SUMMARY
In high transmission regions, we expect parasite lineages within complex malaria infections to be unrelated due to parasite inoculations from different mosquitoes. This project was designed to test this prediction. We generated 485 single-cell genome sequences from fifteen P. falciparum malaria patients from Chikhwawa, Malawi, an area of intense transmission. Patients harbored up to seventeen unique parasite lineages. Surprisingly, parasite lineages within infections tend to be closely related, suggesting that superinfection by repeated mosquito bites is rarer than co-transmission of parasites from a single mosquito. Both closely and distantly related parasites comprise an infection, suggesting sequential transmission of complex infections between multiple hosts. We identified tetrads and reconstructed parental haplotypes which revealed the inbred ancestry of infections and non-Mendelian inheritance. Our analysis suggests strong barriers to secondary infection and outbreeding amongst malaria parasites from a high transmission setting, providing unexpected insights into the biology and transmission of malaria.
eTOC
In high transmission regions, malaria infections are expected to comprise unrelated parasite lineages due to parasite inoculations from different mosquitoes. Using single-cell genome sequencing, Nkhoma et al find parasite lineages within infections to be closely related, indicating that co-transmission predominantly shapes within-host malaria parasite diversity.
Graphical Abstract

INTRODUCTION
Malaria remains a major global health problem, with ~400,000 malaria-related deaths in 2015, and over 200 million clinical cases (WHO, 2016) cases The intensity of malaria transmission is correlated with the complexity of infection (COI), the number of genetically distinct parasites observed within a single infection. Genetically distinct malaria parasites can infect an individual through two routes (Figure 1). A single individual may be bitten by two (or more) infected mosquitoes, each bearing a unique parasite genotype (Figure 1B), or an individual may be bitten by a single mosquito bearing more than one parasite genotype (Figure 1C). Throughout, we refer to these two processes as superinfection and co-transmission respectively. Superinfection of an individual by multiple infectious mosquito bites is often used to explain the high complexity of infection found in high transmission regions(Volkman et al., 2012). Following a bloodmeal, gametocyte stage parasites fuse in the mosquito midgut, and an obligate round of sexual recombination occurs. If only a single parasite genotype is present, all offspring will be identical (Figure 1A, B). When multiple parasite genotypes are present recombinant progeny may arise (Conway et al., 1991, Mu et al., 2005, Nkhoma et al., 2012, Wong et al., 2017, Wong et al., 2018) (Figure 1C). At a population level recombination of parasite genotypes shapes the local decay of linkage disequilibrium and haplotype variation (Mu et al., 2005, Mu et al., 2007, Neafsey et al., 2008).
Figure 1. The within host genetic diversity of malaria parasites is shaped by mosquito transmission.
(A) A simple monoinfection is generated when an uninfected individual is bitten by a mosquito bearing a single parasite genotype. (B) A superinfection occurs when an individual is bitten by two mosquitoes, each bearing a single parasite genotype. (C) Co-transmission of parasites occurs when a single mosquito bearing multiple genetically distinct parasites bites an uninfected individual. As genetic recombination is an obligate stage of mosquito transmission multiple related parasites may infect an individual through this route.
The clinical impact of complex infections has been studied in mouse malaria models. Here, interactions between genetically distinct malaria parasites are known to influence the evolution of parasite virulence, antimalarial drug resistance, immunity, gametocyte sex ratios, and malaria transmission (Bell et al., 2006, de Roode et al., 2005, Wargo et al., 2007a, Wargo et al., 2007b, Reece et al., 2008). However, translating these findings to human malaria has been a major challenge. This is due to the paucity of appropriate tools for resolving infection complexity on a large-scale at the level of single parasitized cells: we cannot directly infer the composition of malaria infections by bulk sequencing of infected blood samples. Complex infections confound most traditional genetic analysis, preventing the accurate inference of allele frequencies and even simple genotype-phenotype associations (Nair et al., 2014, Wong et al., 2018). However, powerful new approaches to analyze individual malaria infections are emerging, aided by recent advances in targeted capture of singly-infected erythrocytes from complex mixtures and improved methods for single-cell sequencing (Nair et al., 2014, Trevino et al., 2017), and computational approaches for interpreting infection complexity (Chang et al., 2017, Zhu et al., 2018).
Using single-cell sequencing and cloning parasites by limiting dilution from a single individual, we previously saw a range of inferred relationships amongst co-infecting parasite haplotypes, including identical clonal lineages, siblings and unrelated individuals (Nair et al., 2014, Nkhoma et al., 2012, Trevino et al., 2017). However, it is unknown to what extent these findings can be generalized across a population. This project was designed to understand the degree to which the genetic diversity of individual infections is driven by superinfection of unrelated strains, or co-transmission of related ones. To do this, we performed whole genome sequencing of bulk infections and single-cell sequencing of parasite-infected cells isolated from malaria patients in Chikhwawa, a high transmission region in Malawi.
RESULTS
Infection complexity in bulk sequenced samples
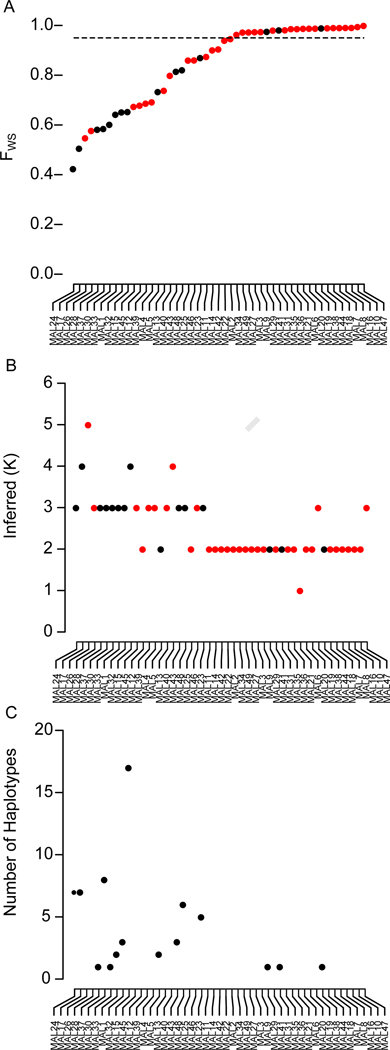
To resolve the within-host population structure of malaria infections, we performed a cross-sectional survey of individuals infected with uncomplicated P. falciparum malaria in Chikhwawa, Malawi, an area of high malaria transmission (entomological inoculation rate 183 infectious bites per person per year (Mzilahowa et al., 2012). In this setting, we might expect that patients will contain a mixture of unrelated parasites, resulting from independent mosquito inoculations (i.e. that superinfections will predominate). We performed bulk parasite genome sequencing of 49 infections to a median read depth of 31 (interquartile range 20.93–48.37). We estimated the complexity of infection from bulk sequence data using 10,997 unfixed SNP positions with a minor allele frequency (MAF) >0.05 using the FWS statistic (Auburn et al., 2012, Manske et al., 2012) and DEploid (Zhu et al., 2018) (Figure 2a,b, Table S1). FWS grades infections on a continuous scale of complexity where infections with an FWS>0.95 are considered clonal and DEploid estimates the number of haplotypes (K) present in sequence data by jointly estimating haplotypes and their abundances. In close agreement with contemporary estimates of within-host diversity (Early et al., 2018) from the same location, 22 of 49 infections (44.9%) were considered clonal by FWS. The within-host allele frequency (WHAF) captured from deep sequencing can be used to infer the presence of related parasites (Pearson et al., 2016). The patterns of unfixed mutations in the remaining 27 infections suggest a simple model of superinfection is insufficient to universally capture all patterns of within-host relatedness (Data S1). Across the genome the WHAF in superinfected patients cluster around three values: fixed to the reference allele; fixed to the alternative allele; or at an intermediate frequency determined by the proportion of the two strains. Analysis of bulk sequencing data could not definitively resolve superinfection and co-transmission, so we selected 15 infections across the range of FWS and inferred K (the number of haplotypes present) for single-cell sequencing, using a recently optimized method capable of near-complete genome capture (Trevino et al., 2017). The malaria parasite undergoes 4–5 rounds of DNA replication within a single-cell producing segmented schizont stage parasites with an average of 16 genome copies (Reilly et al., 2007). We isolate individual schizonts by Fluorescence Activated Cell Sorting (FACs), followed by whole genome amplification (WGA) under highly sterile conditions before sequencing the amplified product.
Figure 2. Complexity of infection inferred from bulk and single-cell sequencing.
(A) FWS scores for 49 bulk sequenced infections. Infections above the dashed line (FWS=0.95) are assumed to be clonal. (B) Inferred number of haplotypes (K) inferred by DEploid, infections are ordered by the FWS score. Black dots in (A) and (B) denote infections also deconvoluted by single-cell sequencing. (C) Number of unique haplotypes inferred by single-cell sequencing. This data is included in Table S1.
Single-cell sequencing of malaria parasites
In total we sequenced the genomes of 485 single-cells subjected to WGA (437 unique to this study), 49 bulk infections and 24 clones isolated from a single patient by limiting dilution (Nkhoma et al., 2012, Rosario, 1981). Prior to genotype filtering we scored 175,543 biallelic SNPs with a VQSLOD>0 across the 558 genome sequences. The highly repetitive and AT-rich P. falciparum genome (Gardner et al., 2002) presents unique challenges with generating an accurate picture of the variation present in a single-cell. We were particularly concerned with capture of DNA from more than one genetic background during the single-cell sequencing protocol and implemented stringent quality checks. Using sequencing data from the 24 clones we estimated the threshold for identifying single-cell sequences where there was potential contamination from exogenous DNA at 1% of mixed base calls. The sequences from the cloned lines were integrated into the single-cell dataset for downstream analysis. After excluding low coverage libraries (<75,000 calls, n=23) and sequences with >1% mixed base calls (n=38) 424 single-cell sequences remained. After including 23 of the sequences from ex vivo expanded clones there were 13–45 sequences per infection (mean 29.9 sequences; Figure S1). The number of haplotypes per sample attempted was estimated by rarefaction analysis (described below).
After quality control we retained 60,002 SNPs scored in at least 90% of the 496 sequences, 10,997 of which had a MAF>0.05 across the 49 bulk sequenced infections. As the 10,997 SNPs were ascertained from population data which did not undergo whole genome amplification this category of SNPs are unlikely to be artifacts from the MDA reaction, nor mutations arising during the course of an infection. Across our dataset parasites classified as clonally identical were identical at 99.97% of the original 60,002 sites, equivalent to a genome-wide error-rate of 1.2×10−6 mutations per basepair per cell, suggesting our approach is comparable (or superior) to leading single-cell sequencing methods (Leung et al., 2015, Hou et al., 2013, Lu et al., 2012). As an initial characterization of our data we estimated the genetic diversity in each infection from the number of unfixed sites from read pileups in bulk sequencing or across called genotypes in single-cell sequencing. For paired bulk/single-cell data from the same infection a mean of 1.6 fold (range 0.7–9.1 fold) more polymorphic sites were discovered by single-cell sequencing than by bulk sequencing (Figure S1). This is likely due to the limits in discovery of very low frequency SNPs by bulk sequencing. By subsampling our single-cell data we saw diminishing returns from sequencing additional cells, with 90% of the observed polymorphic sites captured by sampling a mean of 21.6 cells (range 7–43, Figure S1).
Haplotypic diversity of malaria infections
An important goal in malaria genomics is estimating the number of unique haplotypes (or complexity of infection) within an infection (Volkman et al., 2012). We estimated the number of unique haplotypes directly from the single-cell data. To exclude potential confounding of de novo mutation and sequencing error, we restricted analysis to 10,997 conservatively called sites with a MAF >0.05 in the 49 bulk sequenced infections. We estimated the number of unique haplotypes per infection by collapsing haplotypes from the same infection that were different at <1% of sites. For each infection, we applied individual-based rarefaction to the haplotype abundances and sequenced additional single genomes until a plateau in the rarefaction curve was reached (Figure S2). Using this approach, between 1 and 17 haplotypes were observed in each infection (Figure 1C, Table S1). Rarefaction of haplotype abundance suggested we had captured all haplotypes present in 14/15 infections. For these 14 samples the number of haplotypes captured was within the 95% confidence interval of the Chao I estimator. One infection (MAL15) showed exceptionally high diversity with 17 of an estimated 30.21 (95% CI=19.7–81.7) haplotypes detected. Two infections (MAL37 and MAL33) show a single haplotype from single-cell sequencing, although FWS scores <0.95 and patterns of segregating sites suggest we have incompletely captured all haplotypes (Data S1). Sequencing more cells did not capture additional haplotypes.
Inference of infection composition from bulk sequences
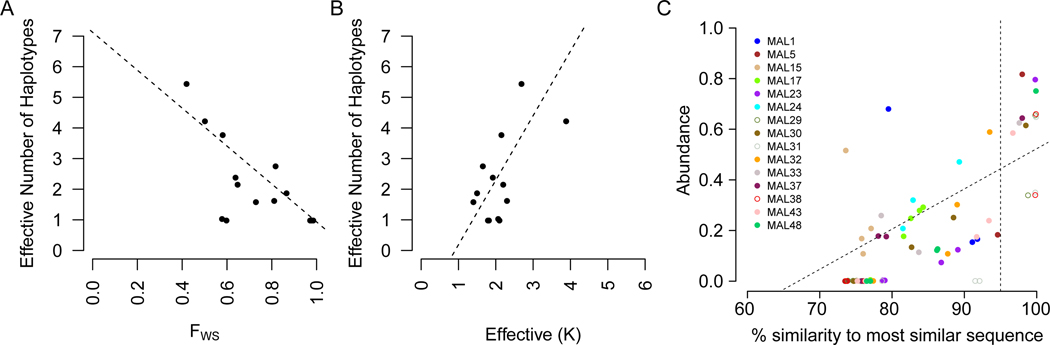
There has been a concerted effort to develop statistical methods for inferring the parasite haplotypes within complex infections using information from bulk sequence data (Chang et al., 2017, Zhu et al., 2018). Such methods aim to phase haplotype data from complex mixtures, extending the methods used to infer haplotypes from diploid genotypes. Our single-cell resolution data from natural infections provides “gold standard” data for comparison with inferences. We found a strong correlation between the effective number of haplotypes (Zhu et al., 2018) observed by single-cell sequencing and the effective K from DEploid (Pearson’s r2=0.61) and FWS (Pearson’s r 2=−0.51, Figure 3A,B). The effective K and effective number of haplotypes normalizes the absolute haplotype number by the abundance of each haplotype and are distinct from the number of haplotypes presented in Figure 2. As within-host abundance is accounted for in these measures (and in FWS) it is not surprising these agree more closely than when comparing absolute haplotype number or Inferred K. However, while DEploid performed well in determining the predominant haplotype present within infections, it was unable to accurately determine the additional haplotypes present within infections (Figure 3C). The poor performance most likely stems from the assumption that haplotypes found within infections are unrelated. We suggest that incorporating relatedness into these models may improve performance of these inference methods. Importantly, our data provide a suitable dataset for optimizing and improving these statistical models.
Figure 3. Inferring the composition of malaria infections from bulk sequencing data.
Correlation between the effective number of haplotypes inferred from single-cell sequencing and either FWS (A) or Effective K inferred from DEploid (B). (C) The maximum similarity between each haplotype inferred by DEploid and single-cell sequences from the same infection. The abundance of the haplotype (as estimated by DEploid) for each haplotype is shown on the y- axis. Majority haplotypes were inferred with high accuracy (73.6–99.9% similarity), with reduced accuracy for minority haplotypes. After exclusion of low abundance (<10%) haplotypes and haplotypes from clonal infections there was a significant relationship between abundance and maximum similarity (adjusted r2=0.32, p=4.6×10−5, linear model). Clonal infections (MAL29, MAL31, MAL38) and simple superinfections (MAL5) perform uniformly well with DEploid, though other infections show variable success.
Recent ancestry of individual infections
The size of chromosomal blocks that are shared identical-by-descent (IBD) between infections provides a metric for assessing parasite relatedness: recent relatives share large blocks, while in distant relatives these blocks are smaller because they have been broken up by recombination events. As recombination occurs in the mosquito (Figure 1) related parasites are likely to have arisen from a single mosquito bite and have been co-transmitted. To better characterize levels of relatedness within infections we identified blocks of chromosomes shared IBD between all paired sequences using a hidden Markov model (Schaffner et al., 2018). IBD sharing between clonal bulk sequenced infections was rare, with a mean of 0.73 blocks shared between infections (range 0–5), encompassing a mean of 88.5kb (range 3.8–342.7kb) of each genome, with a mean block length of 50.8kb (range 3.8–142.4kb). In contrast, within infections parasites shared a mean of 13.0 (range 0–30) IBD blocks between parasite genomes, encompassing a mean of 16,334.2kb (range 3.1–20,577.0kb) of each genome, with a mean shared block length of 1,143.6kb (range 3.1–1469.8kb). As we limit inference of IBD to the ‘core’ genome (Miles et al., 2016) identical parasites share 20,577kb of their genomes IBD in 14 blocks (one per chromosome). The presence of IBD sharing between individuals supports recent shared ancestry. For 10/15 of the infections there was at least one block of IBD shared in all pairwise comparisons. As our filtering of IBD blocks was limited to >2.5cM we are limited to inference of relatedness over the last 25 generations (~6 years (Henden et al., 2018)).
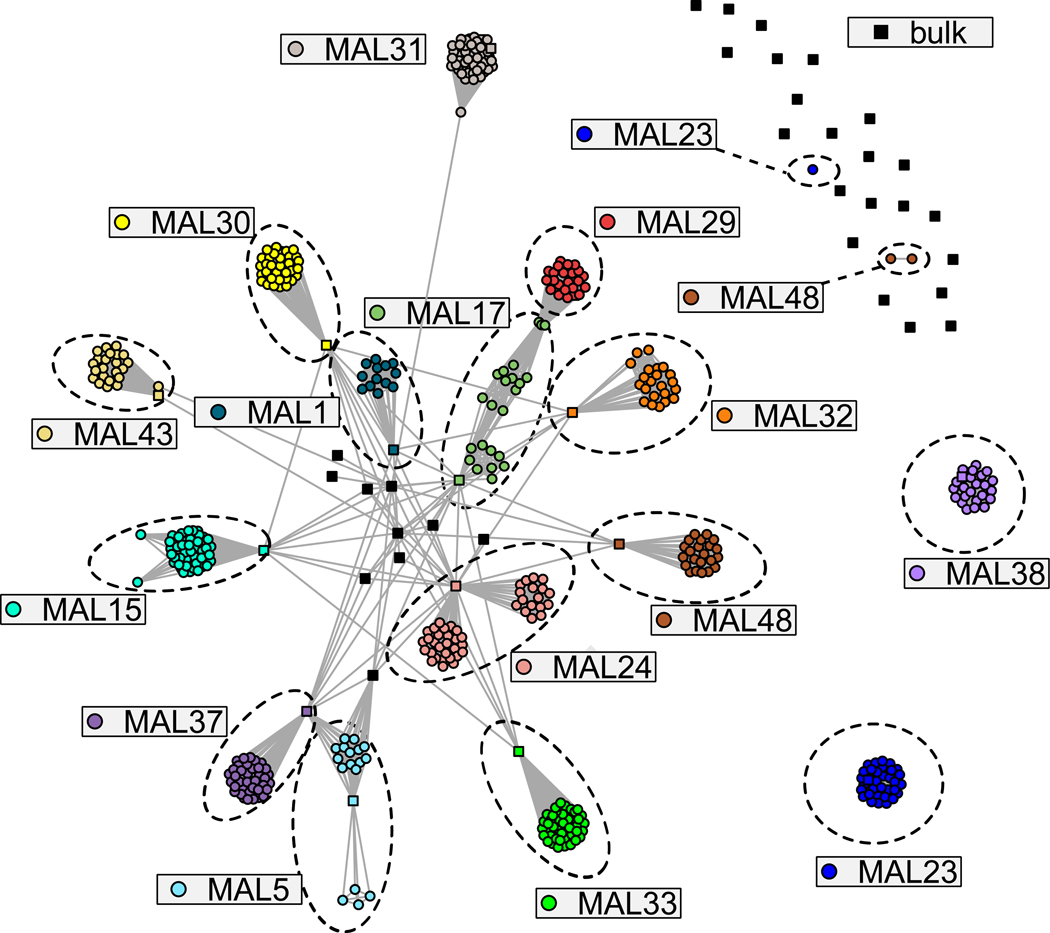
Recent studies have highlighted the power of IBD networks to capture the structure of a parasite population (Henden et al., 2018). We built a network of pairwise shared IBD, creating links between parasites with >15% of their genomes shared IBD (Figure 4, Data S2). This revealed close connectivity between parasites from the same infection, with much sparser connectivity between parasites from different infections. We observed subdivision within individual infections. For instance, MAL5 and MAL24 form two clusters of parasites that were connected by the sequence derived from bulk sequencing to one another in agreement with expectations of superinfection. MAL15 and MAL23 show either direct connections or indirect connections (passing through another genotype) between all parasites in agreement with the expectations of co-transmission. Varying the minimum IBD required to connect genomes allowed us to visualize how relatedness subdivides individual infections across a range of IBD sharing (Data S2).
Figure 4. A network representation of pairwise IBD sharing across the genomes.
Each node represents a single parasite colored by the infection of origin. Nodes are joined if >15% of the genomes are shared IBD. Each node is colored by the infection it was derived from, with bulk sequences denoted by a square and single-cell sequences by a circle. The parasite from a single infection are highlighted by dashed lines. MAL23 and MAL48 both contain multiple unlinked clusters indicative of superinfected parasites.
The distribution of total pairwise shared IBD and the average shared block lengths can be used to infer the relationships between individual genomes (Huff et al., 2011, Li et al., 2014). We inferred the degree of relatedness from our data using the Estimation of Recent Shared Ancestry (ERSA) algorithm. Under this scheme 0 denotes identical clones, 1 denotes parent-sibling, 2 denotes full/half siblings with higher numbers denoting increasingly distant relationships. ERSA estimates relatedness between individuals from distribution of IBD tract lengths (Figure 5A, B, Figure S3) using individuals assumed to be unrelated from the same population as a reference. We see a spectrum of relationships within each infection (Figure 5C). In MAL5 this confirmed the lack of relatedness between the two clusters of parasites, suggesting this infection was the result of a genuine superinfection. However, no other infections can be classified so simply, commonly showing relationships as distant as 4th degree (equivalent to ‘first cousins’). Within our data this suggests that it is not uncommon for parasites to be transmitted through two generations (human-mosquito-human-mosquito-human), with up to four generations of co-transmission seen in our data in infection MAL24 and MAL17. In our data we see only a single unambiguous instance of superinfection of two unrelated parasites with no concurrent co-transmission (MAL5). Across the analysis, the genetic diversity of three infections (MAL17, MAL24 and MAL48) appears to be driven by both superinfection of unrelated parasites and co-transmission of related parasites (in addition to MAL5 where only superinfection is suspected).
Figure 5. Recent ancestry inferred from IBD sharing.
(A) Density plot of the total IBD shared between parasites from a single infection (labeled to the left of the plot). (B) Density plot of the mean IBD block length between parasites from a single infection. The dotted lines in A and B show the expected value for parasites separated by differing number of meiosis, e.g. clonally identical (~21Mb total IBD/~1.5Mb Mean IBD length) and separated by a single meiosis (~10Mb total IBD/~0.6Mb Mean IBD length). The most distant relationship shown is 5 meiosis. (C) Relative frequency of different degrees of relatedness inferred between parasites from the same infection using the ERSA algorithm (ns - no significant relatedness observed). (D) The number of mitochondrial haplotypes identified in this infection.
Mitochondrial inheritance within individual infections
Mitochondria are inherited maternally during malaria parasite meiosis and therefore allow the identification of parasites which share a maternal lineage within infections. We found 93 SNPs in the mitochondrial DNA which varied within our dataset. Across all 498 parasites we were able to capture the genotype of 79.1% (36,645/46,314) of these 93 sites. We expect parasites which are identical across their nuclear chromosomes to also be identical across their mitochondrial genome. We find this to be the case. Within individual hosts, parasites which shared 100% of their genome IBD also shared 100% of their mitochondrial genome sequence. Across the 498 genome sequences for which we had reliable estimates of genome-wide IBD and mitochondrial genotypes there were only 2 of a possible 36,645 sites (0.0055%) where the mitochondrial genotype varied within a group of parasites which were 100% IBD. These arise at position 1,692 in cells 9 and 22 of infection MAL24. Visual inspection of these sites (Figure S4) and the presence of the reads supporting both genotypes in the bulk genome sequence supports these being genuine. However, as these may have arisen as de novo mutations during the current infection, or be artifacts of the WGA and genome sequencing pipeline we excluded this site from further analysis.
We identified a total of 20 unique mitochondrial haplotypes across the entire dataset (Figure S5 and Figure S6). We counted the number of mitochondrial haplotypes present in each infection (Figure 5D). This showed 9 infections contained a single mitochondrial haplotype, 3 infections contained 2 haplotypes, 1 infection contained 3 haplotypes, and 2 infections contained 4 haplotypes. All monoclonal infections contained a single mitochondrial haplotype, as did 4/10 (40%) polyclonal infections. Within polyclonal infections distinct mitochondrial haplotypes were observed between very close relatives (sharing >60% of their genomes IBD) supporting the retention of diversity we observe in the face of recurrent inbreeding. As mitochondria are uniparentally inherited the presence of multiple mitochondrial haplotypes across related parasites from within the same infection demonstrates multiple oocysts within a mosquito are responsible for initiating an infection. We tested if a higher level of shared pair-wise IBD is a predictor of sharing a mitochondrial haplotypes between two parasites. After excluding clonally identical parasites we estimate a 10% increase in IBD shared between parasites from the same infections increasing the odds of sharing a mitochondrial genotype by a factor of 10.082 (p=5.25×10−8, logistic regression).
Identification of meiotic siblings and reconstruction of parental haplotypes
During meiosis a tetrad of four recombinant progeny arise from a single paired set of chromosomes. Genetic characterization of tetrads has enabled precise capture of tracts of gene conversion, cross-overs and non-cross-overs in yeast, mammalian and plant genetics (Cole et al., 2014, Hou et al., 2013, Mancera et al., 2011, Li et al., 2015). Identifying progeny which came from the same tetrad, rather than simply sharing the same parents, allows us to estimate the number of ookinetes giving rise to an infection. As meiotic siblings share reciprocal recombination breakpoints, it also enables a fine-scale measurement of gene conversion and non-Mendelian inheritance in natural infections. Chance identification of potential meiotic siblings in low transmission regions (Dharia et al., 2010) and P. vivax (Bright et al., 2014) has suggested that tetrad analysis may be feasible. We identified potential members of tetrads as those sharing the same mitochondrial genotype with a coefficient of relatedness of 1, which would contain half-sibs, full-siblings, and meiotic siblings. For each group we inferred the parental haplotypes and recombination breakpoints using Hapi (Li et al., 2018). Following this we filtered out parasites which did not share reciprocal breakpoints consistent with shared meiosis.
Across the dataset we were able to capture one complete tetrad, from infection MAL15 (Figure 6). In the absence of complete tetrads it is difficult to definitively conclude parasites came from the same meiosis, or simply share identical parents. However, we found 10 groups of parasites sharing reciprocal breakpoints between 2–4 genotypes (inclusive of the tetrad from MAL15) suggesting they arose from the same meiosis (Data S3). Based on these potential mating’s we suggest the minimum number of ookinetes required in an infected mosquito for each infection as between 1 and 5 (Table S1). Importantly, despite exhaustive capture of genetic diversity in nearly all infections, most members of a tetrad do not reach observable frequencies in the blood stream. This suggests there is a considerable attrition in the representation of progeny between the mosquito midgut and the human blood stage. We estimated crossover locations across the dataset using a total of 570 unique crossovers. As seen in previous studies on P. falciparum genetic crosses (Jiang et al., 2011) chromosome length and crossover count were correlated (r2=0.9, p=8.43×10−6, Pearson’s correlation, Figure S7).
Figure 6. Patterns of recombination in a tetrad formed during meiosis.
Two parental haplotypes (red and blue) were inferred using Hapi and the inheritance of these haplotypes was inferred in the four parasite genotypes shown here. Across the genome there is consistent 1:1 inheritance of parental genomes aside from the proximal end of chromosome 4 and the distal end of chromosome 2 (green boxes, also shown expanded). Inbreeding among the ancestors of the parental genotypes has eroded variation on chromosomes 8 and 9 (orange boxes).
The capture of a complete tetrad allows us to observe non-Mendelian inheritance in a natural malaria genetic cross. We identified 33 recombination events across the genome, with no cross-overs detected on chromosomes 1,7 and 9. We found evidence for 2 major tracts of skewed inheritance resulting from gene conversion. Adjacent to the right-most telomere of chromosome 2 a tract of 872 bp covering 18 markers, and adjacent to the left-most telomere of chromosome 4 a tract of 180,970bp. Within the chromosome 4 region there is a block of 19,432bp from one parent, and 161,538bp from the other. In addition to regions of skewed inheritance we saw 2 large regions (770,168bp on chromosome 8 and 51,426bp on chromosome 9) where variation between the parents had been eliminated by inbreeding. Other regions with no variation distinguishing parents surrounded known hypervariable genes (Miles et al., 2016), and were not included in our analysis.
DISCUSSION
There has been a concerted effort to understand the complexity of malaria infections from either deep sequencing data (Assefa et al., 2014, O’Brien et al., 2016, Zhu et al., 2018), or from genotyping a limited number of markers (Chang et al., 2017, Galinsky et al., 2015). We show here there is considerable depth to complex infections which may be challenging to infer from bulk analysis alone. Through a combination of deep sequencing of bulk infections and single-cell sequencing, we have generated the most comprehensive picture of the within-host diversity of malaria infections to date. This provides a much-needed standard for developing novel tools for probing the complexity of infections from deep sequencing data. By using multiple estimates of relatedness targeting distinct features of the data, we argue that most complex infections result from parasites co-transmitted from single mosquito bites in our dataset. Strikingly, our analysis supports only a single infection where simple superinfection of two unrelated strains has occurred (MAL5), and a further three infections where both superinfection and co-transmission have concurrently contributed to diversity (MAL17, MAL24, MAL48). The remaining infections were either monomorphic or showed strong support for co-transmission of related strains only. Notably, the most diverse infection we studied (MAL15) was explained entirely by co-transmission of related parasites. In the two infections where we were unable to capture the minor strains (MAL33 and MAL37), patterns of unfixed SNPs within the infection suggest the uncaptured strain was related to the captured strain (Data S1). It may be that the minor isolate failed to develop to DNA-rich late stages or was present at a fraction lower than we were able to sample within the constraints of this work.
In this work we have generated a detailed picture of within-host genetic variation across fifteen malaria patients. While this is a modest number of infections, these findings are directly informative about the limits of malaria complexity. We have surveyed a high transmission setting and focused on polyclonal infections. This sampling enriches for infections with the highest likelihood of genetic diversity arising from superinfection rather than co-transmission. In spite of this we find co-transmission to be widespread, and likely underappreciated as a mechanism generating and maintaining genetic diversity in natural malaria populations. There is a pressing need to extend these observations across the range of malaria endemicity to fully capture the transmission network of malaria infections.
These results could not have been obtained by statistical inference of bulk sequence data alone. There have been impressive advances in imputation of individual parasite haplotypes from bulk sequence data with the development of DEploid (Zhu et al., 2018). We identify two to seventeen haplotypes in the infections we dissect here, with 50% of our infections bearing six or more unique haplotypes. As DEploid typically limits inference to five haplotypes, reliance on bulk inference would have discounted information on the additional haplotypes. When we examine the haplotypes inferred by DEploid we see the accuracy of imputation is not equivalent across all haplotypes in an infection (Figure 3C). Majority haplotypes inferred by DEploid share high similarity (median of 97.8%) to single-cell haplotypes, suggesting DEploid is a highly effective at capturing common haplotypes. However, minority haplotypes (5–50% abundance) were captured poorly (median 84.3% similarity). Encouragingly, we see particularly good performance for DEploid in inferring the composition of a single infection with two unrelated haplotypes (MAL5). The relatively poor performance of inference in other infections suggests that incorporating inbreeding and complex relatedness structures may lead to algorithmic improvements. In general, the data we present here provides an ideal training set for improvements in the implementation of statistical tools for understanding polyclonal infections and in generating guidelines for the interpretation of haplotypes inferred from bulk sequence data.
Only parasites which transmit gametes to the same mosquito can produce recombinant offspring. Patterns of parasite diversity and relatedness within individual mosquitoes (Annan et al., 2007) (albeit in a distinct population) are in general agreement with our results – most mating is between related parasites. The mechanisms underlying why inbreeding is common, even in high transmission settings, is less clear. Malaria transmission is intense in Chikhwawa (Mzilahowa et al., 2012) and we expected superinfection to be more prevalent than we observed. A mechanism controlling the outcome of superinfection, perhaps by hepcidin based inhibition of liver development in superinfecting sporozoites (Portugal et al., 2011), could explain why we do not see more superinfection. Alternatively, the low numbers of superinfecting parasites emerging from the liver relative to those present in established infections (which may contain 1011−12 blood stage parasites) may limit establishment of superinfections. This would represent an infectious disease example of the “priority” effects (De Meester et al., 2016) that are important in determining assemblies of ecological communities or microbiomes. Immune-mediated selection of parasite variants sharing alleles at the major antigenic loci (Farnert et al., 2008) could also generate a strong relatedness structure within infections. However, in this study, the effects of host immunity are likely to be limited because only infections from children who have little or no pre-existing malaria immunity were studied. In analyzing why superinfection is less common, it is also worth noting that analysis of parasite diversity is generally limited to single blood draws due to the need to treat symptomatic patients expediently. As this sampling strategy may overlook sub-populations circulating at lower frequencies, there may be additional genetic variation which escapes routine analysis. Most importantly, the strong relatedness structure we observe will limit the amount of outbreeding in malaria parasites, even in regions of intense transmission. Restrained outbreeding amongst malaria parasites could profoundly shape the evolution of parasite virulence, drug resistance and malaria transmission dynamics as shown in mice (Wargo et al., 2007a, Wargo et al., 2007b, Huijben et al., 2010, Huijben et al., 2011, Alizon, 2013).
The depletion of genetic variation during repeated rounds of co-transmission has been previously modelled (Wong et al., 2018), suggesting a substantial decline in the number of clonal lineages and an increase in average relatedness can arise through a single transmission cycle. We directly observe this by reconstructing tetrads. In all but one case we do not observe complete tetrads in our data. Some members of the tetrads were either lost during the infection or were present at too low a frequency to be sampled in this study. Either of these scenarios suggest that genetic variation will be depleted over successive transmission cycles, a prospect directly explored in Figure 5. Our data suggest complex infections comprise parasites which have been co-transmitted longer than two transmission cycles. Due to the lengthy developmental cycle of the parasite, and the propensity for mosquitoes to disperse the potential for these patterns to be driven by local population structure is minimal (Conway and McBride, 1991, Prugnolle et al., 2008). We observe that substantial genetic variation is maintained despite the bottleneck of mosquito transmission with up to 17 unique haplotypes likely inoculated by a single mosquito. Understanding how patterns of transmission and within host dynamics contribute to the diversity and relatedness structure within malaria infections will help inform ongoing elimination and control efforts.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ian Cheeseman (ianc@txbiomed.org). Raw sequence data has been deposited at the sequence read archive (https://www.ncbi.nlm.nih.gov/sra) under study number SRP155167.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Malaria-infected blood samples (5 ml; thin smear parasitaemia: 0.2 to 21.8%) were obtained prior to treatment from children aged 19 to 116 months old presenting to Chikhwawa District Hospital in Malawi with uncomplicated P. falciparum malaria from February to June 2016. Blood samples were collected only from children whose parents or legal guardians provided consent. Ethical approval for this study was obtained from the University of Malawi College of Medicine Research and Ethics Committee (Protocol number P.02/13/1528) and the Liverpool School of Tropical Medicine Research Ethics Committee (Protocol number 14.035). Each research subject was assigned a unique ID number at the time of enrollment. Similarly, each blood sample was assigned a unique barcode at the time of collection. The sample barcode was used to identify and track each sample while being processed and analyzed in the laboratory. Detailed information about all research subjects and blood samples they donated are provided in Table S1.
METHOD DETAILS
Sample Collection
Venous blood (5ml) from each subject was collected directly into an Acid Citrate Dextrose tube (BD, UK). The sample was immediately placed in an ice-cold container and transported to our laboratory in Blantyre within six hours of collection. Half of each blood sample was washed using incomplete RPMI 1640 media (Sigma-Aldrich, UK). Three aliquots of the sample were cryopreserved in glycerolyte 57 solution (Fenwal, Lake Zurich, IL, USA) and stored in liquid nitrogen. Parasites used in fluorescence-activated single-cell sorting were cultured from one of these aliquots. The second half of the sample was filtered using CF11 columns to deplete human leucocytes (Venkatesan et al., 2012), and was stored at −80°C until needed. Parasite DNA was extracted from this sample using a DNA Mini Kit (QIAGEN, USA) and directly sequenced on an lllumina HiSeq instrument.
Selection of samples for single cell capture
Using the FWS metric we stratified samples into monoclonal and polyclonal based on a threshold of 0.95. We randomly selected three monoclonal samples and twelve polyclonal samples for further analysis.
Single-cell capture
We performed single-cell capture, whole genome amplification, sequencing analysis and followed guidelines for preventing contamination using the approaches described in (Nair et al., 2014, Trevino et al., 2017). We outline these approaches below.
Parasite culture
Approximately 1 ml. of cryopreserved blood sample was thawed at 37°C to revive intact cells (~200ul recovered pellet, ~1% parasitemia). The sample was washed twice by adding 10mL of complete media (filter-sterilized incomplete RPMI 1640 media to which 5% w/v of hypoxanthine and 8% w/v of albumax II were added). Following the final centrifugation step (425 × g for 5 minutes) cells were resuspended and grown in 8 ml. complete media in a sealed T25 tissue culture flask flushed with 5% C02, 5% 02 and 90% N2 prior to being sealed. The culture flask was incubated at 37°C for 40 hours to allow for parasite progression to late stages, which generates higher quality genomic data after MDA and library preparation (Trevino et al., 2017). To stain parasitized cells in readiness for FACS, ~8 μl of an infected red blood cell pellet (~108 cells) was resuspended in 10 ml. of 1× PBS (Lonza, USA) which included 5 pi of Vibrant DyeCycle Green at 37°C for 30 minutes with intermittent manual inversion of the tube approximately every 10 minutes. Cells were washed once in 1× PBS and resuspended in 5 – 8 mL of 1× PBS in a foil-covered tube to protect the dye from photobleaching in preparation for FACS sorting. Catalog numbers for the critical reagents used in parasite culture and capture of single parasitized cells are provided in the Key Resources Table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological Samples | ||
| Infected red blood cells | This Study | MAL1 |
| Infected red blood cells | This Study | MAL2 |
| Infected red blood cells | This Study | MAL3 |
| Infected red blood cells | This Study | MAL4 |
| Infected red blood cells | This Study | MAL5 |
| Infected red blood cells | This Study | MAL6 |
| Infected red blood cells | This Study | MAL7 |
| Infected red blood cells | This Study | MAL8 |
| Infected red blood cells | This Study | MAL9 |
| Infected red blood cells | This Study | MAL10 |
| Infected red blood cells | This Study | MAL11 |
| Infected red blood cells | This Study | MAL12 |
| Infected red blood cells | This Study | MAL13 |
| Infected red blood cells | This Study | MAL14 |
| Infected red blood cells | This Study | MAL15 |
| Infected red blood cells | This Study | MAL16 |
| Infected red blood cells | This Study | MAL17 |
| Infected red blood cells | This Study | MAL18 |
| Infected red blood cells | This Study | MAL19 |
| Infected red blood cells | This Study | MAL20 |
| Infected red blood cells | This Study | MAL21 |
| Infected red blood cells | This Study | MAL22 |
| Infected red blood cells | This Study | MAL23 |
| Infected red blood cells | This Study | MAL24 |
| Infected red blood cells | This Study | MAL25 |
| Infected red blood cells | This Study | MAL26 |
| Infected red blood cells | This Study | MAL27 |
| Infected red blood cells | This Study | MAL28 |
| Infected red blood cells | This Study | MAL29 |
| Infected red blood cells | This Study | MAL30 |
| Infected red blood cells | This Study | MAL31 |
| Infected red blood cells | This Study | MAL32 |
| Infected red blood cells | This Study | MAL33 |
| Infected red blood cells | This Study | MAL34 |
| Infected red blood cells | This Study | MAL35 |
| Infected red blood cells | This Study | MAL36 |
| Infected red blood cells | This Study | MAL37 |
| Infected red blood cells | This Study | MAL38 |
| Infected red blood cells | This Study | MAL39 |
| Infected red blood cells | This Study | MAL40 |
| Infected red blood cells | This Study | MAL41 |
| Infected red blood cells | This Study | MAL42 |
| Infected red blood cells | This Study | MAL43 |
| Infected red blood cells | This Study | MAL44 |
| Infected red blood cells | This Study | MAL45 |
| Infected red blood cells | This Study | MAL46 |
| Infected red blood cells | This Study | MAL47 |
| Infected red blood cells | This Study | MAL48 |
| Infected red blood cells | This Study | MAL49 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DyeCycle Green | Vybrant | Cat#: V35004 |
| AccuGENE 1X PBS | Lonza | Cat #: 51225 |
| RPMI 1640 media | Gibco | Cat #: 11875119 |
| Glycerolyte 57 Solution | Fenwal | Cat #: 4A7831 |
| Cellulose Type B powder | Advantec | Cat #: 49020020 |
| HEPES Buffer Solution | Gibco | Cat #: 15630-080 |
| Gentamicin Reagent Solution | Gibco | Cat #: 15710064 |
| AlbuMAXTM II Lipid-Rich BSA | Gibco | Cat #: 11021029 |
| Hypoxanthine | Sigma | Cat #: H9636 |
| Critical Commercial Assays | ||
| Single-Cell FX DNA | QIAGEN | Cat#: 180714 |
| Deposited Data | ||
| Single-cell sequence data | This Paper | Study Number: SRP155167 |
| Software and Algorithms | ||
| moimix | NA | https://github.com/bahlolab/moimix/ |
| DEploid v0.5 | Zhu et al., 2018 | https://github.com/mcveanlab/DEploid/ |
| SeqArray v1.12.9 | Zheng et al., 2017 | http://github.com/zhengxwen/SeqArray/ |
| rareNMtests | Cayuela and Gotelli., 2014 | https://cran.r-project.org/web/packages/rareNMtests/index.html |
| hmmIBD v2.0.0 | Schaffner et al., 2018 | https://github.com/glipsnort/hmmIBD/ |
| ERSA | Huff et al., 2011; Li et al., 2014 | http://www.hufflab.org/software/ersa/ |
| R v3.4.0 | Team., 2017 | https://www.r-project.org/ |
| GATK v3.5 | De Pristo et al., 2011 | https://software.broadinstitute.org/gatk/ |
| BWA MEM v0.7.5a | Li, 2013 | http://bio-bwa.sourceforge.net/ |
| Picard v1.56 | NA | http://broadinstitute.github.io/picard/ |
| Hapi v0.0.1 | Li, 2018 | https://github.com/Jialab-UCR/Hapi/ |
FACS Sorting
Cells were sorted by MoFlo Astrios (Beckman Coulter) by gating the two brightest observed populations according to DNA fluorescence, the sort was run in single-cell sort mode with a drop envelope of 0.5. Individual cells were sorted into 0.2 mL PCR tubes containing 5 pi autoclaved sterile PBS (Lonza), which had been prepared under sterile conditions in a PCR hood. Each event required about 15 seconds to open the tube, place on the sorting rack, recover, and close the tube. Tubes were then immediately stored on dry ice and transferred to −80 °C longer-term storage within an hour.
Generation of single-cell DNA libraries
Library preparation for individually sorted late-stage parasites was carried out using the Qiagen Single-Cell FX DNA kit without library amplification according to manufacturer’s instructions. Whole genome amplification preparation was carried out under a PCR hood and DNA was amplified on a dedicated PCR machine. Library products were analyzed by TapeStation and included off-target peaks typical of MDA DNA inputs. Adapter-ligated DNA products were quantified by KAPA Hyperplus Kits. All sequencing was performed on an Illumina HiSeq 2500. A detailed description of the development of the single-cell sequencing methods used here, and the protocols in place to control for contamination are available (Nair et al., 2014, Trevino et al., 2017). Raw sequence data has been deposited at the sequence read archive (https://www.ncbi.nlm.nih.gov/sra) under study number SRP155167.
Sequence analysis
We aligned raw sequencing reads to v3 of the 3D7 genome reference (http://www.plasmodb.org) using BWA MEM v0.7.5a (Li, 2013). After removing PCR duplicates and reads mapping to the ends of chromosomes (Picard v1.56) we recalibrated base quality scores, realigned around indels and called genotypes using GATK v3.5 (DePristo et al., 2011) in the GenotypeGVCFs mode using QualByDepth, FisherStrand, StrandOddsRatio VariantType, GC Content and max_alterate_alleles set to 6. We recalibrated quality scores and calculated VQSLOD scores using SNP calls conforming to Mendelian inheritance, excluding sites where the VQSLOD score was <0. Median read depth of WGA single-cells was 28.3 (interquartile range (IQR) 12.5–46.4) with median of 90.5% (IQR 78.1–96.0%) of the genome covered by at least one read. In contrast the non-WGA samples had a median read depth of 31.11 (IQR 20.93–48.37) and a median of 95.8% (IQR 93.1–97.4%) of the genome covered by at least one read. A potential source of error in single-cell genomics is the inclusion of exogenous DNA amplified alongside the target genome in downstream analysis. As an initial indication of the potential of non-target DNA being introduced to our analysis we first examined the proportion of reads mapping to the P. falciparum genome (Gardner et al., 2002) in each sequence. We observed a median of 93.3% (IQR 87.0–95.4%) of reads map to the parasite genome for single-cell sequences, compared to 35.7% (IQR 19.7–48.5%) for bulk patient samples and 79.4% (IQR 74.5–86.9%) for clonally expanded samples suggesting our stringent handling protocols were effective at eliminating environmental DNA. For a more rigorous test we identified lines with potential cross contamination based on unfixed basecall frequency. As the parasite genome is haploid during blood stages all variants are expected to be fixed in genome sequencing data. The highly AT-rich and repetitive nature of the parasite genome makes alignment challenging, generating false positive unfixed variants in clonal lines. After excluding highly error-prone genomic regions (calls outside of the “core genome”(Miles et al., 2016) or within microsatellites) we measured the proportion of mixed base calls (>5% of reads at a locus mapping to the minority allele) at high confidence biallelic SNPs (>10 reads mapped, VQSLoD>0, GQ>70). Using the cloned lines and bulk population samples as a guide we estimated 1% as an appropriate threshold for excluding putatively mixed lines (Figure S1).
While each single cell genome sequence is independent, clonally identical cells provide an estimate of how replicable our approach is. Parasites classified as clonally identical in our downstream analysis were identical across an average of 99.97% of high quality genotypes identified above.
QUANTIFICATION AND STATISTICAL ANALYSIS
Estimating the complexity and diversity of bulk sequenced samples
FWS was calculated in moimix (https://github.com/bahlolab/moimix) for all bulk patient samples. We estimated the number of unique haplotypes and their sequence from deep sequence of bulk infections using DEploid (Zhu et al., 2018) v0.5 (https://github.com/mcveanlab/DEploid). We used 10,997 HQ SNPs with a MAF >5%. For a reference panel we used 10 bulk Malawian samples presumed to be clonal (FWS>0.95) and population level allele frequencies from across the complete bulk sequencing data. We inferred the most likely number of haplotypes (K) using the command:
/dEploid -ref sample_reference_allele_counts.txt -alt sample_alternative_allele_counts.txt -plaf population_allele_freq.txt -o sample_out -ibd -noPanel -exclude highly_variable_sites.txt -sigma 7 -seed 2
Estimating relatedness between sequences
SNP data were imported into R using SeqArray (Zheng et al., 2017). Between all samples passing quality control we calculated the proportion of shared alleles and using SNPs which were at >5% MAF in the bulk sequenced samples. We used a distance matrix generated from this data (1-pairwise allele sharing) to estimate the number of unique haplotypes in each infection by collapsing together sequences which differed at <1% of sites. Rarefaction of haplotype abundance was performed using the rareNMtests package (Cayuela and Gotelli, 2014) in R. We called regions of IBD between all samples passing quality control using hmmlBD v2.0.0 (Schaffner et al., 2018) (https://github.com/glipsnort/hmmIBD). We performed maximum-likelihood estimation of recent shared ancestry using ERSA 2.0 (Huff et al., 2011, Li et al., 2014) (http://www.hufflab.org/software/ersa/) using the output from hmmlBD using the flags -- min_cm=1.5 --adjust_pop_dist=true --number_of_chromosomes=14 --rec_per_meioses=19. We converted the basepair positions to a uniform genetic map using the scaling factor 1cM=9.6kb (Jiang et al., 2011) and excluded IBD chunks <1cM in length. As identical clones are not specifically modelled in ERSA we excluded these from analysis, though their abundance is shown in the ‘0’ bar in Figure 5C. All other statistical analysis and visualization was performed in R v3.4.0 (Team, 2017). Inference of parental genotypes and recombination breakpoints was performed using Hapi.
Supplementary Material
Table S1. Haplotype diversity of malaria infections.
Highlights.
485 single genome sequences reveal patterns of relatedness within malaria infections
Co-transmission of related parasites is more common than superinfection
Serial passage of complex infections without loss of diversity is commonplace
Reconstruction of a single meiosis reveals the extent of inbreeding in mosquitoes
ACKNOWLEDGEMENTS
We thank all children and their families who participated in this study in Chikhwawa, Malawi. We also thank Andrew Mtande, Ruth Daiman and Miriam Phiri for their help with participant recruitment and clinical management. We are also grateful to Clement Masesa and Lumbani Makhaza for designing our data collection tool and for managing the study database. This study was supported by a Wellcome Trust Intermediate Fellowship in Tropical Medicine and Public Health (Grant # 099992/Z/12/Z to SCN) and an NIH grant (NIAID AI110941-01A1 to IHC). FACS data were generated in the Flow Cytometry Shared Resource Facility (supported by UTHSCSA, NIH-NCI P30 CA054174, UL1RR025767 (CTSA)).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DATA AND CODE AVAILABILITY
This study did not generate any novel code. The data generated during this study are available from the sequence read archive (https://www.ncbi.nlm.nih.gov/sra) under study number SRP155167.
REFERENCES
- ALIZON S. 2013. Parasite co-transmission and the evolutionary epidemiology of virulence. Evolution, 67, 921–33. [DOI] [PubMed] [Google Scholar]
- ANNAN Z, DURAND P, AYALA FJ, ARNATHAU C, AWONO-AMBENE P, SIMARD F, RAZAKANDRAINIBE FG, KOELLA JC, FONTENILLE D. & RENAUD F. 2007. Population genetic structure of Plasmodium falciparum in the two main African vectors, Anopheles gambiae and Anopheles funestus. Proc Natl Acad Sci U S A, 104, 7987–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ASSEFA SA, PRESTON MD, CAMPINO S, OCHOLLA H, SUTHERLAND CJ & CLARK TG 2014. estMOI: estimating multiplicity of infection using parasite deep sequencing data. Bioinformatics, 30, 1292–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AUBURN S, CAMPINO S, MIOTTO O, DJIMDE AA, ZONGO I, MANSKE M, MASLEN G, MANGANO V, ALCOCK D, MACINNIS B, ROCKETT KA, CLARK TG, DOUMBO OK, OUEDRAOGO JB & KWIATKOWSKI DP 2012. Characterization of within-host Plasmodium falciparum diversity using next-generation sequence data. PLoS One, 7, e32891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BELL AS, DE ROODE JC, SIM D. & READ AF 2006. Within-host competition in genetically diverse malaria infections: parasite virulence and competitive success. Evolution, 60, 1358–71. [PubMed] [Google Scholar]
- BRIGHT AT, MANARY MJ, TEWHEY R, ARANGO EM, WANG T, SCHORK NJ, YANOW SK & WINZELER EA 2014. A high resolution case study of a patient with recurrent Plasmodium vivax infections shows that relapses were caused by meiotic siblings. PLoS Negl Trop Dis, 8, e2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAYUELA L. & GOTELLI NJ 2014. rareNMtests: Ecological and biogeographical null model tests for comparing rarefaction curves. [Google Scholar]
- CHANG HH, WORBY CJ, YEKA A, NANKABIRWA J, KAMYA MR, STAEDKE SG, DORSEY G, MURPHY M, NEAFSEY DE, JEFFREYS AE, HUBBART C, ROCKETT KA, AMATO R, KWIATKOWSKI DP, BUCKEE CO & GREENHOUSE B. 2017. THE REAL McCOIL: A method for the concurrent estimation of the complexity of infection and SNP allele frequency for malaria parasites. PLoS Comput Biol, 13, e1005348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COLE F, BAUDAT F, GREY C, KEENEY S, DE MASSY B. & JASIN M. 2014. Mouse tetrad analysis provides insights into recombination mechanisms and hotspot evolutionary dynamics. Nat Genet, 46, 1072–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CONWAY DJ, GREENWOOD BM & MCBRIDE JS 1991. The epidemiology of multiple-clone Plasmodium falciparum infections in Gambian patients. Parasitology, 103 Pt 1, 1–6. [DOI] [PubMed] [Google Scholar]
- CONWAY DJ & MCBRIDE JS 1991. Genetic evidence for the importance of interrupted feeding by mosquitoes in the transmission of malaria. Trans R Soc Trop Med Hyg, 85, 454–6. [DOI] [PubMed] [Google Scholar]
- DE MEESTER L, VANOVERBEKE J, KILSDONK LJ & URBAN MC 2016. Evolving Perspectives on Monopolization and Priority Effects. Trends Ecol Evol, 31, 136–146. [DOI] [PubMed] [Google Scholar]
- DE ROODE JC, PANSINI R, CHEESMAN SJ, HELINSKI ME, HUIJBEN S, WARGO AR, BELL AS, CHAN BH, WALLIKER D. & READ AF 2005. Virulence and competitive ability in genetically diverse malaria infections. Proc Natl Acad Sci U S A, 102, 7624–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DEPRISTO MA, BANKS E, POPLIN R, GARIMELLA KV, MAGUIRE JR, HARTL C, PHILIPPAKIS AA, DEL ANGEL G, RIVAS MA, HANNA M, MCKENNA A, FENNELL TJ, KERNYTSKY AM, SIVACHENKO AY, CIBULSKIS K, GABRIEL SB, ALTSHULER D. & DALY MJ 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet, 43, 491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DHARIA NV, PLOUFFE D, BOPP SE, GONZALEZ-PAEZ GE, LUCAS C, SALAS C, SOBERON V, BURSULAYA B, KOCHEL TJ, BACON DJ & WINZELER EA 2010. Genome scanning of Amazonian Plasmodium falciparum shows subtelomeric instability and clindamycin-resistant parasites. Genome Res, 20, 1534–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EARLY AM, LIEVENS M, MACINNIS BL, OCKENHOUSE CF, VOLKMAN SK, ADJEI S, AGBENYEGA T, ANSONG D, GONDI S, GREENWOOD B, HAMEL M, ODERO C, OTIENO K, OTIENO W, OWUSU-AGYEI S, ASANTE KP, SORGHO H, TINA L, TINTO H, VALEA I, WIRTH DF & NEAFSEY DE 2018. Host-mediated selection impacts the diversity of Plasmodium falciparum antigens within infections. Nat Commun, 9, 1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FARNERT A, LEBBAD M, FARAJA L. & ROOTH I. 2008. Extensive dynamics of Plasmodium falciparum densities, stages and genotyping profiles. Malar J, 7, 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GALINSKY K, VALIM C, SALMIER A, DE THOISY B, MUSSET L, LEGRAND E, FAUST A, BANIECKI ML, NDIAYE D, DANIELS RF, HARTL DL, SABETI PC, WIRTH DF, VOLKMAN SK & NEAFSEY DE 2015. COIL: a methodology for evaluating malarial complexity of infection using likelihood from single nucleotide polymorphism data. Malar J, 14, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GARDNER MJ, HALL N, FUNG E, WHITE O, BERRIMAN M, HYMAN RW, CARLTON JM, PAIN A, NELSON KE, BOWMAN S, PAULSEN IT, JAMES K, EISEN JA, RUTHERFORD K, SALZBERG SL, CRAIG A, KYES S, CHAN MS, NENE V, SHALLOM SJ, SUH B, PETERSON J, ANGIUOLI S, PERTEA M, ALLEN J, SELENGUT J, HAFT D, MATHER MW, VAIDYA AB,MARTIN DM, FAIRLAMB AH, FRAUNHOLZ MJ, ROOS DS, RALPH SA, MCFADDEN GI, CUMMINGS LM, SUBRAMANIAN GM, MUNGALL C, VENTER JC, CARUCCI DJ, HOFFMAN SL, NEWBOLD C, DAVIS RW, FRASER CM & BARRELL B. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature, 419, 498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HENDEN L, LEE S, MUELLER I, BARRY A. & BAHLO M. 2018. Identity-by-descent analyses for measuring population dynamics and selection in recombining pathogens. PLoS Genet, 14, e1007279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOU Y, FAN W, YAN L, LI R, LIAN Y, HUANG J, LI J, XU L, TANG F, XIE XS & QIAO J. 2013. Genome analyses of single human oocytes. Cell, 155, 1492–506. [DOI] [PubMed] [Google Scholar]
- HUFF CD, WITHERSPOON DJ, SIMONSON TS, XING J, WATKINS WS, ZHANG Y, TUOHY TM, NEKLASON DW, BURT RW, GUTHERY SL, WOODWARD SR & JORDE LB 2011. Maximum-likelihood estimation of recent shared ancestry (ERSA). Genome Res, 21, 768–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUIJBEN S, NELSON WA, WARGO AR, SIM DG, DREW DR & READ AF 2010. Chemotherapy, within-host ecology and the fitness of drug-resistant malaria parasites. Evolution, 64, 2952–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUIJBEN S, SIM DG, NELSON WA & READ AF 2011. The fitness of drug-resistant malaria parasites in a rodent model: multiplicity of infection. J Evol Biol, 24, 2410–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JIANG H, LI N, GOPALAN V, ZILVERSMIT MM, VARMA S, NAGARAJAN V, LI J, MU J, HAYTON K, HENSCHEN B, YI M, STEPHENS R, MCVEAN G, AWADALLA P, WELLEMS TE & SU XZ 2011. High recombination rates and hotspots in a Plasmodium falciparum genetic cross. Genome Biol, 12, R33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEUNG ML, WANG Y, WATERS J. & NAVIN NE 2015. SNES: single nucleus exome sequencing. Genome Biol, 16, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI H, GLUSMAN G, HU H, SHANKARACHARYA, CABALLERO J, HUBLEY R, WITHERSPOON D, GUTHERY SL, MAULDIN DE, JORDE LB, HOOD L, ROACH JC & HUFF CD. 2014. Relationship estimation from whole-genome sequence data. PLoS Genet, 10, e1004144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI HJAPA 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. [Google Scholar]
- LI R, QU H, CHEN J, WANG S, CHATER JM, ZHANG L, WEI J, ZHANG Y-M, XU C, ZHONG W-D, ZHU J, LU J, MA R, FERRANTE SP, ROOSE ML & JIA Z. 2018. Inference of Chromosome-length Haplotypes using Genomic Data of Three to Five Single Gametes. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI X, LI L. & YAN J. 2015. Dissecting meiotic recombination based on tetrad analysis by single-microspore sequencing in maize. Nat Commun, 6, 6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LU S, ZONG C, FAN W, YANG M, LI J, CHAPMAN AR, ZHU P, HU X, XU L, YAN L, BAI F, QIAO J, TANG F, LI R. & XIE XS 2012. Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science, 338, 1627–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MANCERA E, BOURGON R, HUBER W. & STEINMETZ LM 2011. Genome-wide survey of post-meiotic segregation during yeast recombination. Genome Biol, 12, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MANSKE M, MIOTTO O, CAMPINO S, AUBURN S, ALMAGRO-GARCIA J, MASLEN G, O’BRIEN J, DJIMDE A, DOUMBO O, ZONGO I, OUEDRAOGO JB, MICHON P, MUELLER I, SIBA P, NZILA A, BORRMANN S, KIARA SM, MARSH K, JIANG H, SU XZ, AMARATUNGA C, FAIRHURST R, SOCHEAT D, NOSTEN F, IMWONG M, WHITE NJ, SANDERS M, ANASTASI E, ALCOCK D, DRURY E, OYOLA S, QUAIL MA, TURNER DJ, RUANO-RUBIO V, JYOTHI D, AMENGA-ETEGO L, HUBBART C, JEFFREYS A, ROWLANDS K, SUTHERLAND C, ROPER C, MANGANO V, MODIANO D, TAN JC, FERDIG MT, AMAMBUA-NGWA A, CONWAY DJ, TAKALA-HARRISON S, PLOWE CV, RAYNER JC, ROCKETT KA, CLARK TG, NEWBOLD CI, BERRIMAN M, MACINNIS B. & KWIATKOWSKI DP 2012. Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature, 487, 375–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MILES A, IQBAL Z, VAUTERIN P, PEARSON R, CAMPINO S, THERON M, GOULD K, MEAD D, DRURY E, O’BRIEN J, RUANO RUBIO V, MACINNIS B, MWANGI J, SAMARAKOON U, RANFORD-CARTWRIGHT L, FERDIG M, HAYTON K, SU XZ, WELLEMS T, RAYNER J, MCVEAN G. & KWIATKOWSKI D. 2016. Indels, structural variation, and recombination drive genomic diversity in Plasmodium falciparum. Genome Res, 26, 1288–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MU J, AWADALLA P, DUAN J, MCGEE KM, JOY DA, MCVEAN GA & SU XZ 2005. Recombination hotspots and population structure in Plasmodium falciparum. PLoS Biol, 3, e335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MU J, AWADALLA P, DUAN J, MCGEE KM, KEEBLER J, SEYDEL K, MCVEAN GA & SU XZ 2007. Genome-wide variation and identification of vaccine targets in the Plasmodium falciparum genome. Nat Genet, 39, 126–30. [DOI] [PubMed] [Google Scholar]
- MZILAHOWA T, HASTINGS IM, MOLYNEUX ME & MCCALL PJ 2012. Entomological indices of malaria transmission in Chikhwawa district, Southern Malawi. Malar J, 11, 380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NAIR S, NKHOMA SC, SERRE D, ZIMMERMAN PA, GORENA K, DANIEL BJ, NOSTEN F, ANDERSON TJ & CHEESEMAN IH 2014. Single-cell genomics for dissection of complex malaria infections. Genome Res, 24, 1028–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NEAFSEY DE, SCHAFFNER SF, VOLKMAN SK, PARK D, MONTGOMERY P, MILNER DA JR., LUKENS A, ROSEN D, DANIELS R, HOUDE N, CORTESE JF, TYNDALL E, GATES C, STANGE-THOMANN N, SARR O, NDIAYE D, NDIR O, MBOUP S, FERREIRA MU, MORAES SDO L, DASH AP, CHITNIS CE, WIEGAND RC, HARTL DL, BIRREN BW, LANDER ES, SABETI PC & WIRTH DF 2008. Genome-wide SNP genotyping highlights the role of natural selection in Plasmodium falciparum population divergence. Genome Biol, 9, R171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NKHOMA SC, NAIR S, CHEESEMAN IH, ROHR-ALLEGRINI C, SINGLAM S, NOSTEN F. & ANDERSON TJ 2012. Close kinship within multiple-genotype malaria parasite infections. Proc Biol Sci, 279, 2589–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’BRIEN JD, IQBAL Z, WENDLER J. & AMENGA-ETEGO L. 2016. Inferring Strain Mixture within Clinical Plasmodium falciparum Isolates from Genomic Sequence Data. PLoS Comput Biol, 12, e1004824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEARSON RD, AMATO R, AUBURN S, MIOTTO O, ALMAGRO-GARCIA J, AMARATUNGA C, SUON S, MAO S, NOVIYANTI R, TRIMARSANTO H, MARFURT J, ANSTEY NM, WILLIAM T, BONI MF, DOLECEK C, HIEN TT, WHITE NJ, MICHON P, SIBA P, TAVUL L, HARRISON G, BARRY A, MUELLER I, FERREIRA MU, KARUNAWEERA N, RANDRIANARIVELOJOSIA M, GAO Q, HUBBART C, HART L, JEFFERY B, DRURY E, MEAD D, KEKRE M, CAMPINO S, MANSKE M, CORNELIUS VJ, MACINNIS B, ROCKETT KA, MILES A, RAYNER JC, FAIRHURST RM, NOSTEN F, PRICE RN & KWIATKOWSKI DP 2016. Genomic analysis of local variation and recent evolution in Plasmodium vivax. Nat Genet, 48, 959–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PORTUGAL S, CARRET C, RECKER M, ARMITAGE AE, GONCALVES LA, EPIPHANIO S, SULLIVAN D, ROY C, NEWBOLD CI, DRAKESMITH H. & MOTA MM 2011. Host-mediated regulation of superinfection in malaria. Nat Med, 17, 732–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PRUGNOLLE F, DURAND P, JACOB K, RAZAKANDRAINIBE F, ARNATHAU C, VILLARREAL D, ROUSSET F, DE MEEUS T. & RENAUD F. 2008. A comparison of Anopheles gambiae and Plasmodium falciparum genetic structure over space and time. Microbes Infect, 10, 269–75. [DOI] [PubMed] [Google Scholar]
- REECE SE, DREW DR & GARDNER A. 2008. Sex ratio adjustment and kin discrimination in malaria parasites. Nature, 453, 609–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REILLY HB, WANG H, STEUTER JA, MARX AM & FERDIG MT 2007. Quantitative dissection of clone-specific growth rates in cultured malaria parasites. Int J Parasitol, 37, 1599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROSARIO V. 1981. Cloning of naturally occurring mixed infections of malaria parasites. Science, 212, 1037–8. [DOI] [PubMed] [Google Scholar]
- SCHAFFNER SF, TAYLOR AR, WONG W, WIRTH DF & NEAFSEY DE 2018. hmmIBD: software to infer pairwise identity by descent between haploid genotypes. Malar J, 17, 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TEAM, R. C. 2017. R: A Language and Environment for Statistical Computing. [Google Scholar]
- TREVINO SG, NKHOMA SC, NAIR S, DANIEL BJ, MONCADA K, KHOSWE S, BANDA RL, NOSTEN F. & CHEESEMAN IH 2017. High-Resolution Single-Cell Sequencing of Malaria Parasites. Genome Biol Evol, 9, 3373–3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VENKATESAN M, AMARATUNGA C, CAMPINO S, AUBURN S, KOCH O, LIM P, UK S, SOCHEAT D, KWIATKOWSKI DP, FAIRHURST RM & PLOWE CV 2012. Using CF11 cellulose columns to inexpensively and effectively remove human DNA from Plasmodium falciparum-infected whole blood samples. Malar J, 11, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOLKMAN SK, NEAFSEY DE, SCHAFFNER SF, PARK DJ & WIRTH DF 2012. Harnessing genomics and genome biology to understand malaria biology. Nat Rev Genet, 13, 315–28. [DOI] [PubMed] [Google Scholar]
- WARGO AR, DE ROODE JC, HUIJBEN S, DREW DR & READ AF 2007a. Transmission stage investment of malaria parasites in response to in-host competition. Proc Biol Sci, 274, 2629–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WARGO AR, HUIJBEN S, DE ROODE JC, SHEPHERD J. & READ AF 2007b. Competitive release and facilitation of drug-resistant parasites after therapeutic chemotherapy in a rodent malaria model. Proc Natl Acad Sci U S A, 104, 19914–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO 2016. World malaria report 2015, World Health Organization. [Google Scholar]
- WONG W, GRIGGS AD, DANIELS RF, SCHAFFNER SF, NDIAYE D, BEI AK, DEME AB, MACINNIS B, VOLKMAN SK, HARTL DL, NEAFSEY DE & WIRTH DF 2017. Genetic relatedness analysis reveals the cotransmission of genetically related Plasmodium falciparum parasites in Thies, Senegal. Genome Med, 9, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WONG W, WENGER EA, HARTL DL & WIRTH DF 2018. Modeling the genetic relatedness of Plasmodium falciparum parasites following meiotic recombination and cotransmission. PLoS Comput Biol, 14, e1005923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHENG X, GOGARTEN SM, LAWRENCE M, STILP A, CONOMOS MP, WEIR BS, LAURIE C. & LEVINE D. 2017. SeqArray-a storage-efficient high-performance data format for WGS variant calls. Bioinformatics, 33, 2251–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHU SJ, ALMAGRO-GARCIA J. & MCVEAN G. 2018. Deconvolution of multiple infections in Plasmodium falciparum from high throughput sequencing data. Bioinformatics, 34, 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Haplotype diversity of malaria infections.