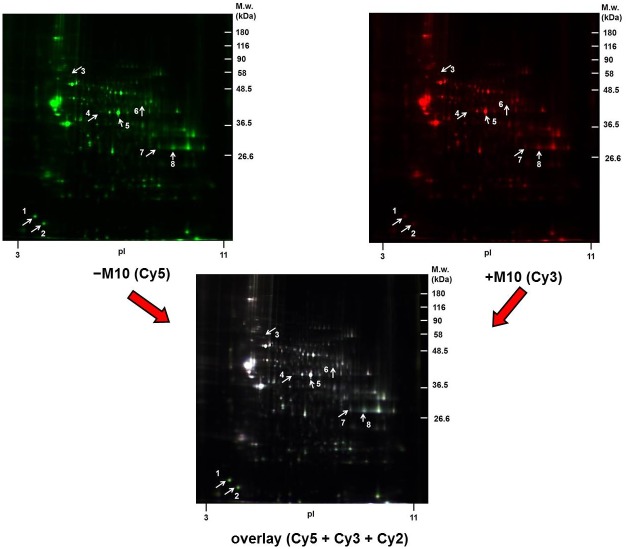
The (+M10) and (−M10) samples of PNS were labeled with Cy3 (+M10) and Cy5 (−M10), respectively, and resolved by 2D electrophoresis (2D-ELFO) together with internal standard (Cy2) as described in Methods. The total number of protein spots labeled with Cy3 (+M10) and Cy5 (−M10) was 307 (Fig 1). Among these, the 8 protein spots were recognized as significantly altered at least 2-fold (p≤0.05) when the difference between Cy3- and Cy5-stained gels was analyzed by PDQuest™ (Bio-Rad) and Student´s t-test (white arrows). These protein spots (1–8) were cut out from the CBB-stained gels (Fig 2A, upper panels) and identified by MALDI-TOF MS/MS (Table 1). Because the same proteins were detected in spots 4, 5 (alpha-enolase) and 7, 8 (glyceraldehyde-3-phosphate dehydrogenase), the number of significantly altered proteins (≥2-fold) in hippocampus by prolonged exposure of rats to morphine was 6 (complete list of peptides used for identification of altered proteins is presented in S1 Table).
Fig 1. The 2D-DIGE comparative proteomic analysis of PNS fractions prepared from rat hippocampus of control (−M10) and morphine-treated rats (+M10) exposed to increasing doses of morphine for 10 days.
PNS fractions were prepared as described in Methods. Staining of PNS samples and resolution of Cy5 (−M10)-, Cy3 (+M10)- and Cy2 (mixed)-labeled proteins in 2D gels (first dimension on pH 3–11 IPG strips and then by SDS-PADE on 10% acrylamide gels) was performed as described in Methods. White arrows indicate the protein spots altered at least 2-fold by morphine (p≤0.05). Comparison of DIGE gels was done by PDQuest™ (Bio-Rad). The significance of the difference between the three sets of (−M10) and (+M10) gels was determined by Student´s t-test. The positions of molecular weight markers are indicated on the right side and pI at the bottom of each gel.
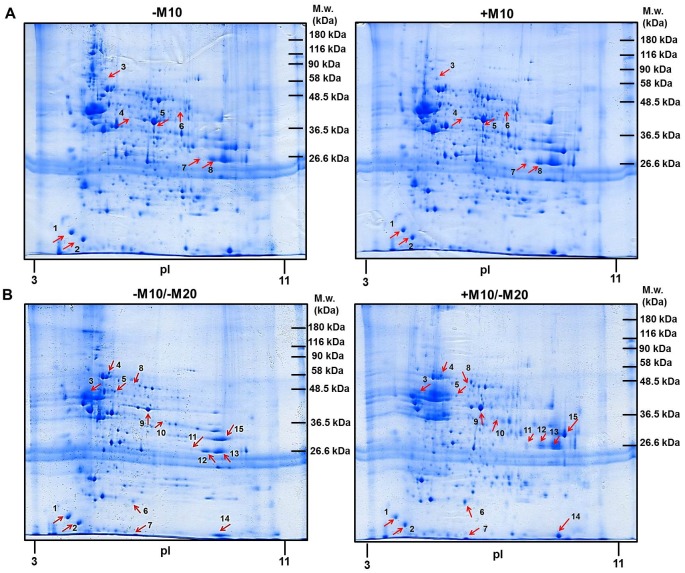
Fig 2. 2D-ELFO of PNS hippocampal samples prepared from experimental groups (−M10) [a, upper left], (+M10) [a, upper right], (−M10/−M20) [b, lower left] and (+M10/−M20) [b, lower right] of control and morphine-treated rats.
PNS fractions (2000 μg of protein) were resolved by 2D-ELFO and stained by CBB as described in Methods. After CBB staining, protein spots 1–8 (A, red arrows) and 1–15 (B, red arrows) were excised from gels and identified by MALDI-TOF MS/MS.
Table 1. MALDI-TOF MS/MS analysis of eight altered protein spots in PNS prepared from hippocampus of rats exposed to morphine for 10 days and sacrificed 24 h after the last dose; difference of protein composition in PNS samples prepared from groups (+M10) and (−M10).
| Spot | Accession number | Protein name | Mascot score | Matched peptides | SCa [%] |
MWb (kDa) |
pIc | Change (fold) |
p value |
|---|---|---|---|---|---|---|---|---|---|
| 1 | SYUB_RAT | Beta-synuclein | 134 | 9 | 42 | 14.5 | 4.48 | ↓ 2.0 | 0.0060 |
| 2 | SYUA_RAT | Alpha-synuclein | 249 | 5 | 36 | 14.5 | 4.74 | ↓ 2.5 | 0.0251 |
| 3 | ACTB_RAT | Actin, cytoplasmic 1 (fragment) | 146 | 8 | 13 | 42.1 | 5.29 | ↑ 2.5 | 0.0068 |
| 4 | ENOA_RAT | Alpha-enolase | 117 | 6 | 11 | 47.4 | 6.16 | ↑ 2.2 | 0.0212 |
| 5 | ENOA_RAT | Alpha-enolase | 679 | 25 | 44 | 47.4 | 6.16 | ↑ 2.2 | 0.0073 |
| 6 | DLDH_RAT | Dihydrolipoyl dehydrogenase, mitochondrial | 180 | 7 | 12 | 54.6 | 7.96 | ↑ 2.0 | 0.0241 |
| 7 | D3ZGY4_RAT | Glyceraldehyde-3-phosphate dehydrogenase | 230 | 7 | 15 | 36.1 | 7.63 | ↓ 2.3 | 0.0106 |
| 8 | D3ZGY4_RAT | Glyceraldehyde-3-phosphate dehydrogenase | 306 | 13 | 18 | 36.1 | 7.63 | ↓ 2.3 | 0.0309 |
a sequence coverage,
b theoretical molecular weight,
c theoretical isoelectric point; complete list of peptides used for identification is presented in S1 Table.
The presence of the same protein in more than just one spot, when resolved by 2D-ELFO, represents according to our previous experience [10,22], evidence for the existence of different subunits of the same protein. In general, this multiplicity occurs due to post-translational modifications of such proteins which result in the production of multiple spots when resolved in 2D gels [25].
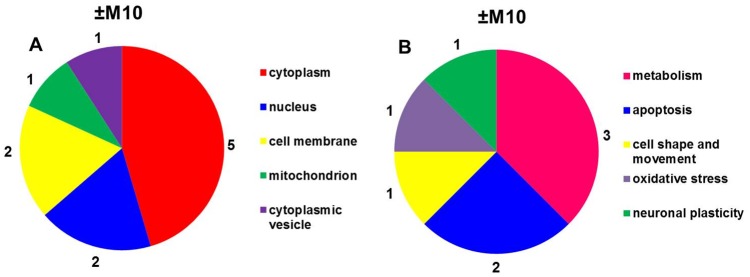
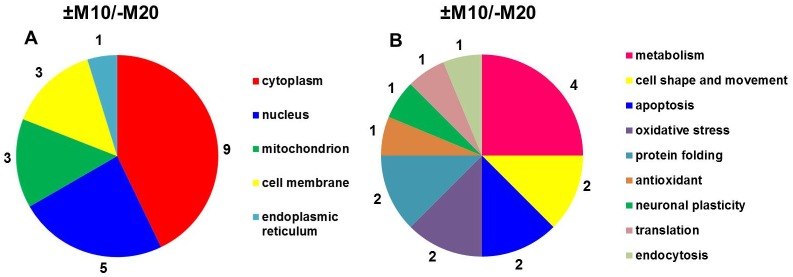
The subcellular localization and functional significance of morphine-altered proteins was determined according to the current annotations in the UniProt database (Table 2, Fig 3). The altered proteins were found to be functionally related to glycolysis, oxidative stress and apoptosis: glyceraldehyde-3-phosphate dehydrogenase (spots 7, ↓2.3-fold and 8, ↓2.3-fold); glycolysis: alpha-enolase (spots 4, ↑2.2-fold and 5, ↑2.2-fold); Krebs cycle: mitochondrial dihydrolipoyl dehydrogenase (spot 6, ↑2.0-fold,); apoptosis: α-synuclein (spot 2, ↓2.5-fold); cell motility: cytoplasmic 1 fragment of actin (spot 3, ↑2.5-fold) and neuronal plasticity: β-synuclein (spot 1, ↓2.0-fold).
Table 2. Subcellular localization and function of morphine-altered proteins in hippocampus of experimental groups (+M10) and (−M10) of rats.
| Spot | Accession number | Protein name | Change (fold) |
Subcellular localization | Molecular functions and biological processes |
|---|---|---|---|---|---|
| 1 | SYUB_RAT | Beta-synuclein | ↓ 2.0 | Cytoplasm | Alpha/beta tubulin-binding, neuronal plasticity |
| 2 | SYUA_RAT | Alpha-synuclein | ↓ 2.5 | Cytoplasm, cell membrane, nucleus | Regulation of dopamine release and transport, anti-apoptotic protein |
| 3 | ACTB_RAT | Actin, cytoplasmic 1 (fragment) | ↑ 2.5 | Cytoplasm, cytoskeleton | ATP binding, cell motility |
| 4,5 | ENOA_RAT | Alpha-enolase | ↑ 2.2, ↑ 2.2 | Cytoplasm, cell membrane | Glycolysis, plasminogen activation, heat-shock protein (HSP48) |
| 6 | DLDH_RAT | Dihydrolipoyl dehydrogenase, mitochondrial | ↑ 2.0 | Mitochondrion matrix, cytoplasmic vesicle | Krebs cycle, glycolysis, aging |
| 7,8 | D3ZGY4_RAT | Glyceraldehyde-3-phosphate dehydrogenase | ↓ 2.3, ↓ 2.3 | Cytoplasm, nucleus | Glycolysis, apoptosis, oxidative stress |
Fig 3. Subcellular localization (A) and functional significance (B) of morphine-altered proteins in experimental groups (+M10) and (−M10) identified by MALDI-TOF MS/MS.
Comparative proteomic analysis of rat hippocampus from animals exposed to morphine for 10 days and sacrificed 20 days since the last dose of morphine (+M10/−M20) and from control animals (−M10/−M20) by 2D-DIGE and MALDI-TOF MS/MS
Samples of PNS were labeled with Cy5 (−M10/−M20) or Cy3 (+M10/−M20) and resolved by 2D-ELFO as described in Methods together with internal standard labeled with Cy2 (Fig 4). The total number of protein spots detected by PDQuest™ (Bio-Rad) was 343. Only spots showing at least 2-fold quantitatively significant difference (p≤0.05) were excised from the CBB-stained gels (Fig 2B, lower panels), digested with trypsin and used for mass spectrometric analysis. Analysis by MALDI-TOF MS/MS identified 15 altered protein spots (Table 3). As the same protein was identified in spots 11, 12, 13 (glyceraldehyde-3-phosphate dehydrogenase), the number of morphine-altered proteins was 13. A complete list of peptides used for recognition of these proteins is presented in S2 Table.
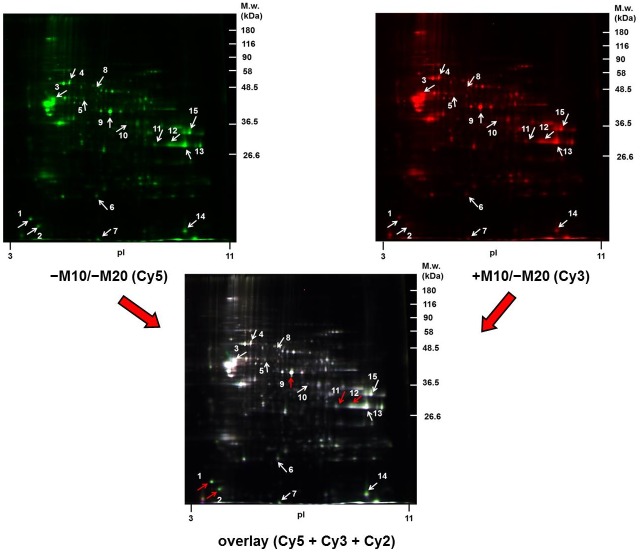
Fig 4. The 2D-DIGE comparative proteomic analysis of PNS fractions prepared from rat hippocampus of control (−M10/−M20) and morphine-treated rats (+M10/−M20) exposed to increasing doses of morphine for 10 days and subsequently nurtured for 20 days without this drug.
The PNS samples were prepared as described in Methods and separated in the first dimension on pH 3–11 IPG strips and then by SDS-PADE on 10% acrylamide gels. The analysis of DIGE gels was done by PDQuest™. Protein spots (1–15) showing quantitatively significant difference at least 2-fold (p≤0.05) were excised from the CBB-stained gels and identified by MALDI-TOF MS/MS (white + red arrows). The red arrows indicate those protein spots which were altered in (±M10) as well as (±M10/−M20) samples of PNS: 1, 2, 9, 11 and 12. The positions of molecular weight markers are indicated on the right side and pI at the bottom of each gel.
Table 3. MALDI-TOF MS/MS analysis of fifteen altered protein spots in PNS prepared from hippocampus of rats exposed to morphine for 10 days and sacrificed 20 days after the last dose; difference of protein composition in PNS samples prepared from groups (+M10/−M20) and (−M10/−M20).
| Spot | Accession number | Protein name | Mascot score | Matched peptides | SCa [%] |
MWb (kDa) |
pIc | Change (fold) |
p value |
|---|---|---|---|---|---|---|---|---|---|
| 1 | SYUB_RAT | Beta-synuclein | 96 | 5 | 29 | 14.5 | 4.48 | ↓ 2.4 | 0.0259 |
| 2 | SYUA_RAT | Alpha-synuclein | 249 | 5 | 36 | 14.5 | 4.74 | ↓ 5.3 | 0.0038 |
| 3 | TBA1A_RAT | Tubulin alpha-1A chain | 298 | 19 | 40 | 50.8 | 4.94 | ↓ 4.5 | 0.0023 |
| 4 | F1M953_RAT | Stress-70 protein, mitochondrial | 423 | 15 | 15 | 74.0 | 5.87 | ↑ 2.1 | 0.0291 |
| 5 | PDIA3_RAT | Protein disulfide-isomerase A3 | 117 | 10 | 13 | 57.0 | 5.88 | ↓ 2.2 | 0.0286 |
| 6 | ATP5H_RAT | ATP synthase subunit d, mitochondrial | 305 | 8 | 31 | 18.8 | 6.17 | ↓ 2.0 | 0.0167 |
| 7 | SODC_RAT | Superoxide dismutase [Cu-Zn] | 81 | 6 | 31 | 16.1 | 5.88 | ↓ 3.1 | 0.0490 |
| 8 | DPYL2_RAT | Dihydropyrimidinase-related protein 2 | 237 | 9 | 17 | 62.6 | 5.95 | ↓ 2.4 | 0.0251 |
| 9 | ENOA_RAT | Alpha-enolase | 694 | 20 | 28 | 47.4 | 6.16 | ↑ 3.1 | 0.0154 |
| 10 | EFTU_RAT | Elongation factor Tu, mitochondrial | 242 | 8 | 13 | 49.9 | 7.23 | ↓ 2.2 | 0.0164 |
| 11 | D3ZGY4_RAT | Glyceraldehyde-3-phosphate dehydrogenase | 109 | 9 | 9 | 36.1 | 7.63 | ↓ 2.4 | 0.0168 |
| 12 | D3ZGY4_RAT | Glyceraldehyde-3-phosphate dehydrogenase | 273 | 6 | 10 | 36.1 | 7.63 | ↓ 2.3 | 0.0087 |
| 13 | D3ZGY4_RAT | Glyceraldehyde-3-phosphate dehydrogenase | 316 | 13 | 20 | 36.1 | 7.63 | ↓ 4.1 | 0.0180 |
| 14 | COF1_RAT | Cofilin-1 | 346 | 9 | 40 | 18.7 | 8.22 | ↓ 2.8 | 0.0115 |
| 15 | ALDOA_RAT | Fructose-bisphosphate aldolase A | 255 | 24 | 53 | 39.8 | 8.31 | ↓ 2.8 | 0.0104 |
a sequence coverage,
b theoretical molecular weight,
c theoretical isoelectric point; complete list of peptides used for identification is presented in S2 Table.
When compared with hippocampus prepared from (±M10) groups of rats (Fig 1 and Table 1), the number of altered protein spots was increased from eight to fifteen (Fig 4 and Table 3). Noticeably, the alteration of five protein spots identified in (±M10) samples of PNS: β-synuclein (spot 1), α-synuclein (spot 2), alpha-enolase (spot 5) and glyceraldehyde-3-phosphate dehydrogenase (spots 7, 8) persisted for 20 days since the last addition of the drug: α-synuclein (spot 1), β-synuclein (spot 2), α-enolase (spot 9) and glyceraldehyde-3-phosphate dehydrogenase (spots 11, 12 and 13).
Protein categorization (subcellular localization and functional significance) performed according to the current annotations in the UniProt database (Table 4, Fig 5) distinguished proteins functionally involved in glycolysis, oxidative stress and apoptosis: glyceraldehyde-3-phosphate dehydrogenase (↓2.4-fold; ↓2.3-fold; ↓4.1-fold, spots 11, 12, 13); glycolysis: α-enolase (↑3.1-fold, spot 9); oxidative phosphorylation: mitochondrial ATPase synthase subunit d (↓2.0-fold, spot 6); cell shape and movement: tubulin α-1A chain (↓4.5-fold, spot 3), cofilin-1 (↓2.8-fold, spot 14); apoptosis: α-synuclein (↓5.3-fold, spot 2), oxidative stress: dihydropyrimidinase-related protein 2 (↓2.4-fold, spot 8), protein folding: mitochondrial stress-70 protein (↑2.1-fold, spot 4), protein disulfide-isomerase A3 (↓2.2-fold, spot 5); detoxification of reactive oxygen species: superoxide dismutase [Cu-Zn] (↓3.1-fold, spot 7); neuronal plasticity: β-synuclein (↓2.4, spot 1); translation: mitochondrial elongation factor Tu (↓2.2-fold, spot 10), and endocytosis: dihydropyrimidinase-related protein 2 (↓2.4-fold, spot 8).
Table 4. Subcellular localization and function of morphine-altered proteins in hippocampus of experimental groups (+M10/−M20) and (−M10/−M20) of rats.
| Spot | Accession number | Protein name | Change (fold) |
Subcellular localization | Molecular functions and biological processes |
|---|---|---|---|---|---|
| 1 | SYUB_RAT | Beta-synuclein | ↓ 2.4 | Cytoplasm | Alpha/beta tubulin-binding, neuronal plasticity |
| 2 | SYUA_RAT | Alpha-synuclein | ↓ 5.3 | Cytoplasm, cell membrane, nucleus | Regulation of dopamine release and transport, anti-apoptotic protein |
| 3 | TBA1A_RAT | Tubulin alpha-1A chain | ↓ 4.5 | Cytoplasm, cytoskeleton | GTP binding, microtubule-based process, cell division |
| 4 | F1M953_RAT | Stress-70 protein, mitochondrial | ↑ 2.1 | Mitochondrion, nucleus | Protein folding, anti-apoptotic protein, erythrocyte differentiation |
| 5 | PDIA3_RAT | Protein disulfide-isomerase A3 | ↓ 2.2 | Endoplasmic reticulum, melanosome | Cell redox homeostasis, protein folding, neurodegenerative processes |
| 6 | ATP5H_RAT | ATP synthase subunit d, mitochondrial | ↓ 2.0 | Mitochondrion | Oxidative phosphorylation, hydrogen ion transport |
| 7 | SODC_RAT | Superoxide dismutase [Cu-Zn] | ↓ 3.1 | Cytoplasm, nucleus | Antioxidant, detoxification of reactive oxygen species |
| 8 | DPYL2_RAT | Dihydropyrimidinase-related protein 2 | ↓ 2.4 | Cytoplasm, cytoskeleton, cell membrane | Neurogenesis, endocytosis, control of oxidative stress |
| 9 | ENOA_RAT | Alpha-enolase | ↑ 3.1 | Cytoplasm, cell membrane | Glycolysis, plasminogen activation, heat-shock protein (HSP48) |
| 10 | EFTU_RAT | Elongation factor Tu, mitochondrial | ↓ 2.2 | Mitochondrion | Elongation factor, translation |
| 11,12,13 | D3ZGY4_RAT | Glyceraldehyde-3-phosphate dehydrogenase | ↓ 2.4, ↓ 2.3, ↓ 4.1 | Cytoplasm, nucleus | Glycolysis, apoptosis, oxidative stress |
| 14 | COF1_RAT | Cofilin-1 | ↓ 2.8 | Cytoplasm, cytoskeleton, nucleus | Actin cytoskeleton, hippocampus development |
| 15 | ALDOA_RAT | Fructose-bisphosphate aldolase A | ↓ 2.8 | Cytoplasm | Glycolysis, gluconeogenesis |
Fig 5. Subcellular localization (A) and functional significance (B) of morphine-altered proteins in experimental groups (+M10/−M20) and (−M10/−M20) identified by MALDI-TOF MS/MS.
Immunoblot analysis of α- and β-synuclein, GAPDH and actin in rat hippocampus of experimental groups (±M10) and (±M10/−M20)
In order to verify the results obtained by comparative 2D-DIGE, PNS hippocampal fractions prepared from groups (−M10), (+M10), (+M10/−M20) and (−M10/−M20) were resolved by 2D-ELFO and the content of α- and β-synuclein, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and actin was determined by immunoblotting with specific antibodies.
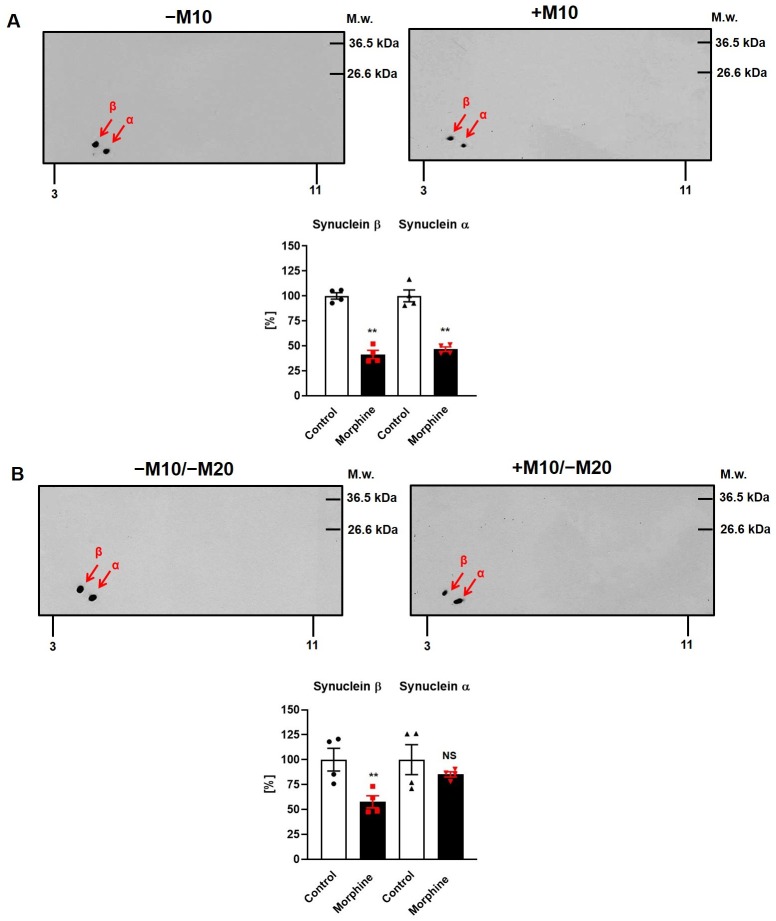
In (±M10) samples of PNS, position of immunoblot signals of α- and β-synuclein in 2D gels (Fig 6A) was the same as in 2D-DIGE gels [compare with Fig 1 and Table 1; spot 1 (β-synuclein, ↓2.0-fold) and spot 2 (α-synuclein, ↓2.5-fold)]. The Mw of these two proteins was ≈ 14 and 16 kDa, and their pI values were ≈ 4.3 and ≈ 4.7, respectively. Intensities of immunoblot signals were decreased to 41.3% (β-synuclein, p<0.01) and 46.6% (α-synuclein, p<0.01) when compared with corresponding controls. Thus, results obtained by 2D-DIGE indicating the decrease of these two proteins by morphine-treatment, were verified by immunoblot analysis of 2D gels. The difference in the magnitude of the decrease may be attributed to the major difference in methodology of detection combined with the necessity to apply largely different amounts of total protein per gel (75 μg versus 2000 μg) to achieve the proper labeling of the sample with Cy probes on the one hand and intensity of immunoblot signals on the other hand [26,27].
Fig 6. 2D immunoblot analysis of α/β-synucleins in PNS prepared from rat hippocampus of experimental groups (±M10) and (±M10/−M20).
(A) The (+M10) and (−M10) or (B) (+M10/−M20) and (−M10/−M20) samples of PNS (2000 μg protein per gel) were extracted in acetone/TCA and resolved by 2D-ELFO as described in Methods. The α- and β-synucleins were recognized by Ab F-11 (sc-514908). The two immunoblot signals with similar Mw of ≈ 14–15 kDa were observed at pI ≈ 4.3 and ≈ 4.7. In group (+M10) (A, upper right), the level of β-synuclein was decreased to 41.3% (p<0.01) when compared with control group (−M10) (A, upper left). The α-synuclein was decreased to 46.6% (p<0.01). In group (+M10/−M20) (B, lower right), a significant down-regulation of β-synuclein (58%, p<0.01) and down-regulation of α-synuclein (85%, p>0.05) was observed as well when compared with intensities of immunoblot signals of these two proteins in group (−M10/−M20) (B, lower left). Data present bar graphs with the individual data points along with the bar with error. The significance of the difference between the (±M10) or (±M10/−M20) samples of PNS was analyzed by Student´s t-test using GraphPadPrism4; *, p<0.05; **, p<0.01; NS, p>0.05.
Accordingly, the densitometric scanning of immunoblot signals of α/β-synucleins in (±M10/−M20) samples of PNS indicated a significant down-regulation to 58% (β-synuclein, p<0.01) and 85% (α-synuclein, p>0.05), respectively (Fig 6B). When combined together, the results of both 2D-DIGE and immunoblot analyses indicated consistently the down-regulation of α- and β-synuclein in hippocampus of rats exposed to morphine for 10 days. Noticeably, this down-regulation persisted for 20 days of abstinence.
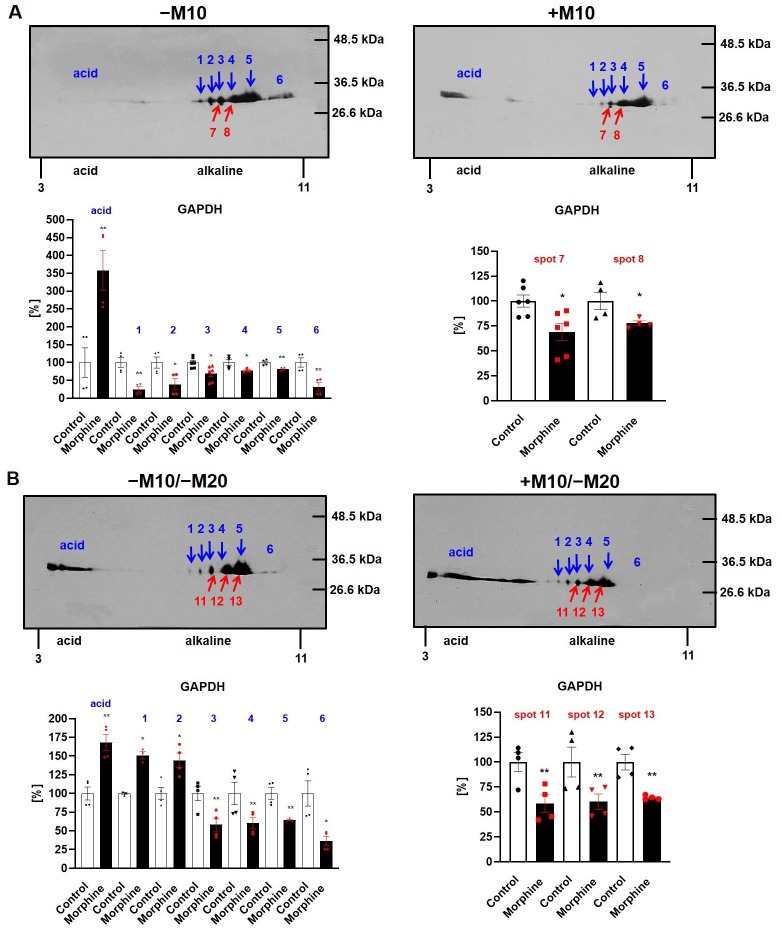
The immunoblot signals of GAPDH detected by Ab FL-335 were distributed over a wide range of pI ≈ 3.5–10, but the Mw ≈ 36 kDa of all these signals was the same in PNS prepared from all four groups of experimental animals, Fig 7. Morphine-treatment resulted in redistribution of GAPDH signals between alkaline and acid region of the gel manifested as a transfer from alkaline to acid region (Fig 7A, upper panels). Consequently, intensities of individual spots in alkaline region (blue arrows) were decreased in (+M10) when compared with (−M10) samples of PNS to: ≈ 25% (spot 1, p<0.01), ≈ 39% (spot 2, p<0.05), ≈ 69% (spot 3, p<0.05), ≈ 78% (spot 4, p<0.05), ≈ 82% (spot 5, p<0.01), and ≈ 32% (spot 6, p<0.01), respectively. Spots 3 and 4 were those identified in DIGE gels as spots 7 and 8 (red arrows; compare with Figs 1 and 2A and Table 1). The signal detected in the acid region was increased to 358% (p<0.01). The sum of signals in alkaline region (1+2+3+4+5+6) was decreased to 67% (p<0.05); the total signal of all spots was decreased to 72% (p<0.05).
Fig 7. 2D immunoblot analysis of GAPDH in PNS prepared from rat hippocampus of experimental groups (±M10) and (±M10/−M20).
(A) The (+M10) and (−M10) or (B) (+M10/−M20) and (−M10/−M20) samples of PNS (2000 μg protein per gel) were extracted in acetone/TCA, resolved by 2D-ELFO and immunobloted with Ab FL-335 (sc-25778) as described in Methods. The seven immunoblot signals (acid + 1–6) exhibited the Mw ≈ 36 kDa were distributed over a wide range of pI (3.5–10) in both groups of experimental animals. In (A), the immunoblot signals 3 and 4 exhibited the same pI as spots 7 and 8 which were recognized as significantly decreased (≥2-fold) by DIGE analysis (compare with Figs 1 and 2A and Table 1). In (B), the immunoblot signals 3, 4 and 5 were identical with spots 11, 12 and 13, which were recognized as significantly decreased (≥2-fold) by DIGE analysis (compare with Figs 4 and 2B and Table 3). Data present bar graphs with the individual data points along with the bar with error. The significance of the difference between the (±M10) or (±M10/−M20) samples of PNS was analyzed by Student´s t-test using GraphPadPrism4; *, p<0.05; **, p<0.01.
Similar results were obtained in (±M10/−M20) samples of PNS. GAPDH signals were distributed over a wide range of pI ≈ 3.5–10, Mw of all isoforms of this enzyme was ≈ 36 kDa and morphine-treatment resulted in the transfer from alkaline to acid region of the gel. However, the relative magnitude of this transfer was less than in (±M10) samples of PNS (Fig 7B, lower panels). Intensities of individual spots in alkaline region (blue arrows) were decreased to ≈ 58% (spot 3, p<0.01), ≈ 60% (spot 4, p<0.01), ≈ 64% (spot 5, p<0.01) and ≈ 36% (spot 6, p<0.05); the two minor spots (1 and 2) were increased to ≈ 151% (p<0.05) and ≈ 145% (spot 2, p<0.05), respectively. The major spots 3, 4 and 5 were those identified in DIGE gels as spots 11, 12 and 13 (red arrows; please compare with Figs 4 and 2B and Table 3). The total signal of GAPDH subunits in alkaline region (1+2+3+4+5+6) was decreased to 72% (p<0.05); the total signal of all spots was unchanged by morphine (NS, p>0.05).
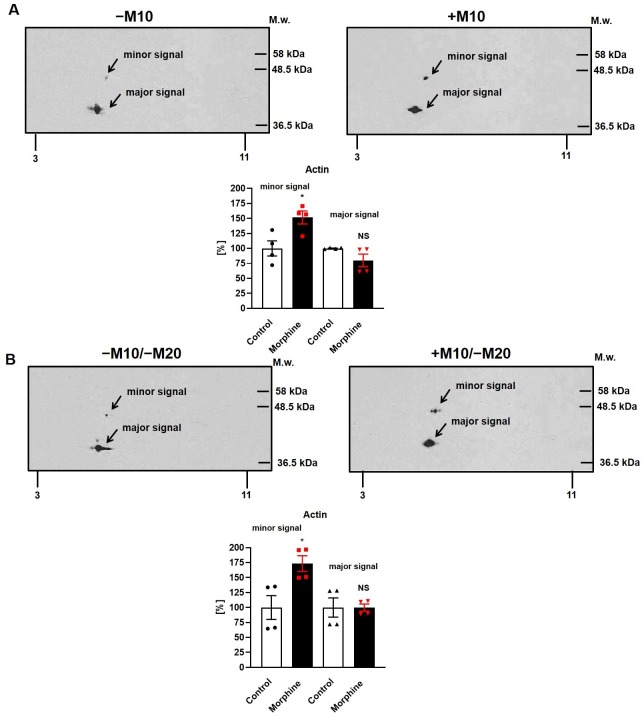
The total signal of actin was unchanged, Fig 8. In (±M10) samples of PNS (Fig 8A), the major signal of actin was decreased to 80%, but this decrease was not significant when averaged in three immunoblots (NS, p>0.05); the minor signal was increased to 152% (*, p<0.05). The total signal of both spots was unchanged (NS, p>0.05). The same result was obtained in (±M10/−M20) groups of rats (Fig 8B): the major signal of actin was unchanged when averaged in three blots (NS, p>0.05), the minor signal was increased to 173% (*, p<0.05). The total signal of both spots was unchanged (NS, p>0.05). As actin is known to be modified by sugars, the increased level of its minor fragment exhibiting the higher molecular weight may represent the glycosylated form detected in both (±M10) and (±M10/−M20) samples of hippocampus.
Fig 8. 2D immunoblot analysis of actin in PNS prepared from rat hippocampus of experimental groups (±M10) and (±M10/−M20).
The (+M10) and (−M10) (A) or (+M10/−M20) and (−M10/−M20) (B) samples of PNS (200 μg protein per gel) were extracted in acetone/TCA, resolved by 2D-ELFO and immunoblotted as described in Methods. The two signals were recognized with Ab I-19 (sc-1616): the major signal with Mw ≈ 42 kDa at pI of ≈ 5.2–5.8 and the minor signal with Mw ≈ 46 kDa at pI of ≈ 5.4. Data present bar graphs with the individual data points along with the bar with error. The significance of the difference between (±M10) or (±M10/−M20) samples of PNS was analyzed by Student´s t-test using GraphPadPrism4; *, p<0.05; NS, p>0.05.
The level of beta-actin commonly used as a loading control was also detected and was unchanged (NS, p>0.05), S1 File. Moreover, 1D immunoblot resolution of GAPDH was performed (S2 File). The difference in the expression level between (±M10) and (±M10/−M20) samples was not significant.
Label-free MS analysis of rat hippocampus from animals exposed to morphine for 10 days (±M10); comparison with animals exposed to morphine and subsequently nurtured for 20 days in the absence of this drug (±M10/−M20)
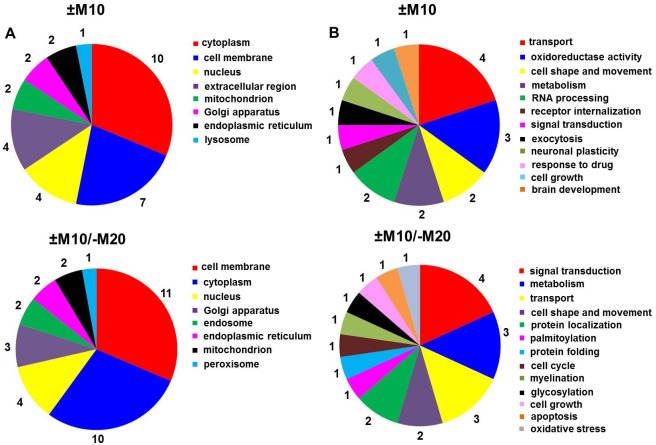
In the last part of our work, we applied the gel-free, label-free proteomic approach denominated as MaxLFQ [21]. The LFQ analysis identified 19 proteins with significantly altered expression level (≥1.7-fold) in group (+M10) when compared with group (−M10), Table 5, Fig 9, upper panels. According to the current annotations in the UniProt database, the identified proteins were functionally related to transport (1-solute carrier family 22 member 23, 4-urea transporter 1, 17-GTP-binding protein SAR1b, 18-peflin), oxidoreductase activity (2-ECSIT, 6-ATP synthase subunit f, 19-NADPH-dependent diflavin oxidoreductase 1), cell shape and movement (8-filamin A, 11-plastin 1), metabolism (10-beta-galactosidase, 12-GMP reductase), RNA processing (15-alanyl-tRNA editing protein Aarsd1, 16-RNA polymerase II-associated protein 3), receptor internalization and brain development (3-CD9 antigen), signal transduction (5-SH2B adapter protein 1), exocytosis (7-exocyst complex component 1), neuronal plasticity (9-pro-MCH), response to drug (13-protein phosphatase 1E), and cell growth (14-alpha-2-HS-glycoprotein).
Table 5. Subcellular localization and function of morphine-altered proteins in groups (+M10) and (−M10) identified by MaxLFQ.
| Accession number | Protein name | Change (fold) |
p value | Subcellular localization | Molecular functions and biological processes |
|---|---|---|---|---|---|
| 1-Q9QZG1 | Solute carrier family 22 member 23 | ↑ 3.1 | 0.0001 | Cell membrane | Membrane integrated transporter, ion transport |
| 2-Q5XIC2 | Evolutionarily conserved signaling intermediate in Toll pathway, mitochondrial | ↑ 3.0 | 0.0012 | Mitochondrion, nucleus, cytoplasm | Assembly of mitochondrial NADH |
| 3-P40241 | CD9 antigen | ↑ 2.7 | 0.0159 | Cell membrane, extracellular region | Brain development, cell adhesion, receptor internalization |
| 4-P97689 | Urea transporter 1 | ↑ 2.2 | 0.0127 | Cell membrane | Urea transport |
| 5-Q62985 | SH2B adapter protein 1 | ↑ 2.2 | 0.0439 | Nucleus, cytoplasm, cell membrane | Signal transduction, tyrosine kinase adaptor activity |
| 6-D3ZAF6 | ATP synthase subunit f, mitochondrial | ↑ 1.7 | 0.0363 | Mitochondrion | Hydrogen ion transport, cristae formation |
| 7-F1LQN9 | Exocyst complex component 1 | ↑ 1.7 | 0.0034 | Cell membrane | Exocytosis |
| 8-C0JPT7 | Filamin A | ↓ 3.8 | 0.0223 | Cytoplasm—cytoskeleton, nucleus, cell membrane | Actin cytoskeleton reorganization |
| 9-P14200 | Pro-MCH | ↓ 2.3 | 0.0002 | Extracellular region | Regulation of synaptic plasticity, glucose homeostasis |
| 10-D3ZUM4 | Beta-galactosidase | ↓ 2.0 | 0.0200 | Golgi apparatus, lysosome, extracellular region | Galactose catabolism |
| 11-A0A0G2QC04 | Plastin 1 | ↓ 1.9 | 0.0231 | Cytoplasm—cytoskeleton | Actin cytoskeleton reorganization |
| 12-Q5FVP6 | GMP reductase | ↓ 1.9 | 0.0002 | Cytoplasm | Purine metabolism |
| 13-Q80Z30 | Protein phosphatase 1E | ↓ 1.9 | 0.0062 | Nucleus, cytoplasm | Negative regulation of protein kinase activity, response to drug, positive regulation of stress fiber assembly |
| 14-P24090 | Alpha-2-HS-glycoprotein | ↓ 1.9 | 0.0005 | Extracellular region | Organ regeneration, negative regulation of cell growth |
| 15-Q5XI97 | Alanyl-tRNA editing protein Aarsd1 | ↓ 1.8 | 0.0005 | Cytoplasm | RNA processing |
| 16-Q68FQ7 | RNA polymerase II-associated protein 3 | ↓ 1.8 | 0.0094 | Cytoplasm | RNA processing |
| 17-Q5HZY2 | GTP-binding protein SAR1b | ↓ 1.7 | 0.0044 | Endoplasmic reticulum, Golgi apparatus | Protein transport, vesicle organization |
| 18-Q641Z8 | Peflin | ↓ 1.7 | 0.0065 | Endoplasmic reticulum, cytoplasm, cell membrane | Response to calcium ion, ER to Golgi vesicle-mediated transport |
| 19-D4ABT4 | NADPH-dependent diflavin oxidoreductase 1 | ↓ 1.7 | 0.0002 | Cytoplasm | Electron transfer chain |
Fig 9. Subcellular localization (A) and functional significance (B) of morphine-altered proteins in experimental groups ±M10 (upper panels) and ±M10/−M20 (lower panels) identified by MaxLFQ.
The LFQ analysis of proteins in PNS prepared from rats sacrificed 20 days since the last dose of morphine (group +M10/−M20) identified 20 proteins with significantly altered expression level (≥1.7-fold) when compared with group (−M10/−M20), Table 6, Fig 9, lower panels. Those 20 proteins were functionally related to signal transduction (10-rho GTPase-activating protein 39, ↑1.7-fold; 15-crk-like protein, ↓1.9-fold; 17-SH2B adapter protein 1, ↓1.9-fold; 19-regulator of G-protein signaling 14, ↓1.7-fold), metabolism (1-aldose 1-epimerase, ↑3.6-fold; 6-citrate lyase subunit beta-like protein, ↑2.0-fold; 18-farnesyl pyrophosphate synthase ↓1.8-fold), transport (2-equilibrative nucleoside transporter 2, ↑3.1-fold; 9-ADP/ATP translocase 2, ↑1.7-fold; 16-AP complex subunit sigma, ↓1.9-fold), cell shape and movement (11-signal-induced proliferation-associated 1-like protein 1, ↑1.7-fold; 7-transmembrane protein 35A, ↑1.8-fold), protein localization (8-A-kinase anchor protein 7 isoforms delta and gamma, ↑1.7-fold; 13-Golgin B1, ↓2.4), palmitoylation (3-alpha/beta hydrolase domain-containing protein 17B, ↑2.7-fold), protein folding (4-alpha-2-macroglobulin receptor-associated protein, ↑2.2-fold), regulation of cell cycle (5-calcium/calmodulin-dependent protein kinase II inhibitor 1, ↑2.1-fold), myelination (12-neurofascin, ↓3.0-fold), glycosylation (14-protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2, ↓2.2-fold), apoptosis and oxidative stress (20-alpha-synuclein, ↓1.7-fold).
Table 6. Subcellular localization and function of morphine-altered proteins in groups (+M10/−M20) and (−M10/−M20) identified by MaxLFQ.
| Accession number | Protein name | Change (fold) |
p value | Subcellular localization | Molecular functions and biological processes |
|---|---|---|---|---|---|
| 1-Q66HG4 | Aldose 1-epimerase | ↑ 3.6 | 0.0073 | Cytoplasm | Carbohydrate metabolism |
| 2-O54699 | Equilibrative nucleoside transporter 2 | ↑ 3.1 | 0.0188 | Cell membrane | Nucleoside transport |
| 3-Q6AY17 | Alpha/beta hydrolase domain-containing protein 17B | ↑ 2.7 | 0.0233 | Cell membrane, endosome | Protein palmitoylation/depalmitoylation |
| 4-Q99068 | Alpha-2-macroglobulin receptor-associated protein | ↑ 2.2 | 0.0140 | Endoplasmic reticulum, endosome, Golgi apparatus | Chaperone for LDL-receptor related proteins |
| 5-Q9JI15 | Calcium/calmodulin-dependent protein kinase II inhibitor 1 | ↑ 2.1 | 0.0306 | Cell membrane | Negative regulation of cell cycle, negative regulation of proteolysis |
| 6-Q5I0K3 | Citrate lyase subunit beta-like protein, mitochondrial | ↑ 2.0 | 0.0214 | Mitochondrion | Vitamin B12 metabolism |
| 7-Q6JAM9 | Transmembrane protein 35A | ↑ 1.8 | 0.0430 | Peroxisome | Modulation of sympathetic neurite outgrowth |
| 8-Q6JP77 | A-kinase anchor protein 7 isoforms delta and gamma | ↑ 1.7 | 0.0017 | Nucleus, cytoplasm, cell membrane | Protein localization |
| 9-Q09073 | ADP/ATP translocase 2 | ↑ 1.7 | 0.0042 | Mitochondrion | ATP:ADP antiporter activity, chromosome segregation |
| 10-P18890 | Rho GTPase-activating protein 39 | ↑ 1.7 | 0.0042 | Cytoplasm | Signal transduction |
| 11-O35412 | Signal-induced proliferation-associated 1-like protein 1 | ↑ 1.7 | 0.0008 | Cell membrane, cytoplasm | Actin cytoskeleton reorganization, regulation of axonogenesis |
| 12-D3ZW56 | Neurofascin | ↓ 3.0 | 0.0057 | Cell membrane | Myelination, axon guidance |
| 13-A0A0G2JWG6 | Golgin B1 | ↓ 2.4 | 0.0234 | Golgi apparatus | Protein localization |
| 14-Q5NDF0 | Protein O-linked-mannose beta-1,4—N-acetylglucosaminyltransferase 2 | ↓ 2.2 | 0.0092 | Endoplasmic reticulum | O-linked glycosylation |
| 15-Q5U2U2 | Crk-like protein | ↓ 1.9 | 0.0174 | Cell membrane, cytoplasm | Signal transduction, hippocampus development, regulation of cell growth |
| 16-B5DFI3 | AP complex subunit sigma | ↓ 1.9 | 0.0188 | Cell membrane, cytoplasm, Golgi apparatus | Protein transport |
| 17-Q62985 | SH2B adapter protein 1 | ↓ 1.9 | 0.0099 | Nucleus, cytoplasm, cell membrane | Signal transduction, tyrosine kinase adaptor activity |
| 18-P05369 | Farnesyl pyrophosphate synthase | ↓ 1.8 | 0.0336 | Cytoplasm | Lipid metabolism, cholesterol biosynthesis |
| 19-O08773 | Regulator of G-protein signaling 14 | ↓ 1.7 | 0.0033 | Cell membrane, cytoplasm, nucleus | Signal transduction, cell division, learning, long-term memory |
| 20-P37377 | Alpha-synuclein | ↓ 1.7 | 0.0075 | Cytoplasm, cell membrane, nucleus | Negative regulation of apoptosis, aging, response to oxidative stress, regulation of dopamine release and transport |
Discussion
2D-DIGE and 2D immunoblot analysis of rat hippocampus from animals exposed to morphine for 10 days (±M10); comparison with animals exposed to morphine and subsequently nurtured for 20 days in the absence of this drug (±M10/−M20)
The number of altered proteins in hippocampus was increased from 6 to 13 after 20 days of abstinence. This finding is just the opposite when compared with that observed in forebrain cortex, where the number of differentially expressed proteins was decreased from 28 (±M10) to 14 (±M10/−M20) when determined in CBB-stained 2D gels or from 113 to 19 when determined by LFQ [10]. Noticeably, the altered level of four proteins identified in (±M10) samples of PNS (β-synuclein, α-synuclein, alpha-enolase and GAPDH) persisted for 20 days since the withdrawal of morphine (Figs 1 and 4, Tables 1 and 3).
The α-synuclein is a pathological protein functionally related to production of “Lewy bodies” in Parkinson´s disease and dementia [28–31]. While its aggregation represents the major risk factor for neurodegeneration, its function under physiological conditions remains poorly understood. It has been suggested that α-synuclein is involved in negative regulation of dopaminergic neurotransmission, synaptic vesicle cycling, synaptic plasticity and neuroprotection [32]. Interestingly, Ziolkowska et al. [33] observed the down-regulation of α-synuclein mRNA in the basolateral amygdala, dorsal striatum, nucleus accumbens and ventral tegmental area of mice withdrawn from chronic morphine treatment for 48 h; on the other hand, protein level of α-synuclein, determined in the same brain regions, was up-regulated. β-synuclein was described as the non-amyloidogenic homolog of α-synuclein with anti-apoptotic effect [34]. The evidence for down-regulation of α/β synucleins persisting for 20 days since morphine withdrawal was confirmed by 2D immunoblot analysis, Fig 6.
In 2010, the group of Dr. Piotr Suder created the Morphinome Database (www.addiction-proteomics.org) in order to facilitate the search for the proteins altered by morphine administration. This database is continuously updated. More than 20% of proteins distinguished in this database are functionally related to regulation of energy metabolism—glycolysis, gluconeogenesis, Krebs cycle and oxidative phosphorylation [35].
In our study, the down-regulation of glycolytic enzyme GAPDH was observed after 10 days of morphine treatment and this decrease was also noticed in hippocampus of rats sacrificed after 20 days of drug withdrawal. Noticeably, the 2D immunoblot analysis recognized the subset of six GAPDH spots (in both ±M10 and ±M10/−M20 samples of PNS) which were present in alkaline region of 2D gels and selectively decreased by morphine (Fig 7). As GAPDH was found to be extensively modified by post-translational modifications [36–40], and post-translational modifications of this enzyme were suggested to play a role in oxidative stress and apoptosis [41,42], it is reasonable to assume that morphine-induced change of six GAPDH subunits reveals different functions fulfilled by individual variants of this enzyme.
One of the functional consequences of manifestation of oxidative stress is the decrease of cellular ATP level and blockade of glycolysis [43]. Accordingly, proteomic analysis of hippocampus from mice exposed to morphine for 10 days indicated a decreased level of three enzymes related to regulation of glycolysis and mitochondria: E2 component of the pyruvate dehydrogenase complex, lactate dehydrogenase 2, and Fe-S protein 1 of NADH dehydrogenase [44]. Further view on literature data dealing with morphine-induced alteration of CNS energy metabolism suggests that chronic morphine treatment impairs the glucose metabolism and ATP production. This effect is associated with morphine withdrawal symptoms and impairment in memory [45].
Our data also indicated an increased level of α-enolase, another glycolytic enzyme participating in a multitude of pathological processes. Increased level of α-enolase was again noticed in (±M10) as well as (±M10/−M20) samples of hippocampus (Tables 1 and 3). Based on comparative proteomic analysis of human, mouse and rat tissues, Díaz-Ramos et al. [46] reported that α-enolase could be regarded as a marker of “pathological stress” proceeding in a range of diseases—cancer, skeletal myogenesis, Alzheimer´s disease, rheumatoid arthritis, inflammatory bowel disease, autoimmune hepatitis and membranous glomerulonephritis. Morphine administration resulted in up-regulation of α-enolase in striatal neuronal cell cultures [47], but the change of this protein was not detected in hippocampus of morphine-treated rats by proteomic analysis of Bierczynska-Krzysik et al. [48].
Apart from protein changes persisting for 20 days after morphine withdrawal, decreased levels of tubulin α-1A chain, cofilin-1 and superoxide dismutase were found in PNS prepared from (±M10/−M20) rats (Table 3). Data of Marie-Claire et al. [49] indicated decreased mRNA and protein level of α-tubulin in rat striatum after chronic morphine treatment, whilst proteomic analysis of Bodzon-Kulakowska et al. [47] indicated the increase of α- and β-chains of tubulin in striatal neuronal cell cultures after morphine exposure. The full understanding of the functional meaning of down- or up-regulation of cytoskeleton protein tubulin α-1A will therefore require further attention.
The large decrease of superoxide dismutase (↓3.1-fold) in hippocampal samples (±M10/−M20) (Table 3) may be interpreted as the evidence of attenuated efficiency of hippocampus recovering from prolonged morphine effect in protection against oxidative stress. Superoxide dismutase, the enzyme catalyzing the conversion of superoxide radical O2─ to O2 and H2O2, represents an important antioxidant system protecting mammalian cells against oxidative damage [50,51]. This protection is necessary as morphine treatment was reported to modulate both enzymatic and non-enzymatic antioxidant defense systems and induce oxidative stress and apoptosis under in vivo conditions [52,53].
Label-free quantification of rat hippocampus from animals exposed to morphine for 10 days (±M10); comparison with animals exposed to morphine and subsequently nurtured for 20 days in the absence of this drug (±M10/−M20)
With the aim to obtain a larger coverage of proteome changes elicited by morphine, we have also used the LFQ technique. The results supported those obtained by gel-based proteomic analysis. The LFQ identified 19 proteins with significantly changed expression level (≥1.7-fold) in (±M10) samples and this number was increased to 20 in (±M10/−M20) samples of hippocampus, Tables 5 and 6, Fig 9. Thus, the lack of reversibility of chronic morphine effect after 20 days of drug withdrawal was confirmed.
What type of underlying mechanism could cause this lack of reversibility? Opioids were reported to cause significant changes of glutamatergic transmission [54], neurogenesis [55], dendritic stability [56] and long-term potentiation [57–59]. Data of Cai et al. [60] indicated that morphine-treatment of mice for 6 days resulted in a decrease of excitatory synapse densities in parallel with an enhancement of densities of inhibitory synapses in hippocampus. This effect was mediated by μ-opioid receptors. In primary hippocampal neurons, the short-term exposure to morphine (0.5–24 h) induced an increase of intracellular levels of reactive oxygen species (ROS) generated by NADPH-oxidase. The increase of ROS was followed by an increase of endoplasmic reticulum stress markers and autophagy. Mattei et al. [12] suggested that i) hippocampus is a vulnerable part of brain susceptible to oxidative damage during aging and chronic stress, ii) prolonged withdrawal may lead to homeostatic changes heading for the restoration of the physiological norm and iii) abuse of morphine may lead to ROS-induced neurodegeneration and apoptosis.
The LFQ analysis of our withdrawal samples (Table 6) also confirmed the results of 2D-DIGE and 2D immunoblot analyses indicating the decrease of α-synuclein level (↓1.7-fold). The α-synuclein was described to participate in negative regulation of apoptosis, aging and oxidative stress [61].
In humans, heroin abuse was shown to increase oxidative stress [62]—plasma concentrations of lipid peroxides (LPO) and nitric oxide were increased in parallel with a decrease of antioxidants, vitamin C, vitamin E and beta-carotene. In erythrocytes, LPO were increased but the activity of antioxidant enzymes superoxide dismutase, catalase and glutathione peroxidase was decreased; in mice [63], heroin treatment resulted in a decrease of total antioxidant capacity in serum and activity of antioxidant enzymes superoxide dismutase, catalase and glutathione peroxidase in brain. Disturbance of pro-antioxidant balance was associated with oxidative damage of brain DNA, proteins and lipids. Addition of exogenous antioxidants attenuated the oxidative stress.
Results presented in our work extend these data as the down-regulation of superoxide dismutase protein level (↓3.1-fold) was detected in hippocampal samples collected from (±M10/−M20) rats (Table 3). As discussed above, the large decrease in the expression level of this anti-oxidant enzyme may be interpreted as a piece of supportive evidence for attenuated efficiency of rat hippocampus recovering from addiction to morphine. Protection against oxidative damage is decreased and return to physiological norm obscured.
Conclusions
The 2D-DIGE analysis identified in the hippocampus of rats exposed to morphine for 10 days (+M10) six altered proteins when compared with samples prepared from control animals (−M10). In rats treated with morphine for 10 days and subsequently nurtured for 20 days in the absence of the drug (+M10/−M20), thirteen altered proteins were detected when compared with control animals (−M10/−M20). Thus, the number of protein spots with changed expression level (≥2-fold) was increased 2-fold after 20 days of abstinence.
This result is just the opposite when compared with that detected in the forebrain cortex, where the number of altered proteins was decreased from 28 (±M10) to 14 (±M10/−M20) when determined in CBB-stained 2D gels or from 113 to 19 when determined by LFQ [10].
Noticeably, the altered expression level of four proteins identified in (±M10) samples (α-synuclein, β-synuclein, α-enolase and GAPDH) persisted for 20 days since the withdrawal of morphine.
Immunoblot analysis of 2D gels by specific antibodies oriented against α/β-synucleins and GAPDH confirmed data obtained by comparative 2D-DIGE and MALDI-TOF MS/MS.
When using LFQ analysis, nineteen proteins with significantly changed expression level (≥1.7-fold) were identified in group (+M10) when compared with group (−M10) and the number of altered proteins was increased to twenty in hippocampal samples collected from rats after 20 days of morphine withdrawal (±M10/−M20).
We conclude that the morphine-induced alteration of protein composition in rat hippocampus after cessation of drug supply proceeds in a different manner when compared with the forebrain cortex. In the forebrain cortex, the total number of altered proteins was decreased after 20 days without morphine, whilst in the hippocampus, it was increased. Thus, the two functionally distinct parts of CNS respond to the disturbance of the homeostatic balance caused by drug addiction in a different manner with the aim to restore the physiological norm.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(PDF)
Acknowledgments
The authors thank to Karel Harant and Pavel Talacko (from the Proteomics Core Facility, BIOCEV, Faculty of Science, Charles University in Prague) for performing MALDI-TOF MS/MS and label-free quantification.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by the Czech Science Foundation GA CR (17-05903S and 19-03295S) and by the institutional project of the Institute of Physiology of the Czech Academy of Sciences (RVO:67985823).
References
- 1.Kosten TR, George TP. The neurobiology of opiod dependence: implications for treatment. Sci Pract Perspect.2002;1:13–20. 10.1151/spp021113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kosten TR, Baxter LE. Review Article: effective management of opioid withdrawal symptoms: A gateway to opioid dependence treatment. Am J Addict.2019;28:55–62. 10.1111/ajad.12862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sim LJ, Selley DE, Dworkin SI, Childers SR. Effects of chronic morphine administration on mu opioid receptor-stimulated [35S] GTPgammaS autoradiography in rat brain. J Neurosci.1996;16:2684–2692. 10.1523/JNEUROSCI.16-08-02684.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sim-Selley LJ, Selley DE, Vogt LJ, Childers SR, Martin TJ. Chronic heroin self-administration desensitizes mu opioid receptor-activated G-proteins in specific regions of rat brain. J Neurosci.2000;20:4555–4562. 10.1523/JNEUROSCI.20-12-04555.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maher CE, Martin TJ, Childers SR. Mechanisms of mu opioid receptor/G-protein desensitization in brain by chronic heroin administration. Life Sci.2005;77:1140–1154. 10.1016/j.lfs.2005.03.004 [DOI] [PubMed] [Google Scholar]
- 6.Bourova L, Vosahlikova M, Kagan D, Dlouha K, Novotny J, Svoboda P. Long-term adaptation to high doses of morphine causes desensitization of μ-OR- and δ-OR-stimulated G-protein response in forebrain cortex but does not decrease the amount of G-protein alpha subunit. Med Sci Monit.2010;16:260–270. [PubMed] [Google Scholar]
- 7.Berger AC, Whister JL. How to design an opioid drug that causes reduced tolerance and dependence. Ann Neurol.2010;67:559–569. 10.1002/ana.22002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ujcikova H, Dlouha K, Roubalova L, Vosahlikova M, Kagan D, Svoboda P. Up-regulation of adenylylcyclases I and II induced by long-term adaptation of rats to morphine fades away 20 days after morphine withdrawal. Biochim Biophys Acta.2011;1810:1220–1229. 10.1016/j.bbagen.2011.09.017 [DOI] [PubMed] [Google Scholar]
- 9.Ujcikova H, Brejchova J, Vosahlikova M, Kagan D, Dlouha K, Sykora J et al. Opioid-receptor (OR) signaling cascades in rat cerebral cortex and model cell lines: the role of plasma membrane structure. Physiol Res. 2014a; 63(Suppl 1):S165–176. [DOI] [PubMed] [Google Scholar]
- 10.Ujcikova H, Vosahlikova M, Roubalova L, Svoboda P. Proteomic analysis of protein composition of rat forebrain cortex exposed to morphine for 10 days; comparison with animals exposed to morphine and subsequently nurtured for 20 days in the absence of this drug. J Proteomics.2016;145:11–23. 10.1016/j.jprot.2016.02.019 [DOI] [PubMed] [Google Scholar]
- 11.Morón JA, Abul-Husn NS, Rozenfeld R, Dolios G, Wang R, Devi LA. Morphine administration alters the profile of hippocampal postsynaptic density-associated proteins. Mol Cell Proteomics.2007;6:29–42. 10.1074/mcp.M600184-MCP200 [DOI] [PubMed] [Google Scholar]
- 12.Mattei V, Martellucci S, Santilli F, Manganelli V, Garofalo T, Candelise N et al. Morphine withdrawal modifies prion protein expression in rat hippocampus. PloS One.2017;12:e0169571 10.1371/journal.pone.0169571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kutlu MG, Gould TJ. Effects of drugs of abuse on hippocampal plasticity and hippocampus-dependent learning and memory: contributions to development and maintenance of addiction. Learn Mem.2016;23:515–533. 10.1101/lm.042192.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malberg JE, Eisch AJ, Nestler EJ, Duman RS. Chronic antidepressant treatment increases neurogenesis in adult rat hippocampus. J Neurosci.2000;20:9104–9110. 10.1523/JNEUROSCI.20-24-09104.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boldrini M, Fulmore CA, Tartt AN, Simeon LR, Pavlova I, Popovska V et al. Human hippocampal neurogenesis persists throughout aging. Cell Stem Cell.2018;22:589–599. 10.1016/j.stem.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eriksson PS, Perfilieva E, Bjӧrk-Eriksson T, Alborn AM, Nordborg C, Peterson DA et al. Neurogenesis in the adult human hippocampus. Nat Med.1998;4:1313–1317. 10.1038/3305 [DOI] [PubMed] [Google Scholar]
- 17.Ernst A, Alkass K, Bernard S, Salehpour M, Perl S, Tisdale J et al. Neurogenesis in the striatum of the adult human brain. Cell.2014;156:1072–1083. 10.1016/j.cell.2014.01.044 [DOI] [PubMed] [Google Scholar]
- 18.Ponti G, Peretto P, Bonfanti L. Genesis of neuronal and glial progenitors in the cerebrall cortex of peripuberal and adult rabbits. PloS One.2008;3:e2366 10.1371/journal.pone.0002366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tannu NS, Hemby SE. Two-dimensional fluorescence difference gel electrophoresis for comparative proteomics profiling. Nat Protoc.2006;1:1732–1742. 10.1038/nprot.2006.256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jágr M, Eckhardt A, Pataridis S, Foltan R, Mysak J, Miksik I. Proteomic analysis of human tooth pulp proteomes–comparison of caries-resistant and caries-susceptible persons. J Proteomics.2016;145:127–136. 10.1016/j.jprot.2016.04.022 [DOI] [PubMed] [Google Scholar]
- 21.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Moll Cell Proteomics.2014;13:2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ujcikova H, Eckhardt A, Kagan D, Roubalova L, Svoboda P. Proteomic analysis of post-nuclear supernatant and percoll-purified membranes prepared from brain cortex of rats exposed to increasing doses of morphine. Proteome Sci. 2014b;12:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fountoulakis M, Takács MF, Berndt P, Langen H, Takács B. Enrichment of low abundance proteins of Escherichia coli by hydroxyapatite chromatography. Electrophoresis.1999;20:2181–2195. [DOI] [PubMed] [Google Scholar]
- 24.Ujcikova H, Hlouskova M, Cechova K, Stolarova K, Roubalova L, Svoboda P. Determination of μ-, δ- and κ-opioid receptors in forebrain cortex of rats exposed to morphine for 10 days: Comparison with animals after 20 days of morphine withdrawal. PloS One.2017; 12:e0186797 10.1371/journal.pone.0186797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Person MD, Shen J, Traner A, Hensley SC, Lo HH, Abbruzzese JL et al. Protein fragment domains identified using 2D gel electrophoresis/MALDI-TOF. J Biomol Tech.2006;17:145–156. [PMC free article] [PubMed] [Google Scholar]
- 26.Moravcova Z, Rudajev V, Stohr J, Novotny J, Cerny J, Matousek P et al. Long-term agonist stimulation of IP prostanoid receptor depletes the cognate Gsα protein from membrane domains but does not affect the receptor level. Biochim Biophys Acta.2004;1691:51–65. 10.1016/j.bbamcr.2003.12.004 [DOI] [PubMed] [Google Scholar]
- 27.Rudajev V, Novotny J, Hejnova L, Milligan G, Svoboda P. Thyrotropin-releasing hormone receptor is excluded from lipid domains. Detergent-resistant and detergent-sensitive pools of TRH receptor and Gqα/G11α protein. J Biochem.2005;138:111–125. [DOI] [PubMed] [Google Scholar]
- 28.Stefanis L. Alpha-synuclein in Parkinson´s disease. Cold Spring Harb Percpect Med.2012; 2(2):a009399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bendor J, Logan T, Edwards RH. The function of α-synuclein. Neuron.2013;79:1044–1066 10.1016/j.neuron.2013.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taguchi K, Watanabe Y, Tsujimura A, Tatebe H, Miyata S, Tokuda T et al. Differential expression of alpha-synuclein in hippocampal neurons. PLoS One.2014;9(2):e89327 10.1371/journal.pone.0089327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taguchi K, Watanabe Y, Tsujimura A, Tanaka M. Brain region-dependent differential expression of alpha-synuclein. J Comp Neurol.2016;524:1236–1258. 10.1002/cne.23901 [DOI] [PubMed] [Google Scholar]
- 32.Monti B, Polazzi E, Batti L, Crochemore C, Virgili M, Contestabile A. Alpha-synuclein protects cerebellar granule neurons against 6-hydroxydopamine-induced death. J Neurochem.2007;103:518–530. 10.1111/j.1471-4159.2007.04778.x [DOI] [PubMed] [Google Scholar]
- 33.Ziolkowska B, Gieryk A, Bilecki W, Wawrzcak-Bargiela A, Wedzony K, Chocyk A et al. Regulation of α-synuclein expression in limbic and motor brain regions of morphine-treated rats. J Neurosci.2005;25:4996–5003. 10.1523/JNEUROSCI.4376-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.da Costa CA, Masliah E, Checler F. β-synulein displays an antiapoptotic p53-dependent phenotype and protects neurons from 6-hydroxydopamine-induced caspase 3 activation. J Biol Chem.2003;278:37330–37335. 10.1074/jbc.M306083200 [DOI] [PubMed] [Google Scholar]
- 35.Antolak A, Bodzon-Kułakowska A, Cetnarska E, Pietruszka M, Marszałek-Grabska M, Kotlińska J et al. Proteomic data in morphine addiction versus real protein activity: metabolic enzymes. J Cell Biochem.2017;118:4323–4330. 10.1002/jcb.26085 [DOI] [PubMed] [Google Scholar]
- 36.Kim SY, Chudapongse N, Lee SM, Levin MC, Oh JT, Park HJ et al. Proteomics analysis of phosphotyrosyl proteins in morphine-dependent rat brains. Brain Res Mol Brain Res.2005;133:58–70. 10.1016/j.molbrainres.2004.09.018 [DOI] [PubMed] [Google Scholar]
- 37.Huang Q, Lan F, Zheng Z, Xie F, Han J, Dong L et al. AKT2 kinase suppresses glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mediated-apoptosis in ovarian cancer via phosphorylating GAPDH at threonine 237 and decreasing its nuclear translocation. J Biol Chem.2011;286:42211–42220. 10.1074/jbc.M111.296905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tristan C, Shahani N, Sedlak TW, Sawa A. The diverse functions of GAPDH: views from different subcellular compartments. Cell Signal.2011;23:317–323. 10.1016/j.cellsig.2010.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li T, Liu M, Feng X, Wang Z, Das I, Xu Y et al. Glyceraldehyde-3-phosphate is activated by lysine 254 acetylation in response to glucose signal. J Biol Chem.2014;289:3775–3785. 10.1074/jbc.M113.531640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kunjithapatham R, Geschwind JF, Devine L, Boronina TN, O’Meally RN, Cole RN et al. Occurence of a multimer high-molecular-weight glyceraldehyde-3-phosphate dehydrogenase in human serum. J Proteome Res.2015;14:1645–1656. 10.1021/acs.jproteome.5b00089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chuang DM, Hough C, Senatorov VV. Glyceraldehyde-3-phosphate dehydrogenase, apoptosis, and neurodegenerative diseases. Annu Rev Pharmacol Toxicol.2005;45:269–290. 10.1146/annurev.pharmtox.45.120403.095902 [DOI] [PubMed] [Google Scholar]
- 42.Bell RAV, Smith JC, Storey KB. Purification and properties of glyceraldehyde-3-phosphate dehydrogenase from the skeletal muscle of the hibernating ground squirrel, Ictidomys tridecemlineatus. PeerJ.2014;2:e634 10.7717/peerj.634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hwang NR, Yim SH, Kim YM, Jeong J, Song EJ, Lee Y et al. Oxidative modifications of glyceraldehyde-3-phophate dehydrogenase play a key role in its multiple cellular functions. Biochem J.2009;423:253–264. 10.1042/BJ20090854 [DOI] [PubMed] [Google Scholar]
- 44.Chen XL, Lu G, Gong YX, Zhao LC, Chen J, Chi ZQ et al. Expression changes of hippocampal energy metabolism enzymes contribute to behavioral abnormalities during chronic morphine treatment. Cell Res.2007;17:689–700. 10.1038/cr.2007.63 [DOI] [PubMed] [Google Scholar]
- 45.Jiang X, Li J, Ma L. Metabolic enzymes link morphine withdrawal with metabolic disorder. Cell Res.2007;17:741–743. 10.1038/cr.2007.75 [DOI] [PubMed] [Google Scholar]
- 46.Díaz-Ramos A, Borrellas AR, García-Melero A, López-Alemany R. α-enolase, a multifunctional protein: its role on pathophysiological situations. J Biomed Biotechnol.2012; 2012:156795 10.1155/2012/156795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bodzon-Kułakowska A, Suder P, Mak P, Bierczynska-Krzysik A, Lubec G, Walczak B et al. Proteomic analysis of striatal neuronal cell cultures after morphine administration. J Sep Sci.2009;32:1200–1210. 10.1002/jssc.200800464 [DOI] [PubMed] [Google Scholar]
- 48.Bierczynska-Krzysik A, Pradeep John JP, Silberring J, Kotlinska J, Dylag T, Cabatic M et al. Proteomic analysis of rat cerebral cortex, hippocampus and striatum after exposure to morphine. Int J Mol Med.2006;18:775–784. [PubMed] [Google Scholar]
- 49.Marie-Claire C, Courtin C, Roques BP, Noble F. Cytoskeletal genes regulation by chronic morphine treatment in rat striatum. Neuropsychopharmacology.2004;29:2208–2215. 10.1038/sj.npp.1300513 [DOI] [PubMed] [Google Scholar]
- 50.Popa-Wagner A, Mitran S, Sivanesan S, Chang E, Buga AM. OS and brain diseases: the good, the bad, and the ugly. Oxid Med Cell Longev.2013;2013:963520 10.1155/2013/963520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skrabalova J, Drastichova Z, Novotny J. Morphine as a potential oxidative stress-causing agent. Mini Rev Org Chem.2013;10:367–372. 10.2174/1570193X113106660031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Payabvash S, Behehstian A, Salmazi AH, Kihumehr S, Ghahremani MH, Tavangar SM et al. Chronic morphine treatment induces oxidant and apoptotic damage in the mice liver. Life Sci.2006;79:972–980. 10.1016/j.lfs.2006.05.008 [DOI] [PubMed] [Google Scholar]
- 53.Ma J, Yuan X, Qu H, Zhang J, Wang D, Sun X et al. The role of reactive oxygen species in morphine addiction of SH-SY5Y cells. Life Sci.2015;124:128–135. 10.1016/j.lfs.2015.01.003 [DOI] [PubMed] [Google Scholar]
- 54.Xu NJ, Bao L, Fan HP, Bao GB, Pu L, Lu YJ et al. Morphine withdrawal increases glutamate uptake and surface expression of glutamate transporter GLT1 at hippocampal synapses. J Neurosci.2003;23:4775–4784. 10.1523/JNEUROSCI.23-11-04775.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eisch AJ, Barrot M, Schad CA, Self DW, Nestler EJ. Opiates inhibit neurogenesis in the adult rat hippocampus. Proc Natl Acad Sci U S A.2000;97:7579–7584. 10.1073/pnas.120552597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liao D, Lin H, Law PY, Loh HH. Mu-opioid receptors modulate the stability of dendritic spines. Proc Natl Acad Sci U S A.2005;102:1725–1730. 10.1073/pnas.0406797102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mansouri FA, Motamedi F, Fathollahi Y, Atapour N, Semnanian S. Augmentation of LTP induced by primed-bursts tetanic stimulation in hippocampal CA1 area of morphine dependent rats. Brain Res.1997;769:119–124. 10.1016/s0006-8993(97)00608-2 [DOI] [PubMed] [Google Scholar]
- 58.Mansouri FA, Motamedi F, Fathollahi Y. Chronic in vivo morphine administration facilitates primed-bursts-induced long-term potentiation of Schaffer collateral-CA1 synapses in hippocampal slices in vitro. Brain Res.1999;815:419–423. 10.1016/s0006-8993(98)01148-2 [DOI] [PubMed] [Google Scholar]
- 59.Pu L, Bao GB, Xu NJ, Ma L, Pei G. Hippocampal long-term potentiation is reduced by chronic opiate treatment and can be restored by re-exposure to opiates. J Neurosci.2002;22:1914–1921. 10.1523/JNEUROSCI.22-05-01914.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cai Y, Yang L, Hu G, Chen X, Niu F, Yuan L et al. Regulation of morphine-induced synaptic alterations: role of oxidative stress, ER stress, and autophagy. J Cell Biol.2016;215:245–258. 10.1083/jcb.201605065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maries E, Dass B, Collier TJ, Kordower JH, Steece-Collier K. The role of α-synuclein in Parkinson´s disease: insights from animal models. Nat Rev Neurosci.2003;4:727–738. 10.1038/nrn1199 [DOI] [PubMed] [Google Scholar]
- 62.Zhou JF, Yan XF, Ruan ZR, Peng FY, Cai D, Yuan H et al. Heroin abuse and nitric oxide, oxidation, peroxidation, lipoperoxidation. Biomed Envir Sci.2000;13:131–139. [PubMed] [Google Scholar]
- 63.Xu B, Wang Z, Li G, Li B, Lin H, Zheng R et al. Heroin-administered mice involved in oxidative stress and exogenous antioxidant-alleviated withdrawal syndrome. Basic Clin Pharmacol Toxicol.2006;99:153–161. 10.1111/j.1742-7843.2006.pto_461.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(PDF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.