Abstract
The mitochondrial genomes of flowering plants are well known for their large size, variable coding-gene set and fluid genome structure. The available mitochondrial genomes of the early angiosperms show extreme genetic diversity in genome size, structure, and sequences, such as rampant HGTs in Amborella mt genome, numerous repeated sequences in Nymphaea mt genome, and conserved gene evolution in Liriodendron mt genome. However, currently available early angiosperm mt genomes are still limited, hampering us from obtaining an overall picture of the mitogenomic evolution in angiosperms. Here we sequenced and assembled the draft mitochondrial genome of Magnolia biondii Pamp. from Magnoliaceae (magnoliids) using Oxford Nanopore sequencing technology. We recovered a single linear mitochondrial contig of 967,100 bp with an average read coverage of 122 × and a GC content of 46.6%. This draft mitochondrial genome contains a rich 64-gene set, similar to those of Liriodendron and Nymphaea, including 41 protein-coding genes, 20 tRNAs, and 3 rRNAs. Twenty cis-spliced and five trans-spliced introns break ten protein-coding genes in the Magnolia mt genome. Repeated sequences account for 27% of the draft genome, with 17 out of the 1,145 repeats showing recombination evidence. Although partially assembled, the approximately 1-Mb mt genome of Magnolia is still among the largest in angiosperms, which is possibly due to the expansion of repeated sequences, retention of ancestral mtDNAs, and the incorporation of nuclear genome sequences. Mitochondrial phylogenomic analysis of the concatenated datasets of 38 conserved protein-coding genes from 91 representatives of angiosperm species supports the sister relationship of magnoliids with monocots and eudicots, which is congruent with plastid evidence.
Introduction
Plant mitochondrial (mt) genomes are about 100–10,000 times larger than those of animals and are structurally more complex due to frequent ongoing recombinations [1]. The notably large size of plant mt genomes is shaped by a combination of several factors, including a rich gene set along with the abundant introns that carries, and the capability of uptaking and integrating intracellular transferred sequences from the chloroplast [2] and nucleus [3], and horizontally transferred genes from foreign donors [4, 5]. Based on the database of the available plant mt genomes (https://www.ncbi.nlm.nih.gov/genome/organelle/), species from each of the three bryophyte lineage hold rather stable mt genome size, conserved gene content, and similar gene order [6–8], whereas the mt genomes of vascular plants demonstrate significant genome size variation, gene set variability, and structural dynamics [9–11]. In particular, vascular plant mt genomes range in size from 66 Kb [12] to 11.3 Mb [13] with encoded genes from 13 to 64 [14]. Neither of the two vascular plant mt genomes sequenced to date, even those of different accessions from the same species, share the same gene order [8], which is in stark contrast to the conserved structural evolution of the plastid genomes of the land plants [15].
The structural fluidity of vascular plant mt genomes is associated with the recombination activity of the repeated sequences [9, 16], such as, it has been proposed that intragenomic homologous recombination via inverted repeats would lead to an inversion, and direct repeats lead to a subdivision of the main genome into sub-circles [17]. As a result, vascular plant mt genome generally contains, in coexistence of the master circle conformation, a variety of rearranged molecules (alternative conformation) in substoichiometric levels. If one of those structural variants is passed on to the progeny, then the gene order might be changed within two generations [18], as observed in a few species with large DNA insert libraries [3, 9, 13, 18–25] or/and third-generation sequencing reads [26, 27]. These studies also suggested repeat recombination frequency to be associated with the repeat length and identity. Generally, large repeats (>1000 bp) with high sequence similarity tend to recombine more frequently, medium repeats (100–1000 bp) recombine occasionally, and small repeats (<100 bp) rarely. Some studies have also suggested the adaptive value of repeat recombination against desiccation in vascular plant mitochondrion [28]. However, it remains a big challenge to predict the specific repeats recombining and to study the functional consequences of the mitochondrial repeat recombination, due to limited data.
Angiosperms, with nearly 250,000 species, represent the most diverse of all major lineages of land plants and the dominant vegetation in earth’s terrestrial ecosystems [29]. It would be of considerable interest to understand the genome evolution of the nuclear, plastid and mitochondrial of angiosperms, especially those of the early diverging lineages. The Four available mt genomes of the early angiosperms show extreme diversities on many aspects: The enormous, 3.9-Mb mt genome of Amborella trichopoda contains six genome equivalents of foreign mtDNAs, acquired from green algae, mosses, and other angiosperms [5]. The 617-Kb mt genome of Nymphaea colorata holds the most abundant repeats (~50% of genome, 83,705 repeats) among that of the land plants whereas only a few of these show recombinational evidence [27]. The 1.1-Mb mt genome of Schisandra sphenanthera (NC_042758, unpublished) holds huge portions of promiscuous sequences (656 Kb, 60%), but small amount of repeats (49 Kb, 4%). The 553-Kb mt genome of Liriodendron tulipifera is conserved in gene content and gene order with extraordinarily low mutation rate [30]. Expanded sampling of the mt genomes of early angiosperms would allow more insights into the mitogenomic diversity and evolution of angiosperms.
As a phylogenetically early assemblage of angiosperms, magnoliids contain remnants of many of the oldest lineages of angiosperms and occupy a pivotal position in the phylogeny of angiosperms. Recently, two independent phylogenomic analyses including each of the two newly reported nuclear genomes of magnoliids have led to controversial taxonomic placements of magnoliids [31–33]. Specifically, magnoliids (with Cinnamomum kanehirae as the only representative) is resolved as the sister to eudicots with relatively strong support [32], which is consistent with the result of the phylotranscriptomic analysis of the 1–kp data [34] and of 20 representative transcriptomes [35]. Alternatively, magnoliids (with Liriodendron as the only representative) is resolved as the sister to eudicots and monocots with weak support [33], which is congruent with the plastome phylogenomic analysis of land plants and Viridiplantae [36, 37]. The controversial taxonomic placements of magnoliids relative to monocots and eudicots between plastid and nuclear evidence need to be further tested with mitochondrial phylogenomic analyses. The slow-evolving, uniparentally-inherited, non-recombining mitochondrial genome sequences are less suffered from the effects of substitution saturation, incomplete lineage sorting, and hybridization commonly seen in nuclear markers, therefore are more suitable for phylogenetic inferences of higher taxonomic categories [38]. In addition to sequence level, plant mt genomes can also provide phylogenetic information on the structure level. The accumulation of the mt genomes of angiosperms, especially those of early diverging lineages would provide us a good opportunity to examine the phylogenetic position of magnoliids using mitochondrial phylogenomic analyses.
Magnolia biondii Pamp. (Magnoliaceae, magnoliids) is a deciduous tree species widely grown and cultivated in the north-temperate regions of China for its ornamental and pharmaceutical values. The dried flower buds of M. biondii (herbal name, Xin-Yi) are a traditional Chinese medicine with a long history of clinical use in the treatment of allergic rhinitis and sinusitis [39]. Modern phytochemical studies have characterized the chemical constitutes of the volatile oil [40], lignans [41], and alkaloids [42] from different parts of the plant M. biondii, whereas the genetic background of this species is still understudied with only the plastid genome (KY085894, Unpublished) deposited in the GenBank. Here we sequenced and assembled the draft mt genome of M. biondii using the Oxford Nanopore sequencing technology to study the mitogenomic diversity and the evolution of the early flowering plants.
Materials and methods
Mitochondrial genome assembly and annotation
The mt genome of M. biondii was obtained from the genome project of M. biondii led by Shouzhou Zhang (unpublished data). The total genomic DNA of M. biondii was extracted using a modified CTAB method [43] and quality controlled using Agarose gel electrophoresis and Nanodrop 2000 Spectrophotometer (Thermo Fisher Scientific, USA). Single molecule sequencing of the Magnolia genomic DNA was performed on the Oxford Nanopore PromethION sequencing platform in Nextomics (Nextomics Biosciences Co., Ltd., Wuhan, China). The raw reads in fastq format were corrected, trimmed, and de novo assembled using Canu [44]. One mt contig of 967,100 bp, with an average read coverage of 122 × (SRR9720304, S1A Fig), was retrieved from the genome assembly results with Blastn using the 41 protein-coding genes of Liriodendron tulipifera (KC821969) as the reference. This mt contig was further polished with 10X genomics reads (S1 Table) generated by BGI-SEQ500 (BGI, Shenzhen) using software Pilon [45] for three rounds of error correction. The resultant mt contig was elongated in both ends with Canu corrected long reads using BWA [46], yielding a circular molecule of 995,279 bp (S1B Fig). We mapped all the corrected genome reads to the circular molecule, but observed uneven read coverage of this putative mt genome. The newly elongated region received very low coverage (~7 ×) in the reads mapping file (SRR9720674, S1B Fig). Therefore, to be cautious, our subsequent analyses were based on the corrected mt contig of 967,100 bp.
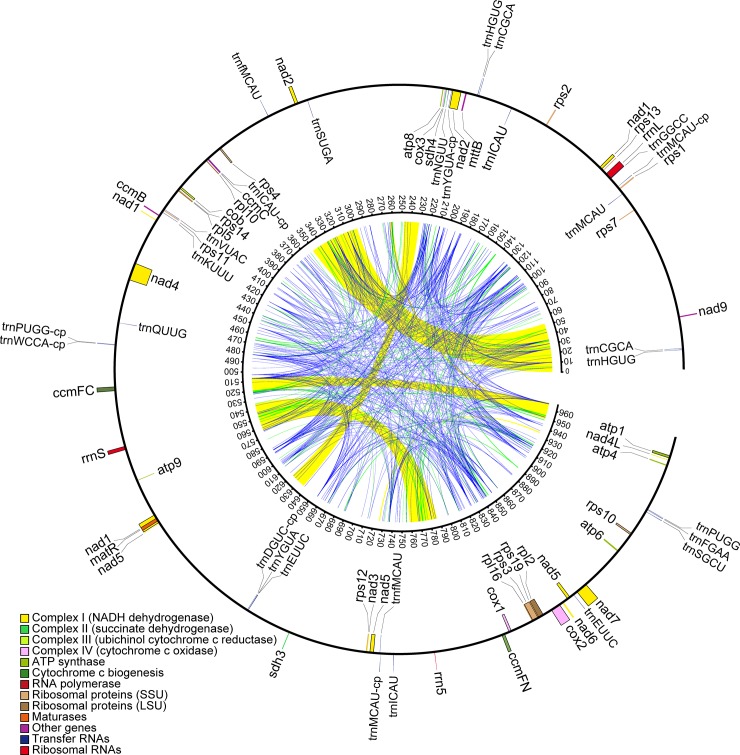
The annotation for the draft mt genome of Magnolia was performed as previously described [6, 47]. Protein coding genes and rRNA genes were annotated by Blastn searches of the non-redundant database at National Center for Biotechnology Information (NCBI). The exact gene and exon/intron boundaries were confirmed in Geneious software (v.10.0.2, Biomatters, www.geneious.com) by mapping the RNA-seq reads (S1 Table) to the mt genome of Magnolia using Bowtie2 [48] and further validated by aligning each gene to its orthologs from available annotated plant mitochondrial genomes at the NCBI website (www.ncbi.nlm.nih.gov/genome/organelle). The tRNA genes were detected using tRNAscan-SE 2.0 [49]. Nuclear and plastid homologous sequences were annotated by searching the Magnolia mt genome against the nuclear (unpublished) and the chloroplast genome of M. biondii (KY085894, unpublished) using Blastn with an e-value cut-off of 1e-6. The mtDNA sharing of Magnolia with the mt genomes of other angiosperms was also performed with Blastn with the same parameters using the intergenic spacer regions of Magnolia mt genome as the query. The annotated Magnolia mt genome was submitted to GenBank under the accession number of MN206019 and visualized using OGDraw 1.2 [50] to generate the genome map (Fig 1).
Fig 1. Draft mitochondrial genome map of Magnolia biondii.
The total length of the Magnolia draft mt genome is 967,100 bp. Genes (exons are shown as closed boxes) shown outside the curve are transcribed clockwise, whereas those inside are transcribed counter-clockwise. Genes from the same protein complex are colored the same, introns are indicated in white boxes, and tRNAs of chloroplast origin are noted with a ‘-cp’ suffix. Repeat distributions and occurrences are show inside the gene map. Large repeats >1,000 bp in length are indicated in yellow, medium-sized repeats in the range of 100–1,000 bp in length are indicated in green, and small repeats <100bp in length are colored blue. Numbers on the inner curve represent genome coordinates (Kb).
Repeats and repeat-mediated homologous recombinations
Repeat identification of Magnolia and other angiosperm plant mt genomes was carried out using the python tools as described by Wynn & Christensen [28]. Repeats were counted in three categories, large repeats above 1000 bp, medium repeats in the range of 100–1000 bp, and small repeats between 50 and 100 bp. For the detection of the ongoing repeat-mediated intragenomic recombinations, we set up a mt read database from all the corrected Nanopore reads. We used the Magnolia mt genome sequence as the reference to blast the total genomic read database with an e-value cut-off of 1e-6 for the extraction of mt reads. Finally, we got a mt read database of 174,003 reads with an average length of 22,527 bp, and a total length of 3,919,721,246 bp.
Repeat-mediated homologous recombinations were evaluated for those repeat pairs ranging from 50 to 29,306 bp with blast identity > 85% following Dong et al. [27]. Specifically, for each repeat pair, we built four or eight reference sequences, each with 1000 bp up- and down-stream of the two template sequences (original sequences), and two (for repeat pair with identity equals100%) or six (for repeat pair with identity less than100%) recombinant sequences (alternative conformations) constructed from the putative recombination products, respectively (S3 Fig). Then, we searched these recombinant sequences against the Magnolia mt genome sequence to remove those located in the genome. After that, we blasted the remaining reference sequences against the Magnolia mt reads database, and count the number of matching reads with blast identity > 95%, and a hit coverage of at least 200 bp in both flanking regions of each repeat sequence.
Phylogenetic analysis
For mitochondrial phylogenomic analyses of angiosperms, we downloaded 82 representative mt genomes of vascular plants from the NCBI Organelle Genome Resources database (http://www.ncbi.nlm.nih.gov/genome/organelle/), including two gymnosperm outgroups and 81 angiosperm ingroups with only one representative per genus. These representative mt genomes were selected based on the quality of annotation and the number of encoded-genes. These mt genomes comprise 24 angiosperm orders with an emphasis on eudicots (16 orders) and monocots (4 orders). The early angiosperms were represented by only five species: Liriodendron tulipifera, Magnolia biondii, Schisandra sphenanthera, Nymphaea colorata, and Amborella trichopoda. To have a good representation of magnoliids and early angiosperms, we downloaded 10 SRA accessions of whole genome sequencing sequences from magnoliids (5), Austrobaileyales (2), Nymphaeales (2), and Chloranthaceae (1). Overall, the mitochondrial phylogenomic analyses in our study comprised 91 representatives of angiosperms, which represented 91 genera, 43 families, 28 orders of APG IV [51]. Our sampling covers all the three so-called ANA grade (Amborelllales, Nymphaeales, Austrobaileyales) [52], and all the five mesangiosperms (Ceratophyllum, Chloranthales, magnoliids, eudicots, and monocots) lineages but Ceratophyllum for which the available sequencing data is from the targeted sequencing of the nuclear genes and yielded no mt genes for our study. We included 7 representatives from magnoliids, covering all the four orders, Canellales (representated by Warburgia ugandensis), Laurales (representated by Cinnamomum micranthum f. kanehirae and Persea americana), Magnoliales (representated by Magnolia biondii and Liriodendron tulipifera), and Piperales (representated by Peperomia macraeana and Piper auritum). This taxonomic sampling scheme was designed to reconstruct an overall angiosperm phylogeny, and to infer the phylogenetic relationship of magnoliids relative to monocots and eudicots.
For the downloaded mt genomes, we extracted 38 conserved mitochondrial protein-coding genes in Geneious 10.0.0 (www.geneious.com) for subsequent phylogenetic analysis, including, atp1, atp4, atp6, atp8, atp9, ccmB, ccmC, ccmFC, ccmFN, cob, cox1, cox2, cox3, matR, mttB, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, nad7, nad9, rpl10, rpl16, rpl2, rpl5, rps1, rps3, rps4, rps7, rps10, rps12, rps13, rps14, rps19, and sdh4. For the SRA sequencing reads, we extract the conserved mt genes using bioinformatics pipeline Hybpiper [53] with the protein sequences of 38 conserved mt genes from 40 representative angiosperms as the bait references. All the gene matrices were parsed with custom Perl script to remove those harboring premature stop codons and blasted against NCBI nucleotide database to remove potentially HGTs. All the genes were firstly evaluated for substitution saturation using DAMBE5 [54] for three codon positions, respectively. As substitution saturation was not detected for any of these mt genes, we included all the mt genes in our phylogenetic analyses.
Each mitochondrial gene was aligned using a local version of TranslatorX [55]. The program first translates the nucleotide sequence into an amino acids sequence using the standard genetic code, and then uses MAFFT [56] to create an amino acid alignment. The alignment is further trimmed for ambiguous portions by GBLOCKS [57] with the least stringent settings. The cleaned amino acid alignment is then used as a guide to generate the nucleotide sequence alignment. The resulted individual mitochondrial alignments were concatenated into combined datasets using the software FASconCAT-G [58]. The concatenated datasets for amino acids (AA) and nucleotides (NT) were analyzed using Partitionfinder [59] for best-fit models and partition schemes, and RAxML v7.2.3 [60] for subsequent phylogenetic tree reconstruction with the maximum likelihood (ML) method with 500 bootstrap replicates. Bayesian inferences were performed in MrBayes [61] using the same partition schemes and best-fit models as estimated by Partitionfinder [59]. In both cases, two independent analyses were run for a total of 10,000,000 generations of Monte Carlo Markov chains and a sampling frequency of 1000 generation. After discarding the first 25% of the trees as burn-in, maximum credibility trees were constructed using TreeAnnnotator v.1.7.5 [62], visualized and rooted in Figtree v1.4.1 [63].
Results and discussion
Genome sequencing and assembly
Nanopore sequencing of the total genomic DNA produced 12,836,970 reads with an average read length of 13,492 bp (S1 Table). After the correction step, we got 5,858,689 reads with an average length of 14,839 bp. The corrected reads were trimmed and de novo assembled in Canu [44]. After that, we retrieved from the genome assembly a single linear mt contig of 967,100 bp with an average read coverage of 122 × (S1A Fig). This mt contig was further corrected using paired-end reads generated by BGI-SEQ500 (BGI, Shenzhen) using the software Pilon [45]. There are several large repeats in the corrected mt contig, including one direct repeat of 52 Kb (in the position of 1–52,072 bp and 298,360–350,756 bp), which is too long even for Nanopore reads to bridge across. The extension of the left side of the mt contig would lead to the duplication of the region between positions 52,073 and 298,359 bp (S2A Fig). Therefore, the mt contig was elongated only in right end and the extended sequence revealed overlaps with the region of 95,120–155,401 bp, yielding a circular molecule of 995,279 bp (S1 Fig). However, the elongated region received very low coverage (~7 ×) in the whole genome read mapping file (S1B Fig), indicating that this putative circular molecule might be an alternative conformation in substoichiometric level. Although in vivo existences of the linear or/and branched mt genomes were proposed [64] and could also take place in Magnolia mitochondrion, we prudently decided to refer to the original mt contig as the draft mt genome of Magnolia.
Genome size and gene content
The draft mt genome of Magnolia is a linear molecule of 967,100 bp with a GC ratio of 46.6% (Genbank accession: MN206019; Fig 1). This is nearly twice the size of Liriodendron with a genome size of 554 Kb (Table 1). The relatively large genome size of Magnolia is associated with the expansion of the intergenic spacers that reached 890 Kb, accounting for 92% of the genome size. The amount and the proportion of the intergenic spacers of Magnolia are notably larger than that of the Nymphaea (519 Kb, 84%) and Liriodendron (479 Kb, 85%). The intergenic spacer regions are usually packed with repeated sequences, nuclear and plastid transferred sequences, horizontally transferred sequences, and promiscuous sequences of unknown origin. The Magnolia mitogenomic spacers contain a total of 1,145 identified repeat sequences, accumulating to 262 Kb (30% of the spacers), which is slightly less than that of the highly repetitive Nymphaea, but two times larger than that of the Liriodendron and four times larger than that of Schisandra. The nuclear and plastid homologous sequences of Magnolia mitogenomic spacers add up to 288 Kb (32% of the spacer regions), and 26 Kb (3% of spacer regions), respectively. In contrast to the relatively large amount of nuclear homologous sequences in other three early angiosperms, Magnolia nuclear genome sequence transfers might not play such a significant role in the spacer expansion of its mt genome.
Table 1. General features of the mitochondrial genomes of the five early angiosperms.
| Genome feature | Amborella trichopoda | Nymphaea colorata | Schisandra sphenanthera | Liriodendron tulipifera | Magnolia biondii |
|---|---|---|---|---|---|
| Accession | KF754799–KF754803 | KY889142 | NC_042758 | KC821969 | MN206019 |
| Size (bp) | 3,866,039 | 617,195 | 1,101,768 | 553,721 | 967,100 |
| GC% | 45.90% | 45.10% | 46.40% | 47.70% | 46.60% |
| Genes | 63 | 64 | 64 | 64 | 64 |
| tRNAs | 20 | 20 | 20 | 20 | |
| rRNAs | 3 | 3 | 3 | 3 | 3 |
| Protein coding genes | 40 | 41 | 41 | 41 | 41 |
| Cis-spliced introns | 19 | 19 | 19 | 20 | 20 |
| Trans-spliced introns | 6 | 6 | 6 | 5 | 5 |
| Gross length of repeats (Kb) | 914 (24%) | 302 (49%) | 49 (4%) | 86 (16%) | 264 (27%) |
| Plastid-derived (Kb) | 138 (4%) | 13 (4%) | 43 (4%) | 25 (4%) | 26 (3%) |
| Nuclear-homologous sequences of the spacers (Kb) | 1,486 (38%) | 429 (70%) | — | 427 (77%) | 289 (30%) |
| Total gene length (Kb) | 78 (2%) | 98 (16%) | 78 (7%) | 75 (14%) | 77 (8%) |
| Protein exons (Kb) | 34 (1%) | 36 (6%) | 36 (3%) | 35 (6%) | 35 (4%) |
| Cis-spliced intron length (Kb) | 39 (1%) | 55 (9%) | 33 (3%) | 34 (6%) | 37 (4%) |
Magnolia shares 80% (595 Kb) of its spacer region with the other sequenced plant mt genomes. Among early angiosperms, Magnolia mt genome shares its intergenic spacers the most with that of the Liriodendron (358 Kb), followed by Amborella (203 Kb), Schisandra (148 Kb), and finally, Nymphaea (45 Kb). The high mtDNA sharing level between Magnolia and Liriodendron might reflect their relatively recent divergence time (ca. 55 Mya, www.timetree.org) and lower sequence turnover rate [23]. Angiosperm mt genomes are highly divergent because rapid structural evolution induced by recombinations could frequently result in losses of gene synteny as well as the mtDNA sequence fragments [19]. For example, in Fabaceae, the average amount of mtDNA sharing among species with a divergence time of 50 Mya is ca. 170 Kb [65], which is only half that between Magnolia and Liriodendron.
The Magnolia mt genome encodes 64 unique genes, including 41 protein coding genes, 20 tRNAs (14 mitochondrial native and 6 plastid derived), and 3 rRNAs (rrn5, rrn18, and rrn26) (Table 1). Total gene length adds up to 8% of the total mt genome length, with protein-coding regions comprising only 4% (35 Kb) of the genome length. In general, the gene content of Magnolia is very similar to the other published mt genomes of early angiosperms, especially to Liriodendron [66], Schisandra (NC_042758) and Nymphaea [27]. The Magnolia mt genome contains 25 group II introns disrupting 10 genes, including 20 cis-spliced and five trans-spliced introns (nad1i394g2, nad1i669g2, nad2i542g2, nad5i1455g2, nad5i1477g2), which is identical to the intron set of Liriodendron, but differs from Schisandra, Nymphaea and Amborella by its cis-spliced nad1i728g2, which is a trans-spliced intron in the latter three. Overall, the draft genome of Magnolia retains a similarly rich gene and intron set as that of the available four early angiosperms, suggesting that the mt genomes of the last common ancestor of flowering plants might possess a rich 41 protein-coding gene set with 25 group II introns, 20 tRNAs, and 3 rRNAs, which is followed by subsequent lineage specific losses of genes and introns in different lineages, alternatively, these earliest angiosperms might have independently gone through similar processes in gene losses and gains in the mt genome evolution.
Repeats and recombination rate
The draft mt genome of Magnolia contains 1,145 repeated sequences that are longer than 50 bp, accounting for 27% of the genome. The repeated sequences in Magnolia mt genome contain large proportions (54%) of the medium and large repeats, suggesting potentially more frequent recombinations in the Magnolia mitochondrion. We have checked all these repeats for recombination evidences with our long reads database. Surprisingly, no evidence of recombination is detected other than the 17 repeats shown in Table 2. The recombination equilibrium is detected in three largest repeats, including two inverted repeats of 16 Kb and 3 Kb and one direct repeats of 29 Kb. Longer repeat sequences show higher recombination rate, and inverted repeats are more prone to recombination than direct repeats.
Table 2. Recombination frequency of the 17 recombinationally active repeats in the mitochondrial genome of Magnolia biondii.
| Repeat no. | Repeat length | Identity | Start | End | Start | End | Direction | Reads supporting alternative conformation | Reads supporting master conformation |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 29,306 | 99.56 | 757,889 | 787,141 | 533,954 | 563,196 | Direct | 8 (47.06%) | 9 (52.94%) |
| 2 | 15,874 | 99.45 | 620,421 | 636,259 | 231,809 | 247,637 | Inverted | 27 (49.09%) | 28 (50.91%) |
| 3 | 2,822 | 99.33 | 949,369 | 952,184 | 259,868 | 262,681 | Inverted | 86 (51.50%) | 81 (48.50%) |
| 4 | 1,029 | 98.74 | 933,397 | 934,421 | 725,278 | 726,297 | Inverted | 19 (7.98%) | 219 (92.02%) |
| 5 | 746 | 99.46 | 520,696 | 521,439 | 259,868 | 260,611 | Direct | 2 (0.74%) | 269 (99.26%) |
| 6 | 651 | 99.23 | 121,635 | 122,282 | 94,466 | 95,114 | Inverted | 8 (3.05%) | 254 (96.95%) |
| 7 | 495 | 99.39 | 947,714 | 948,208 | 919,248 | 919,739 | Inverted | 4 (1.76%) | 223 (98.24%) |
| 8 | 463 | 99.14 | 465,328 | 465,787 | 325,427 | 325,888 | Inverted | 1 (0.35%) | 284 (99.65%) |
| 9 | 462 | 99.13 | 465,328 | 465,787 | 26,800 | 27,259 | Inverted | 1 (0.35%) | 283 (99.65%) |
| 10 | 268 | 100 | 736,243 | 736,510 | 545,526 | 545,793 | Direct | 3 (0.93%) | 319 (99.07%) |
| 11 | 268 | 99.25 | 769,472 | 769,737 | 736,243 | 736,510 | Direct | 3 (0.94%) | 318 (99.06%) |
| 12 | 180 | 95 | 950,890 | 951,065 | 543,270 | 543,448 | Inverted | 1 (0.25%) | 396 (99.75%) |
| 13 | 162 | 97.53 | 950,906 | 951,065 | 767,221 | 767,382 | Inverted | 1 (0.26%) | 392 (99.74%) |
| 14 | 160 | 100 | 543,270 | 543,429 | 260,987 | 261,146 | Direct | 1 (0.25%) | 400 (99.75%) |
| 15 | 154 | 100 | 521,585 | 521,738 | 521,534 | 521,687 | Direct | 1 (0.46%) | 219 (99.54%) |
| 16 | 103 | 100 | 604,952 | 605,054 | 597,457 | 597,559 | Direct | 1 (0.38%) | 260 (99.62%) |
| 17 | 103 | 100 | 521,636 | 521,738 | 521,534 | 521,636 | Direct | 1 (0.45%) | 221 (99.55%) |
The length of the repeats and recombination rate are clearly correlated, with no recombination evidence detected for repeat sequences of shorter than 100 bp. Recombination between direct repeats in a master-circle conformation of mtDNA could produce two sub-circles, while recombination between inverted repeats, an inversion. With recombination between these and other repeats, Magnolia mtDNAs contain predominant existence of master conformation along with many other alternative conformations with inversions and/or subcircles. This mtDNA heteroplasmy may potentially provide more genetic materials for evolutionary selection [67], hence conferring on Magnolia some ecological and genetic fitness during its evolution.
Plastid derived mitogenomic sequences
Magnolia mt genome contains 54 plastid insertions from 54 bp to 4 kb (Table 3) with the total length adding up to 26 Kb, comprising 3% of the genome, which is rather uniform in angiosperm mt genome in terms of both quantity and ratio. The transfer of plastid DNA to the mitochondrion most likely occurred in the ancestor of vascular plants [27]. These plastid transferred DNAs sometimes carries plastid genes, and the transferred protein-coding genes usually became nonfunctional, whereas the tRNA genes mostly remain functional. In Magnolia mt genome, we annotated six plastid derived tRNAs, including trnDGUC-cp, trnMCAU-cp, trnNGUU-cp, trnICAU-cp, trnPUGG-cp, and trnWCCA-cp. The transfer of these tRNAs could be dated back to different evolutionary stages of vascular plants. For example, the transfers of trnHGUG-cp and trnMCAU-cp might have happened in the common ancestor of the seed plants with their earliest occurrence in some gymnosperms [30]. The plastid-derived trnDGUC-cp mostly occurs in the mt genomes of some dicots but not in monocots and gymnosperms, therefore the presence of this tRNA in Magnolia and Liriodendron might represent their earliest emergence in time [30]. This suggests either paralleled gains of this tRNA once in Magnoliaceae and then once again in the ancestor of dicots, or as a single-gain event before the split of magnoliids from the rest of angiosperms, followed by subsequent lineage specific losses in monocots.
Table 3. Plastid insertions in the mitochondrial genome of Magnolia biondii.
| Plastid insertion | Minimum | Maximum | Length | Plastid genes carried |
|---|---|---|---|---|
| 1 | 400,085 | 404,363 | 4,279 | ndhB (partial)–rps7–rps12–trnVGAC |
| 2 | 485,860 | 488,764 | 2,905 | trnPUGG–trnWCCA–petG–petL–psbE–psbF–psbL–psbJ (partial) |
| 3 | 745,633 | 747,636 | 2,004 | ycf2 (partial)–trnICAU–rpl23-rpl2 (partial) |
| 4 | 409,616 | 411,176 | 1,561 | psbD (partial)–psbC (partial) |
| 5 | 394,605 | 395,943 | 1,339 | ndhJ–ndhK (partial) |
| 6 | 659,032 | 661,065 | 1,245 | psbM–trnDGUC–trnYGUA–trnEUUC |
| 7 | 837,610 | 838,785 | 1,176 | infA (partial)–rps8–rpl14 |
| 8 | 757,400 | 758,558 | 1,159 | trnAUGC–trnIGAU |
| 9 | 657,536 | 658,679 | 1,144 | petN |
| 10 | 543,561 | 544,412 | 852 | rrn16 (partial) |
| 11 | 767,513 | 768,361 | 849 | rrn16 (partial) |
| 12 | 754,944 | 755,663 | 720 | rpl2 (partial) |
| 13 | 533,955 | 534,619 | 665 | trnIGAU (partial) |
| 14 | 96,326 | 96,851 | 526 | rrn23 (partial) |
| 15 | 879,912 | 880,391 | 480 | ndhB (partial) |
| 16 | 262,147 | 262,584 | 438 | atpA (partial) |
| 17 | 949,466 | 949,903 | 438 | atpA (partial) |
| 18 | 96,995 | 97,431 | 437 | rrn23 (partial) |
| 19 | 101,192 | 101,562 | 371 | rrn23 (partial) |
| 20 | 544,905 | 545,182 | 278 | rrn16 (partial) |
| 21 | 269,458 | 269,702 | 245 | clpP (partial) |
| 22 | 98,321 | 98,528 | 208 | rrn23 (partial) |
| 23 | 190,158 | 190,337 | 180 | ycf2 (partial) |
| 24 | 752,648 | 752,805 | 158 | rrn23 (partial) |
| 25 | 872,552 | 872,694 | 143 | rrn23 (partial) |
| 26 | 14,492 | 14,627 | 136 | rrn16 (partial) |
| 27 | 29,848 | 29,980 | 133 | ycf2 (partial) |
| 28 | 221,881 | 221,966 | 86 | trnNGUU |
| 29 | 71,527 | 71,612 | 86 | rrn23 (partial) |
| 30 | 205,622 | 205,703 | 82 | trnHGUG |
| 31 | 98,988 | 99,067 | 80 | rrn23 (partial) |
| 32 | 366,960 | 367,036 | 77 | trnICAU |
| 33 | 110,706 | 110,781 | 76 | trnMCAU |
| 34 | 885,128 | 885,203 | 76 | rrn23 (partial) |
| 35 | 738,972 | 739,045 | 74 | trnfMCAU |
| 36 | 99,634 | 99,707 | 74 | rrn23 (partial) |
| 37 | 739,435 | 739,507 | 73 | trnMCAU |
| 38 | 577,233 | 577,303 | 71 | None |
| 39 | 465,334 | 465,404 | 71 | trnfMCAU (partial) |
| 40 | 27,182 | 27,252 | 71 | trnfMCAU (partial) |
| 41 | 818,833 | 818,898 | 66 | atpE (partial) |
| 42 | 686,509 | 686,572 | 64 | trnPUGG (partial) |
| 43 | 213,524 | 213,587 | 64 | trnPUGG (partial) |
| 44 | 329,994 | 330,056 | 63 | rrn16 (partial) |
| 45 | 31,362 | 31,422 | 61 | rrn16 (partial) |
| 46 | 950,996 | 951,056 | 61 | rrn16 (partial) |
| 47 | 260,996 | 261,056 | 61 | rrn16 (partial) |
| 48 | 163,196 | 163,255 | 60 | rrn16 (partial) |
| 49 | 583,586 | 583,641 | 56 | ndhH (partial) |
| 50 | 917,652 | 917,706 | 55 | trnFGAA (partial) |
| 51 | 675,036 | 675,088 | 53 | ndhI (partial) |
| 52 | 202,238 | 202,290 | 53 | atpB (partial) |
| 53 | 473,683 | 473,735 | 53 | trnQUUG (partial) |
| 54 | 545,277 | 545,327 | 51 | rrn16 (partial) |
Genome structure and conserved gene clusters
Vascular plant mt genomes are featured by structural dynamics with 31 rearrangements needed to reconcile the gene order of any two mt genomes [8]. The comparison of the mt gene orders of the five early angiosperms (S2 Table) in UniMoG [68] indicates that the gene order of Magnolia mt genome requires 31, 34, 34, and 44 rearrangements to get collinearity with that of the Liriodendron, Schisandra, Amborella, and Nymphaea, respectively. The repeat number in each mt genome and the divergence time of the three species related to Magnolia appear to be correlated with the number of rearrangement events [8, 65]. Despite structural fluidity, we observed several conserved gene clusters (e.g., rpl2–rps19–rps3–rpl16, atp8–cox3–sdh4, nad3–rps12, rpl5–rps14–cob, rps13–nad1.x2.x3, trnSGCU–trnFGAA–trnPUUG, trnYGUA–nad2.x3.x4.x5, <nad5.x4.x5><trnETTC–nad7>) in Magnolia mt genome compared with the gene order of that of the other angiosperms [69]. The retention of these gene orders across angiosperms [27, 30] despite the fast structural evolution over hundreds of millions of years might suggest certain selection forces and constraints upon the retention of these conserved gene clusters.
Phylogenetic inference
The NT dataset is comprised of 38 protein-coding genes, adding up to 30,903 bp (missing data, ~9.8%), with 9,541 parsimony-informative sites (30.9%), which corresponds to 10,301 characters with 4,153 parsimony-informative sites in AA dataset. Partitionfinder recognized 15 and 9 subsets for NT and AA datasets, respectively.
Our phylogenetic reconstruction based on the NT dataset is largely congruent with the phylogeny of angiosperms reconstructed from four mitochondrial genes [70]. The corresponding AA dataset produced otherwise a novel topology (S4 Fig) with a paraphyletic magnoliids, and a polyphyletic Austrobaileyales, which might be explained by the amino-acid level homoplasy induced by strong selection for high hydrophobicity of the mitochondrial amino acids [70]. Therefore, nucleotide datasets might better reflect the organismal phylogeny in mitochondrial phylogenomic studies. The NT dataset generally produced better BS and PP support than AA in most of the nodes. Our analyses recovered strong BS and PP support for the majority of nodes in the current sampling scope. In all of our analyses, serial divergences of ANA grade (Amborelllales, Nymphaeales, Austrobaileyales) occurred at the base of angiosperm phylogeny, before the diversification of mesangiosperms. With the exception of Ceratophyllum that is not sampled in our study due to insufficient high-quality reads available in NCBI SRA database, the relationships among the four mesangiosperm clades (Chloranthales, magnoliids, eudicots, and monocots) sample here have weak to moderate BS support.
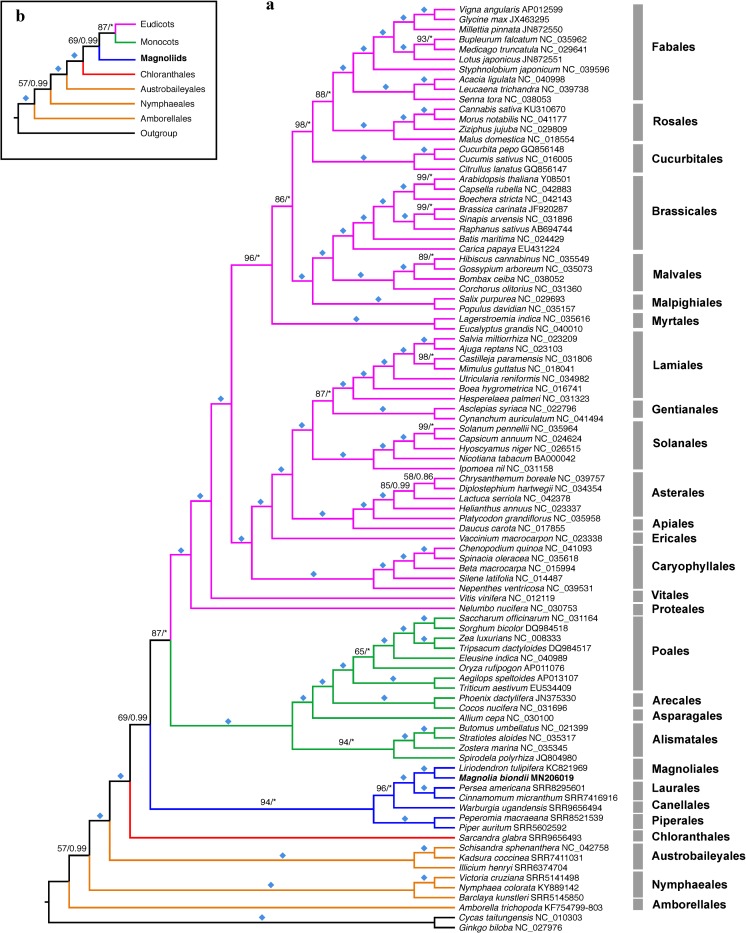
In NT dataset analyses (Fig 2), both monocots and eudicots receive 100% BS and 1.00 PP support. The sister relationship of monocots and eudicots has 87% BS and 1.00 PP support. All magnoliid taxa form a monophyletic group with 94% BS and 1.00 PP support, which is consistent with previous studies [70], albeit with stronger supports in our study. Within magnoliids, Canellales is strongly resolved as the sister to a clade containing Magnoliales and Laurales, rather than the sister to Piperales as in previous analyses [70, 71]. However, extended samplings might be needed to resolve the interordinal relationships within magnoliids. In contrast to the robust sister relationship of magnoliids with eudicots based on nuclear evidence [32, 34, 35], our study recovered a moderately-supported sister relationship of magnoliids with monocots and eudicots with 69% BS and 0.99 PP support in the nucleotide data analyses, which is also congruent with the plastid evidence [36, 37]. Therefore, organellar phylogenomic analyses tend to support the sister relationship of magnoliids with eudicots and monocots.
Fig 2. Phylogenetic tree inferred from the concatenated nucleotide dataset (NT).
Asterisks indicate either BS of 100% or PP of 1.00. Diamonds indicate both BS of 100% and PP of 1.00. a) A detailed phylogeny of 93 taxa. Newly sequenced Magnolia biondii is highlighted in bold. b) An abbreviated tree showing the relationships of major lineages of early angiosperms. Branches representing eudicots, monocots, magnoliids, Chloranthales, and ANA grade are indicated in magenta, green, blue, red, and orange, respectively. Those branches with both BS and PP support below 50% were collapsed.
Our study shows that the mitochondrial phylogenomics are informative tools for resolving relationships among families, orders, or higher taxonomic ranks across angiosperms, especially for reconstruction of ancient phylogenetic relationships. However, some deep nodes, such as the phylogenetic divergence order of Nymphaeales and Amborellales, the relationship among the five mesangiosperm lineages, were not well resolved in the current analysis. Extended samplings of more representatives of the early angiosperms and the comparison of mt phylogeny with those of the plastid [36, 37], nuclear [34, 72], morphology, and non-molecular data would be essential to confidently revolve the phylogenetic relationships of magnoliids relative to monocots and eudicots.
Conclusions
We assembled the draft mt genome of Magnolia using the Oxford Nanopore sequencing technology. The gene and intron content of Magnolia mt genome is similar to that of the Nymphaea and Liriodendron mt genomes, with Magnolia standing out by a relatively larger genome size packed with abundant repeated sequences, ancestrally retained sequences, and nuclear homologous sequences in its intergenic spacers. Despite high proportions of medium and large sized repeats, recombination activity is rather inert with only 17 recombinationally active repeats in the Magnolia mitochondrion. Repeat recombinations in the Magnolia mitochondrion could result in mtDNA heteroplasmy, hence contributing to dynamic structural evolution. Despite that, the Magnolia mt genome retains similar conserved gene clusters as Liriodendron, Nymphaea, Schisandra, and Amborella, suggesting unrecognized selection constraints on the retention of these gene clusters. This study allows new insight on the diversity and evolution of mitochondrial genomes in early flowering plants and repeat-mediated recombination patterns in plant mt genomes. Our study also provides mitochondrial evidences for the sister relationship of magnoliids with a clade comprising eudicots and monocots.
Supporting information
(PDF)
'>'s indicate lines of organism names. Chromosomes are circular if ended with ')', otherwise they are linear if without ')'s. Genes (name) start with '-'. indicate minus strand encoded genes, otherwise positive strand encoded genes.
(PDF)
The reads mapping files in bam format is visualized in Geneious and exported as the image files shown above.
(PDF)
The line plot and the genome map of the circular molecule of the putative mitochondrial genome of Magnolia biondii: (a) the line plot of the generation of the circular molecule; and (b) the genome map of the putative circular mitochodrial genome generated by OGDraw V1.2. Genes outside of the circle are transcribed clockwise, whereas those inside are transcribed counter-clockwise. Genes from the same protein complex are colored the same, introns are indicated in white boxes.
(PDF)
(PDF)
Asterisks indicate either BS of 100% or PP of 1.00. Diamonds indicate both BS of 100% and PP of 1.00. a) A detailed phylogeny of 93 taxa. Newly sequenced Magnolia biondii is highlighted in bold. b) An abbreviated tree showing the relationships of major lineages of early angiosperms. Eudicots, monocots, magnoliids, Chloranthales, and ANA grade are marked in magenta, green, blue, red, and orange, respectively. Branches with both BS and PP support below 50% were collapsed.
(PDF)
Acknowledgments
Ms Yang Peng and Na Li have been extremely helpful in lab work and computer network. We would like to thank two anonymous reviewers for their constructive comments on the manuscript. We would also like to thank Ernest Wu from University of British Columbia, Vancouver, Canada for academic writing improvement.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This project is funded by the Public Welfare Forestry Industry Project of State Forestry Administration of China (No. 201504322) granted to Zhang SZ and National Natural Science Foundation of China (No. 31600171) granted to Dong SS. The Funders provided support in the form of the salaries for authors (Zhang SZ and Dong SS), but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section. The commercial company 'Fairy Lake Botanical Garden, Shenzhen & Chinese Academy of Sciences, Shenzhen 518004, China' did not play a role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript and only provided financial support in the form of authors' salaries and/or research materials.
References
- 1.Knoop V, Volkmar U, Hecht J, Grewe F. Mitochondrial genome evolution in the plant lineage In: Kempken F. ed. Plant Mitochondria, Advances in Plant Biology 1 Springer Science + Business Media LCC; 2011. pp. 3–29. [Google Scholar]
- 2.Alverson AJ, Wei X, Rice DW, Stern DB, Barry K, Palmer JD. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol Biol Evol. 2010;27(6):1436–1448. 10.1093/molbev/msq029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alverson AJ, Rice DW, Dickinson S, Barry K, Palmer JD. Origins and recombination of the bacterial-sized multichromosomal mitochondrial genome of cucumber. Plant Cell. 2011;23(7):2499–2513. 10.1105/tpc.111.087189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bock R. Witnessing genome evolution: experimental reconstruction of endosymbiotic and horizontal gene transfer. Annu Rev Genet. 2017;(51):1–22. [DOI] [PubMed] [Google Scholar]
- 5.Rice DW, Alverson AJ, Richardson AO, Young GJ, Sanchezpuerta MV, Munzinger J, et al. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science. 2013;342(6165):1468–1473. 10.1126/science.1246275 [DOI] [PubMed] [Google Scholar]
- 6.Xue JY, Liu Y, Li L, Wang B, Qiu YL. The complete mitochondrial genome sequence of the hornwort Phaeoceros laevis: retention of many ancient pseudogenes and conservative evolution of mitochondrial genomes in hornworts. Curr Genet. 2009;56(1):53–61. 10.1007/s00294-009-0279-1 [DOI] [PubMed] [Google Scholar]
- 7.Wang B, Xue J, Li L, Yang L, Qiu YL. The complete mitochondrial genome sequence of the liverwort Pleurozia purpurea, reveals extremely conservative mitochondrial genome evolution in liverworts. Curr Genet. 2009;55(6):601–609. 10.1007/s00294-009-0273-7 [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, Medina R, Goffinet B. 350 my of mitochondrial genome stasis in mosses, an early land plant lineage. Mol Biol Evol. 2014;31(10):2586–2591. 10.1093/molbev/msu199 [DOI] [PubMed] [Google Scholar]
- 9.Mower JP, Sloan DB, Alverson AJ. Plant mitochondrial genome diversity: The genomics revolution. Wendel JF, editor. Heidelberg: Springer Vienna; 2012. pp. 123–144. [Google Scholar]
- 10.Sloan DB. One ring to rule them all? Genome sequencing provides new insights into the 'master circle' model of plant mitochondrial DNA structure. New Phytol. 2013;200(4):978–985. 10.1111/nph.12395 [DOI] [PubMed] [Google Scholar]
- 11.Gualberto JM, Mileshina D, Wallet C, Niazi AK, Weber-Lotfi F, Dietrich A. The plant mitochondrial genome: dynamics and maintenance. Biochimie. 2014;100:107–120. 10.1016/j.biochi.2013.09.016 [DOI] [PubMed] [Google Scholar]
- 12.Skippington E, Barkman TJ, Rice DW, Palmer JD. Miniaturized mitogenome of the parasitic plant Viscum scurruloideum is extremely divergent and dynamic and has lost all nad genes. Proc Natl Acad Sci USA. 2015;112(27):E3515–E3524. 10.1073/pnas.1504491112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sloan DB, Alverson AJ, Chuckalovcak JP, Wu M, McCauley DE, Palmer JD, et al. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 2012;10(1):e1001241 10.1371/journal.pbio.1001241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petersen G, Cuenca A, Moller IM, Seberg O. Massive gene loss in mistletoe (Viscum, Viscaceae) mitochondria. Sci Rep. 2015;5:17588 10.1038/srep17588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JM, Cho CH, Park SI, Choi JW, Song HS, West JA, et al. Parallel evolution of highly conserved plastid genome architecture in red seaweeds and seed plants. BMC Biol. 2016;14(1):75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marechal A, Brisson N. Recombination and the maintenance of plant organelle genome stability. New Phytol. 2010;186(2):299–317. 10.1111/j.1469-8137.2010.03195.x [DOI] [PubMed] [Google Scholar]
- 17.André C, Levy A, Walbot V. Small repeated sequences and the structure of plant mitochondrial genomes. Trends Genet. 1992;8:128–132. 10.1016/0168-9525(92)90370-J [DOI] [PubMed] [Google Scholar]
- 18.Sanchez-Puerta MV, Zubko MK, Palmer JD. Homologous recombination and retention of a single form of most genes shape the highly chimeric mitochondrial genome of a cybrid plant. New Phytol. 2015;206(1):381–396. 10.1111/nph.13188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sloan DB, Muller K, McCauley DE, Taylor DR, Storchova H. Intraspecific variation in mitochondrial genome sequence, structure, and gene content in Silene vulgaris, an angiosperm with pervasive cytoplasmic male sterility. New Phytol. 2012;196(4):1228–1239. 10.1111/j.1469-8137.2012.04340.x [DOI] [PubMed] [Google Scholar]
- 20.Naito K, Kaga A, Tomooka N, Kawase M. De novo assembly of the complete organelle genome sequences of azuki bean (Vigna angularis) using next-generation sequencers. Breed Sci. 2013;63(2):176–182. 10.1270/jsbbs.63.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogihara Y, Yamazaki Y, Murai K, Kanno A, Terachi T, Shiina T, et al. Structural dynamics of cereal mitochondrial genomes as revealed by complete nucleotide sequencing of the wheat mitochondrial genome. Nucleic Acids Res. 2005;33(19):6235–6250. 10.1093/nar/gki925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hecht J, Grewe F, Knoop V. Extreme RNA editing in coding islands and abundant microsatellites in repeat sequences of Selaginella moellendorffii mitochondria: the root of frequent plant mtDNA recombination in early tracheophytes. Genome Biol Evol. 2011;3:344–358. 10.1093/gbe/evr027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo W, Grewe F, Fan W, Young GJ, Knoop V, Palmer JD, et al. Ginkgo and Welwitschia mitogenomes reveal extreme contrasts in gymnosperm mitochondrial evolution. Mol Biol Evol. 2016;33(6):1448–1460. 10.1093/molbev/msw024 [DOI] [PubMed] [Google Scholar]
- 24.Guo W, Zhu A, Fan W, Mower JP. Complete mitochondrial genomes from the ferns Ophioglossum californicum and Psilotum nudum are highly repetitive with the largest organellar introns. New Phytol. 2017;213(1):391–403. 10.1111/nph.14135 [DOI] [PubMed] [Google Scholar]
- 25.Sloan DB, Alverson AJ, Storchova H, Palmer JD, Taylor DR. Extensive loss of translational genes in the structurally dynamic mitochondrial genome of the angiosperm Silene latifolia. BMC Evol Biol. 2010;10(1):274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shearman JR, Sonthirod C, Naktang C, Pootakham W, Yoocha T, Sangsrakru D, et al. The two chromosomes of the mitochondrial genome of a sugarcane cultivar: assembly and recombination analysis using long PacBio reads. Sci Rep. 2016;6:31533 10.1038/srep31533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong S, Zhao C, Chen F, Liu Y, Zhang S, Wu H, et al. The complete mitochondrial genome of the early flowering plant Nymphaea colorata is highly repetitive with low recombination. BMC Genomics. 2018;19(1):614 10.1186/s12864-018-4991-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wynn EL, Christensen AC. Repeats of unusual size in plant mitochondrial genomes: identification, incidence and evolution. G3 (Bethesda). 2019;9(2):549–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herendeen PS, Friis EM, Pedersen KR, Crane PR. Palaeobotanical redux: revisiting the age of the angiosperms. Nat Plants. 2017;3(3):17015. [DOI] [PubMed] [Google Scholar]
- 30.Richardson AO, Rice DW, Young GJ, Alverson AJ, Palmer JD. The "fossilized" mitochondrial genome of Liriodendron tulipifera: ancestral gene content and order, ancestral editing sites, and extraordinarily low mutation rate. BMC Biol. 2013;11(1):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soltis DE, Soltis PS. Nuclear genomes of two magnoliids. Nat Plants. 2019;5(1):6–7. 10.1038/s41477-018-0344-1 [DOI] [PubMed] [Google Scholar]
- 32.Chaw SM, Liu YC, Wu YW, Wang HY, Lin CYI, Wu CS, et al. Stout camphor tree genome fills gaps in understanding of flowering plant genome evolution. Nat Plants. 2019;5(1):63–73. 10.1038/s41477-018-0337-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J, Hao Z, Guang X, Zhao C, Wang P, Xue L, et al. Liriodendron genome sheds light on angiosperm phylogeny and species–pair differentiation. Nat Plants. 2018;5(1):18–25. 10.1038/s41477-018-0323-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wickett NJ, Mirarab S, Nguyen N, Warnow T, Carpenter E, Matasci N, et al. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc Natl Acad Sci U S A. 2014;111(45):4859–4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng L, Zhang Q, Sun R, Kong H, Zhang N, Ma H. Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergence times. Nat Commun. 2014;5:4956 10.1038/ncomms5956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gitzendanner MA, Soltis PS, Wong GK-S, Ruhfel BR, Soltis DE. Plastid phylogenomic analysis of green plants: a billion years of evolutionary history. Am J Bot. 2018;105(3):291–301. 10.1002/ajb2.1048 [DOI] [PubMed] [Google Scholar]
- 37.Ruhfel BR, Gitzendanner MA, Soltis PS, Soltis DE, Burleigh JG. From algae to angiosperms–inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evol Biol. 2014;14(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y, Cox CJ, Wang W, Goffinet B. Mitochondrial phylogenomics of early land plants: mitigating the effects of saturation, compositional heterogeneity, and codon-usage bias. Syst Biol. 2014;63(6):862–878. 10.1093/sysbio/syu049 [DOI] [PubMed] [Google Scholar]
- 39.China Pharmacopoeia Committee, Pharmacopoeia of the People's Republic of China, The first Division of 2000 English Edition, China Chemical Industry Press, Beijing, 2000, pp. 143. [Google Scholar]
- 40.Qu L, Qi Y, Fan G, Wu Y. Determination of the volatile oil of Magnolia biondii pamp by GC–MS combined with chemometric techniques. Chromatographia. 2009;70(5–6):905–914. [Google Scholar]
- 41.Zhao W, Zhou T, Fan G, Chai Y, Wu Y. Isolation and purification of lignans from Magnolia biondii pamp by isocratic reversed-phase two-dimensional liquid chromatography following microwave-assisted extraction. J Sep Sci. 2015;30(15):2370–2381. [DOI] [PubMed] [Google Scholar]
- 42.Chen Y, Gao BC, Qiao L, Han GQ. Study on the hydrophilic components of Magnolia biondii Pamp. Acta Pharmaceutica Sinica. 1994;07. [Google Scholar]
- 43.Porebski S, Bailey LG, Bernard RB. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Reporter. 1997;15(1):8–15. [Google Scholar]
- 44.Koren S, Walenz BP, Berlin KM, Jason R, Bergman NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive\\r, k\\r, -mer weighting and repeat separation. Genome Res. 2017;27(5):722 10.1101/gr.215087.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 2014;9(11):e112963 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li L, Wang B, Liu Y, Qiu YL. The complete mitochondrial genome sequence of the hornwort Megaceros aenigmaticus shows a mixed mode of conservative yet dynamic evolution in early land plant mitochondrial genomes. J Mol Evol. 2009;68(6):665–678. 10.1007/s00239-009-9240-7 [DOI] [PubMed] [Google Scholar]
- 48.Langmead B. Fast gapped-read alignment with Bowtie2. Nat Methods. 2012;9(4):357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lowe TM, Chan PP. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016;44(W1):W54–W57. 10.1093/nar/gkw413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Greiner S, Lehwark P, Bock R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019;47:W59–W64. 10.1093/nar/gkz238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chase MW, Christenhusz MJM, Fay MF, Byng JW, Judd WS, Soltis DE, et al. An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 2016;181(1):1–20. [Google Scholar]
- 52.Qiu YL, Lee J, Bernasconi-Quadroni F, Soltis DE, Soltis PS, Zanis M, et al. The earliest angiosperms: evidence from mitochondrial, plastid and nuclear genomes. Nature. 1999;402(6760):404–407. 10.1038/46536 [DOI] [PubMed] [Google Scholar]
- 53.Johnson MG, Gardner EM, Liu Y, Medina R, Goffinet B, Shaw AJ, et al. HybPiper: extracting coding sequence and introns for phylogenetics from high-throughput sequencing reads using target enrichment. Appl Plant Sci. 2016;4(7):1600016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xia X. DAMBE5: a comprehensive software package for data analysis in molecular biology and evolution. Mol Biol Evol. 2013;30(7):1720–1728. 10.1093/molbev/mst064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abascal F, Zardoya R, Telford MJ. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010;38(suppl_2):W7–W13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33(2):511–518. 10.1093/nar/gki198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56(4):564–577. 10.1080/10635150701472164 [DOI] [PubMed] [Google Scholar]
- 58.Kück P, Longo GC. FASconCAT-G: extensive functions for multiple sequence alignment preparations concerning phylogenetic studies. Front Zool. 2014;11(1):1–8. 10.1186/1742-9994-11-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lanfear R, Calcott B, Ho SYW, Guindon S. Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 2012;29(6):1695–1701. 10.1093/molbev/mss020 [DOI] [PubMed] [Google Scholar]
- 60.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- 61.Huelsenbeck JP, Ronquist F. Mrbayes: bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–765. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- 62.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rambaut A. FigTree 1.4.2. http://tree.bio.ed.ac.uk/. 2014.
- 64.Burger G, Gray MW, Lang BF. Mitochondrial genomes: anything goes. Trends Genet. 2003;19(12):709–716. 10.1016/j.tig.2003.10.012 [DOI] [PubMed] [Google Scholar]
- 65.Shi Y, Liu Y, Zhang S, Zou R, Tang J, Mu W, et al. Assembly and comparative analysis of the complete mitochondrial genome sequence of Sophora japonica 'JinhuaiJ2'. PLoS ONE. 2018;13(8):e0202485 10.1371/journal.pone.0202485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Richardson AO, Rice DW, Young GJ, Alverson AJ, Palmer JD. The "fossilized" mitochondrial genome of Liriodendron tulipifera: ancestral gene content and order, ancestral editing sites, and extraordinarily low mutation rate. BMC Biol. 2013;11(1):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang J, Liu G, Zhao N, Chen S, Liu D, Ma W, et al. Comparative mitochondrial genome analysis reveals the evolutionary rearrangement mechanism in Brassica. Plant Biol (Stuttg). 2016;18(3):527–536. [DOI] [PubMed] [Google Scholar]
- 68.Hilker R, Sickinger C, Pedersen CNS, Stoye J. UniMoG–a unifying framework for genomic distance calculation and sorting based on DCJ. Bioinformatics. 2012;28(19):2509 10.1093/bioinformatics/bts440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dong S, Zhao C, Zhang S, Zhang L, Wu H, Liu H, et al. Mitochondrial genomes of the early land plant lineage liverworts (Marchantiophyta): conserved genome structure, and ongoing low frequency recombination. BMC Genomics. 2019;20(1):953 10.1186/s12864-019-6365-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qiu YL, Li L, Wang B, Xue JY, Hendry TA, Li RQ, et al. Angiosperm phylogeny inferred from sequences of four mitochondrial genes. J Syst Evol. 2010;48(6):391–425. [Google Scholar]
- 71.Hilu KW, Borsch T, Muller K, Soltis DE, Soltis PS, Savolainen V, et al. Angiosperm phylogeny based on matK sequence information. Am J Bot. 2003;90:1758–1776. 10.3732/ajb.90.12.1758 [DOI] [PubMed] [Google Scholar]
- 72.Strijk JS, Hinsinger DD, Roeder MM, Chatrou LW, Couvreur TLP, Erkens RHJ, et al. The soursop genome and comparative genomics of basal angiosperms provide new insights on evolutionary incongruence. bioRxiv. 2019;63915. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
'>'s indicate lines of organism names. Chromosomes are circular if ended with ')', otherwise they are linear if without ')'s. Genes (name) start with '-'. indicate minus strand encoded genes, otherwise positive strand encoded genes.
(PDF)
The reads mapping files in bam format is visualized in Geneious and exported as the image files shown above.
(PDF)
The line plot and the genome map of the circular molecule of the putative mitochondrial genome of Magnolia biondii: (a) the line plot of the generation of the circular molecule; and (b) the genome map of the putative circular mitochodrial genome generated by OGDraw V1.2. Genes outside of the circle are transcribed clockwise, whereas those inside are transcribed counter-clockwise. Genes from the same protein complex are colored the same, introns are indicated in white boxes.
(PDF)
(PDF)
Asterisks indicate either BS of 100% or PP of 1.00. Diamonds indicate both BS of 100% and PP of 1.00. a) A detailed phylogeny of 93 taxa. Newly sequenced Magnolia biondii is highlighted in bold. b) An abbreviated tree showing the relationships of major lineages of early angiosperms. Eudicots, monocots, magnoliids, Chloranthales, and ANA grade are marked in magenta, green, blue, red, and orange, respectively. Branches with both BS and PP support below 50% were collapsed.
(PDF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.