Abstract
Canine parvovirus type 2 (CPV‐2) emerged as dog pathogen in the late 1970s, causing severe and often fatal epizootics of gastroenteritis in the canine population worldwide. Although to date CPV‐2 is circulating in all continents, most of the current studies have analysed the amino acid changes accounted in the VP2 gene sequence, with limited information on virus introductions from other countries. The aim of this study was to analyse the genetic features of CPV‐2c strains currently spreading in Italy. Swabs and tissue samples were collected from dogs suspected of CPV infection. The nearly complete genome sequence from the CPV‐positive samples was obtained. The co‐circulation of two different but related CPV‐2c strains, with amino acid changes characteristic of CPV strains of Asian origin (NS1: 60V, 544F, 545F, 630P – NS2: 60V, 151N, 152V ‐ VP2: 5A/G, 267Y, 297A, 324I, 370R), were observed. The phylogenetic analyses inferred from the NS1 and VP2 gene sequences confirmed the relationship with Asian CPV‐2c strains. This study reports the spread of novel CPV‐2c mutants in Italy and supports further studies to evaluate the coexistence of genetically divergent CPV strains in the same geographical environment.
Keywords: Asia, canine parvovirus, carnivore protoparvovirus, dogs, molecular characterization, sequence analysis
1. INTRODUCTION
Canine parvovirus type 2 (CPV‐2) is a small, non‐enveloped single‐stranded DNA virus, recently included in the specie Carnivore protoparvovirus 1, a member of the Protoparvovirus genus (family Parvoviridae, subfamily Parvovirinae) (Cotmore et al., 2019). Its genome consists of an approximately 5,200 nucleotide (nt) DNA molecule containing two large open reading frames (ORFs), encoding two nonstructural (NS1 and NS2) and two structural (VP1 and VP2) proteins, generated through alternative splicing of the same mRNAs (Decaro & Buonavoglia, 2012; Reed, Jones, & Miller, 1988).
In susceptible non‐immunized dogs, CPV‐2 causes an acute and often lethal disease, whose clinical signs are characterized by vomiting, enteritis and acute lymphopenia (Decaro & Buonavoglia, 2012). CPV‐2 emerged as dog pathogen in the late 1970s, most likely as host variant of feline parvovirus (FPV) or a related strain (Truyen, 2006), displaying an intrinsic high rate of nucleotide changes (Decaro et al., 2009; Pereira, Leal, & Durigon, 2007; Shackelton, Parrish, Truyen, & Holmes, 2005). After its emergence, the original type CPV‐2 was replaced by three antigenic variants termed CPV‐2a, CPV‐2b and CPV‐2c (Buonavoglia et al., 2001; Parrish et al., 1991; Parrish, O’Connell, Evermann, & Carmichael, 1985). During the years, several amino acid (aa) changes were accounted in the VP2 gene sequence (Battilani et al., 2001; Geng et al., 2015; Jeoung, Ahn, & Kim, 2008; Nakamura et al., 2004; Truyen, 1999) and, only recently, the analysis of the NS1 gene sequence was included in the CPV phylogenies (Canuti, Rodrigues, Whitney, & Lang, 2017; Grecco et al., 2018; Han et al., 2015; Li et al., 2018; Mira et al., 2019; Pérez et al., 2014; Zhuang et al., 2019).
Previous studies provided information on the CPV strains spreading in Italy (Decaro, Desario, et al., 2007; Decaro et al., 2013, 2006; Dei Giudici et al., 2017; Mira, Dowgier, et al., 2018; Purpari et al., 2018; Tucciarone et al., 2018), suggesting the need of a continuous epidemiological survey to evaluate the CPV circulation and evolution, whereas limited data are available on the spread of novel strains imported from other continents (Mira, Purpari, Lorusso, et al., 2018). The aim of this study was the detection and molecular analysis of CPV strains displaying genetic features of Asian viruses spreading in southern Italy.
2. MATERIALS AND METHODS
During an epidemiological survey, rectal swabs (n = 3) and tissue samples (n = 19) from seven dogs suspected of CPV infection (Table 1), collected in southern Italy (Sicily) from August 2018 to March 2019, were analysed at the Istituto Zooprofilattico Sperimentale della Sicilia “A. Mirri” (Palermo, Italy) for diagnostic purposes. DNA and RNA were extracted from swab/organ homogenates, obtained as previously described (Purpari et al., 2018), using the DNeasy Blood & Tissue Kit (Qiagen S.p.A.) and QIAmp Viral RNA Mini Kit (Qiagen S.p.A.), respectively, according to the manufacturer's instructions. Presence of CPV DNA was evaluated by a diagnostic PCR using a primer pair targeting the VP2 gene (Touihri et al., 2009), as previously described (Mira, Purpari, Lorusso, et al., 2018), and each amplicon was analysed by electrophoresis on a 3% agarose gel supplemented with ethidium bromide.
Table 1.
Details on collected/tested samples of dogs
| Strain | Date of sampling | Place of sampling | Breed | Origin | Age | Sample | Acc. number |
|---|---|---|---|---|---|---|---|
| IZSSI_PA24478/18_id3184 | 20 Aug 2018 | Marsala | Mixed breed | Stray dog | 2 months | Brain, heart, lung, intestine, mesenteric lymph node, spleen | MK802679 |
| IZSSI_PA24478/18_id3230 | 20 Aug 2018 | Marsala | Mixed breed | Stray dog | 2 months | Brain, heart, lung, intestine, mesenteric lymph node, spleen | MK806280 |
| IZSSI_PA31342/18 | 31 Oct 2018 | Castelvetrano | Mixed breed | Stray dog | 3 months | Lung, intestine, spleen | MK806281 |
| IZSSI_PA5455/19 | 13 Nov 2018 | Partanna | Labrador retriever | Pet | 7 months | Rectal swab | MK806282 |
| IZSSI_PA5446/19 | 26 Nov 2018 | Marsala | Epagneul breton | Pet | 50 days | Rectal swab | MK806283 |
| IZSSI_RG3408/19 | 27 Feb 2019 | Comiso | Mixed breed | Pet | 3 months | Rectal swab | MK806284 |
| IZSSI_PA5632 | 04 Mar 2019 | Mazara del Vallo | Mixed breed | Stray dog | 2 years | Lung, heart, intestine, spleen | MK806285 |
Sequencing encompassing both ORFs (NS and VP genes) was carried out using primer pairs developed by Pérez et al. (2014), as previously described (Mira et al., 2019). Sequences were assembled according to an overlapping strategy and analysed using BioEdit ver 7.0.5.3 software (Hall, 1999). Assembled nucleotide sequences were submitted to nBLAST program (Zhang, Schwartz, Wagner, & Miller, 2000) to search related sequences in public domain databases. These sequence data have been submitted to the DDBJ/EMBL/GenBank databases under accession numbers MK802679‐85. The obtained sequences were aligned with reference sequences retrieved from the GenBank database, which included the sequence (accession number MF510157) of a CPV‐2c strain collected from the same region and previously analysed (Mira, Purpari, Lorusso, et al., 2018).
To elucidate the genetic relationships of the analysed CPV strains, two phylogenetic trees, based on the full‐length VP2 and NS1 gene sequences, were constructed with the MEGA X software (Kumar, Stecher, Li, Knyaz, & Tamura, 2018), using the maximum‐likelihood method according to the Tamura 3‐parameter (T92) and Hasegawa–Kishino–Yano (HKY) models, with discrete Gamma distribution (five rate categories) (G) and invariant sites (I) (bootstrap analyses with 1,000 replicates). The models selection was performed using the best‐fit model of nucleotide substitution with MEGA X software (VP2 gene: T92+G+I; NS1 gene: HKY+G).
Extracted DNA/RNA were also amplified using a set of PCR assays for the detection of canine distemper virus (CDV) (Elia et al., 2006), canine adenovirus (CAdV) type 1 and type 2 (Dowgier et al., 2016), canine herpesvirus (CaHV‐1) (Decaro et al., 2010), canine coronavirus (CCoV) (Decaro et al., 2004) and canine rotavirus (CRoV) (Freeman, Kerin, Hull, McCaustland, & Gentsch, 2008).
3. RESULTS AND DISCUSSION
All tissue samples tested positive for CPV by conventional PCR assay and negative for CDV, CAdVs, CaHV‐1, CCoV and CRoV by gel‐based or real‐time (RT) PCR assays. By sequence amplifications, the nearly complete CPV‐genome sequences including both ORFs (4,269 nt) were obtained. Based on the VP2‐426 amino acid residue, all detected strains were characterized as CPV‐2c. NS1 and VP2 sequences of CPV strains analysed in this study showed 100%–99.95% and 100%–99.82% reciprocal nucleotide identities, respectively. The complete genome sequences showed 99.95%–99.93% and 99.95%–99.91% nucleotide identities with CPV strains of Asian origin, such as CPV_IZSSI_2743_17 (Italy, 2017; accession number MF510157), CPV‐SH1516 (China, 2017; acc. no. MG013488) and Canine/China/14/2017 (China, 2017; acc. no. MH476583).
Sequence analysis revealed amino acid changes previously described in Asian CPV‐2c strains (NS1: 60V, 544F, 545F, 630P—NS2: 60V, 151N, 152V—VP2: 5A/G, 267Y, 297A, 324I, 370R) (Table S1). CPV strain IZSSI_PA5632/19 evidenced an additional change at residue 492 of NS1 protein (Table S1). Only one mutation (A/G) was observed among the analysed strains at residue 5 of the VP2 protein, which suggests the circulation of two different but related CPV‐2c strains in southern Italy.
Amino acid change I60V in NS1 also lies at the same residue in the NS2‐encoding sequence, while change at codon 630 of NS1 sequences did not result in any changes in the encoded NS2 protein. Additional two amino acid changes in the NS2‐encoding sequences were observed among the analysed strains: D151N and M152V (Table S1). These changes resulted in silent mutations in the corresponding encoded NS1 protein.
Phylogenetic analysis inferred from VP2 sequences indicated that analysed strains are more related to Asian than to European CPV strains, clustering in a separate clade (Figure 1). Phylogenetic tree inferred from NS1 gene sequences shows that strains clustered within the phylogeny according to the geographical origin and the year of collection rather than to the CPV variant (Figure 2).
Figure 1.
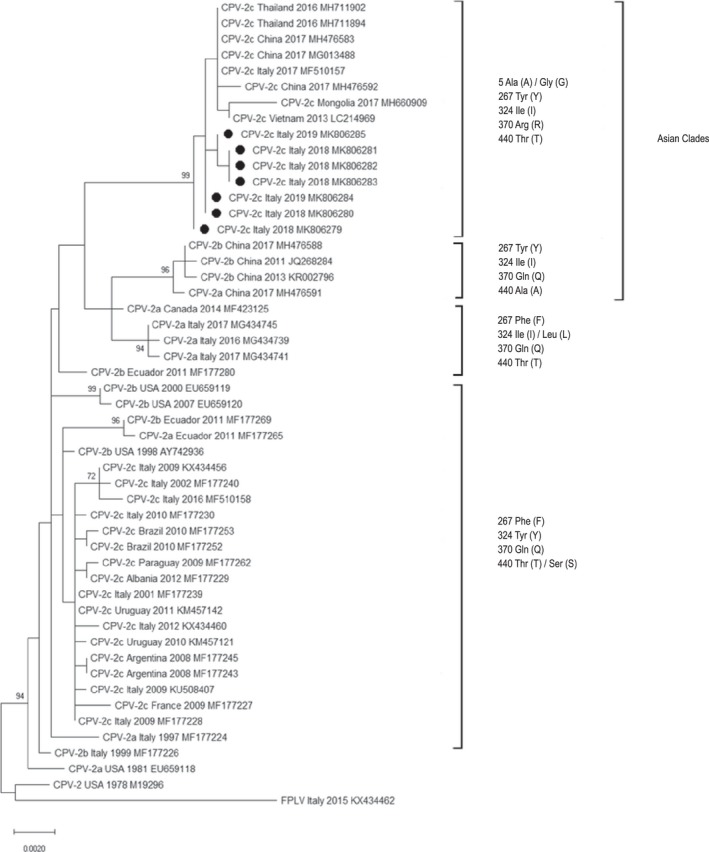
Maximum‐likelihood tree based on 50 full‐length VP2 gene sequences of canine parvovirus type 2 strains (bootstrap 1,000 replicates; bootstrap values greater than 65 are shown). Black dots markings (●) indicate CPV strains analysed in this study. Each sequence is indicated with virus type (FPLV: feline panleukopenia virus—CPV: canine parvovirus) or variant (CPV‐2, CPV‐2a, CPV‐2b, CPV‐2c), country and year of collection, and accession number
Figure 2.

Maximum‐likelihood tree based on 50 full‐length NS1 gene sequences of canine parvovirus type 2 strains (bootstrap 1,000 replicates; bootstrap values greater than 65 are shown). Black dots markings (●) indicate CPV strains analysed in this study. Each sequence is indicated with virus type (FPLV: feline panleukopenia virus—CPV: canine parvovirus) or variant (CPV‐2, CPV‐2a, CPV‐2b, CPV‐2c), country and year of collection, and accession number
The present molecular analysis of CPV strains detected in southern Italy provides new data about the viral spread and dynamics of CPV mutants circulating in Italy. In the last decades, several studies analysed the spread of CPV strains in Italy, and in 2001, the emergence of the CPV‐2c variant was firstly reported (Buonavoglia et al., 2001). In the following years, all three CPV variants were described in Italy, with a slightly higher prevalence of the CPV‐2a and CPV‐2c variants (Decaro, Desario, et al., 2007; Decaro et al., 2013, 2006; Tucciarone et al., 2018). More recently, a CPV‐2c strain displaying genetic signatures typical of Asian viruses was detected in southern Italy (Mira, Purpari, Lorusso, et al., 2018), thus suggesting the introduction of the virus from other countries, as reported for other canine viruses (Decaro, Campolo, et al., 2007; Martella et al., 2006; Mira, Purpari, Bella, et al., 2018). Therefore, a continuous molecular survey was assessed to point out eventual introduction and spread of CPV strains originated from other geographic areas in the Italian canine population.
Since August 2018, CPV‐2c strains with specific molecular signatures were detected from stray and owned dogs. Despite the close genetic relationship with the previous Asian CPV‐2c strain reported in Italy, its absence in the following months suggests a second introduction of Asian CPV‐2c strains. Alternatively, the silent circulation of the original Asian strain in the field with accumulation of few point mutation should be hypothesized. The evidence in rescued stray dogs and in dogs without anamnesis of previous movements, as well as the different dates and places of collection, suggested the active spread of these strains in the field. It remains unclear how these strains have been introduced in Italy, if directly through infected animals or indirectly due to the extreme stability of the CPV in the environment (Hoelzer & Parrish, 2010). According to this study, the spread of Asian CPV strains in a separate geographical area different from Asian countries could be suggested, as previously described in South America (Grecco et al., 2018; Maya et al., 2013).
Whereas CPV‐2a and CPV‐2b are the prevalent variants circulating in Asia (Yi, Tong, Cheng, Song, & Cheng, 2016), and more recently, CPV‐2c has been described in the same continent (Chiang, Wu, Chiou, Chang, & Lin, 2016; Geng et al., 2015; Nakamura et al., 2004; Wang et al., 2016; Zhao et al., 2017; Zhou, Zeng, Zhang, & Li, 2017; Zhuang et al., 2019), showing molecular signatures different from those of other continents. Indeed, the Asian CPV‐2c variant shows specific amino acids in NS1 (60V, 544F, 545V, 630P) and VP2 (5A/G, 267Y, 297A, 324I, 370R) gene sequences. Most of these amino acids have been described in the VP2 of CPV‐2a/2b/2c strains collected in China, Vietnam, India, Taiwan, South Korea, Thailand and Japan (Chiang et al., 2016; Geng et al., 2015; Han et al., 2015; Jeoung et al., 2008; Lin et al., 2014; Mukhopadhyay et al., 2014; Nakamura et al., 2004; Phromnoi, Sirinarumitr, & Sirinarumitr, 2010; Soma, Taharaguchi, Ohinata, Ishii, & Hara, 2013; Xu et al., 2015; Yi et al., 2016; Zhang et al., 2010). In particular, CPV‐2c strains displaying the amino acid glycine (G) instead of the highly conserved alanine (A) at residue 5 of the VP2 have been previously detected in China (Wang et al., 2016) and Italy (Mira, Purpari, Lorusso, et al., 2018). More recently, molecular analyses including the NS1 gene sequence showed the presence of molecular signatures of the Asian CPV strains even in this region (Han et al., 2015; Mira, Purpari, Lorusso, et al., 2018; Zhuang et al., 2019). Indeed, it results critical to extend the analysis to other genomic regions to properly infer the spread of the genetic CPV variants (Grecco et al., 2018).
Moreover, the evidence in the NS1/NS2 gene sequences of amino acid mutations with respect to the other CPV strains spreading worldwide (NS1: 60V, 544F, 545F, 630P ‐ NS2: 60V, 151N, 152V) could contribute to further elucidate its evolution (Mira et al., 2019).
The classification system based on single amino acids (426 and 297) of the VP2 protein does not clearly reflect the phylogenetic relationships of the strains, better supported to proposed “clade” or “lineage/sub‐lineage” new classification criteria (Grecco et al., 2018; Zhuang et al., 2019; Mira et al., 2019). As observed also in this study, phylogeny lacks any clustering based on the single VP2 aa residue 426 (CPV‐2a/2b/2c), as well as on the geographic origin and period of sample collection. Therefore, a wider evolutionary analysis further supports the thesis to consider the CPV antigenic variants as variants of CPV‐2a rather than distinct subtypes (Organtini, Allison, Lukk, Parrish, & Hafenstein, 2015) and could be considered as a more reliable tool in outbreak tracing.
This study reported the early evidence and spread of CPV‐2c strains of Asian origin in the Italian canine population. As observed in South America (Grecco et al., 2018), studies based on the complete coding genome are useful to monitor the spread of CPV strains with Asian origin in a different continent and highlight the need of further studies to evaluate the CPV evolution due to the coexistence of genetically divergent strains in the same geographical environment.
CONFLICT OF INTEREST
The authors of this manuscript declare that there are no conflicts of interest.
Supporting information
ACKNOWLEDGEMENTS
The Authors would like to thank the Centro Veterinario Darwin (Castelvetrano, Italy), Clinica Veterinaria Animal Care (Marsala, Italy), Dr. Alessandra Statelli (Ragusa, Italy) and the veterinary public service of Alcamo‐Castelvetrano, Marsala and Mazara del Vallo (Italy) for samples collection. The Authors also grateful to Rita Profeta, Maria Teresa Todaro and Maria Laura Rizzuto of the Istituto Zooprofilattico Sperimentale della Sicilia ‘A.Mirri’ for their skilful technical assistance. This work was funded by the Ministero della Salute (Italy), Ricerca Corrente IZS SI 03/18 RC ‘Studio del potenziale zoonosico e caratterizzazione genomica dei virus enterici del cane'.
Mira F, Purpari G, Di Bella S, et al. Spreading of canine parvovirus type 2c mutants of Asian origin in southern Italy. Transbound Emerg Dis. 2019;66:2297–2304. 10.1111/tbed.13283
REFERENCES
- Battilani, M. , Scagliarini, A. , Tisato, E. , Turilli, C. , Jacoboni, I. , Casadio, R. , & Prosperi, S. (2001). Analysis of canine parvovirus sequences from wolves and dogs isolated in Italy. Journal of General Virology, 82, 1555–1560. 10.1099/0022-1317-82-7-1555 [DOI] [PubMed] [Google Scholar]
- Buonavoglia, C. , Martella, V. , Pratelli, A. , Tempesta, M. , Cavalli, A. , Buonavoglia, D. , … Carmichael, L. (2001). Evidence for evolution of canine parvovirus type 2 in Italy. Journal of General Virology, 82, 3021–3025. 10.1099/0022-1317-82-12-3021 [DOI] [PubMed] [Google Scholar]
- Canuti, M. , Rodrigues, B. , Whitney, H. G. , & Lang, A. S. (2017). Introduction of canine parvovirus 2 into wildlife on the Island of Newfoundland, Canada. Infection, Genetics and Evolution, 55, 205–208. 10.1016/j.meegid.2017.09.018 [DOI] [PubMed] [Google Scholar]
- Chiang, S. Y. , Wu, H. Y. , Chiou, M. T. , Chang, M. C. , & Lin, C. N. (2016). Identification of a novel canine parvovirus type 2c in Taiwan. Virology Journal, 13, 160 10.1186/s12985-016-0620-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotmore, S. F. , Agbandje‐McKenna, M. , Canuti, M. , Chiorini, J. A. , Eis‐Hubinger, A.‐M. , Hughes, J. , … Ictv Report Consortium . (2019). ICTV virus taxonomy profile: parvoviridae. Journal of General Virology, 100, 367–368. 10.1099/jgv.0.001212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro, N. , Amorisco, F. , Desario, C. , Lorusso, E. , Camero, M. , Bellacicco, A. L. , … Buonavoglia, C. (2010). Development and validation of a real‐time PCR assay for specific and sensitive detection of canid herpesvirus 1. Journal of Virological Methods, 169, 176–180. 10.1016/j.jviromet.2010.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro, N. , & Buonavoglia, C. (2012). Canine parvovirus—A review of epidemiological and diagnostic aspects, with emphasis on type 2c. Veterinary Microbiology, 155, 1–12. 10.1016/j.vetmic.2011.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro, N. , Campolo, M. , Elia, G. , Buonavoglia, D. , Colaianni, M. L. , Lorusso, A. , … Buonavoglia, C. (2007). Infectious canine hepatitis: An “old” disease reemerging in Italy. Research in Veterinary Science, 83, 269–273. 10.1016/j.rvsc.2006.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro, N. , Desario, C. , Addie, D. D. , Martella, V. , Vieira, M. J. , Elia, G. , … Buonavoglia, C. (2007). The study molecular epidemiology of canine parvovirus, Europe. Emerging Infectious Diseases, 13, 1222–1224. 10.3201/eid1308.070505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro, N. , Desario, C. , Amorisco, F. , Losurdo, M. , Elia, G. , Parisi, A. , … Buonavoglia, C. (2013). Detection of a canine parvovirus type 2c with a non‐coding mutation and its implications for molecular characterisation. The Veterinary Journal, 196, 555–557. 10.1016/j.tvjl.2012.12.017 [DOI] [PubMed] [Google Scholar]
- Decaro, N. , Desario, C. , Parisi, A. , Martella, V. , Lorusso, A. , Miccolupo, A. , … Buonavoglia, C. (2009). Genetic analysis of canine parvovirus type 2c. Virology, 385, 5–10. 10.1016/j.virol.2008.12.016 [DOI] [PubMed] [Google Scholar]
- Decaro, N. , Elia, G. , Martella, V. , Campolo, M. , Desario, C. , Camero, M. , … Buonavoglia, C. (2006). Characterisation of the canine parvovirus type 2 variants using minor groove binder probe technology. Journal of Virological Methods, 133, 92–99. 10.1016/j.jviromet.2005.10.026 [DOI] [PubMed] [Google Scholar]
- Decaro, N. , Pratelli, A. , Campolo, M. , Elia, G. , Martella, V. , Tempesta, M. , & Buonavoglia, C. (2004). Quantitation of canine coronavirus RNA in the faeces of dogs by TaqMan RT‐PCR. Journal of Virological Methods, 119, 145–150. 10.1016/j.jviromet.2004.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dei Giudici, S. , Cubeddu, T. , Giagu, A. , Sanna, G. , Rocca, S. , & Oggiano, A. (2017). First molecular characterization of canine parvovirus strains in Sardinia, Italy. Archives of Virology, 162, 3481–3486. 10.1007/s00705-017-3457-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowgier, G. , Mari, V. , Losurdo, M. , Larocca, V. , Colaianni, M. L. , Cirone, F. , … Decaro, N. (2016). A duplex real‐time PCR assay based on TaqMan technology for simultaneous detection and differentiation of canine adenovirus types 1 and 2. Journal of Virological Methods, 234, 1–6. 10.1016/j.jviromet.2016.03.011 [DOI] [PubMed] [Google Scholar]
- Elia, G. , Decaro, N. , Martella, V. , Cirone, F. , Lucente, M. S. , Lorusso, E. , … Buonavoglia, C. (2006). Detection of canine distemper virus in dogs by real‐time RT‐PCR. Journal of Virological Methods, 136, 171–176. 10.1016/j.jviromet.2006.05.004 [DOI] [PubMed] [Google Scholar]
- Freeman, M. M. , Kerin, T. , Hull, J. , McCaustland, K. , & Gentsch, J. (2008). Enhancement of detection and quantification of rotavirus in stool using a modified real‐time RT‐PCR assay. Journal of Virological Methods, 80, 1489–1496. 10.1002/jmv.21228 [DOI] [PubMed] [Google Scholar]
- Geng, Y. , Guo, D. , Li, C. , Wang, E. , Wei, S. , Wang, Z. , … Sun, D. (2015). Co‐Circulation of the Rare CPV‐2c with Unique Gln370Arg Substitution, New CPV‐2b with Unique Thr440Ala Substitution, and New CPV‐2a with High Prevalence and Variation in Heilongjiang Province, Northeast China. PLoS ONE, 10(9), e0137288 10.1371/journal.pone.0137288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grecco, S. , Iraola, G. , Decaro, N. , Alfieri, A. , Alfieri, A. , Gallo Calderón, M. , … Pérez, R. (2018). Inter‐ and intracontinental migrations and local differentiation have shaped the contemporary epidemiological landscape of canine parvovirus in South America. Virus Evolution, 4, vey011. 10.1093/ve/vey011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, T. A. (1999). BioEdit: A user‐friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95–98. [Google Scholar]
- Han, S. , Guo, H. , Sun, S. , Shu, L. , Wei, Y. , Sun, D. , … Liu, X. (2015). Full‐length genomic characterizations of two canine parvoviruses prevalent in Northwest China. Archives of Microbiology, 197, 621–626. 10.1007/s00203-015-1093-4 [DOI] [PubMed] [Google Scholar]
- Hoelzer, K. , & Parrish, C. R. (2010). The emergence of parvoviruses of carnivores. Veterinary Research, 41(6), 39 10.1051/vetres/2010011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeoung, S.‐Y. , Ahn, S.‐J. , & Kim, D. (2008). Genetic analysis of VP2 gene of canine parvovirus isolates in Korea. Journal of Veterinary Medical Science, 70, 719–722. 10.1292/jvms.70.719 [DOI] [PubMed] [Google Scholar]
- Kumar, S. , Stecher, G. , Li, M. , Knyaz, C. , & Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35, 1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. , Tang, J. , Chen, Z. , Li, Q. , Huang, Z. , Wang, Q. , … Liu, G. (2018). Genetic characterization of the complete genome of a mutant canine parvovirus isolated in China. Archives of Virology, 163, 521–525. 10.1007/s00705-017-3586-8 [DOI] [PubMed] [Google Scholar]
- Lin, C. N. , Chien, C.‐H. , Chiou, M. T. , Chueh, L. L. , Hung, M. Y. , & Hsu, H. S. (2014). Genetic characterization of type 2a canine parvoviruses from Taiwan reveals the emergence of an Ile324 mutation in VP2. Virology Journal, 11, 10.1186/1743-422X-11-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martella, V. , Cirone, F. , Elia, G. , Lorusso, E. , Decaro, N. , Campolo, M. , … Buonavoglia, C. (2006). Heterogeneity within the hemagglutinin genes of canine distemper virus (CDV) strains detected in Italy. Veterinary Microbiology, 116, 301–309. 10.1016/j.vetmic.2006.04.019 [DOI] [PubMed] [Google Scholar]
- Maya, L. , Calleros, L. , Francia, L. , Hernández, M. , Iraola, G. , Panzera, Y. , … Pérez, R. (2013). Phylodynamics analysis of canine parvovirus in Uruguay: Evidence of two successive invasions by different variants. Archives of Virology, 158, 1133–1141. 10.1007/s00705-012-1591-5 [DOI] [PubMed] [Google Scholar]
- Mira, F. , Canuti, M. , Purpari, G. , Cannella, V. , Di Bella, S. , Occhiogrosso, L. , … Guercio, A. (2019). Molecular characterization and evolutionary analyses of carnivore protoparvovirus 1 NS1 gene. Viruses, 11, 308 10.3390/v11040308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira, F. , Dowgier, G. , Purpari, G. , Vicari, D. , Di Bella, S. , Macaluso, G. , … Guercio, A. (2018). Molecular typing of a novel canine parvovirus type 2a mutant circulating in Italy. Infection, Genetics and Evolution, 61, 67–73. 10.1016/j.meegid.2018.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira, F. , Purpari, G. , Di Bella, S. , Vicari, D. , Schirò, G. , Di Marco, P. , … Guercio, A. (2018). Update on canine distemper virus (CDV) strains of Arctic‐like lineage detected in dogs in Italy. Veterinaria Italiana, 54, 225–236. 10.12834/VetIt.1455.7862.2 [DOI] [PubMed] [Google Scholar]
- Mira, F. , Purpari, G. , Lorusso, E. , Di Bella, S. , Gucciardi, F. , Desario, C. , … Guercio, A. (2018). Introduction of Asian canine parvovirus in Europe through dog importation. Transboundary and Emerging Diseases, 65, 16–21. 10.1111/tbed.12747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay, H. K. , Matta, S. L. , Amsaveni, S. , Antony, P. X. , Thanislass, J. , & Pillai, R. M. (2014). Phylogenetic analysis of canine parvovirus partial VP2 gene in India. Virus Genes, 48, 89–95. 10.1007/s11262-013-1000-5 [DOI] [PubMed] [Google Scholar]
- Nakamura, M. , Tohya, Y. , Miyazawa, T. , Mochizuki, M. , Phung, H. T. T. , Nguyen, N. H. , … Akashi, H. (2004). A novel antigenic variant of Canine parvovirus from a Vietnamese dog. Archives of Virology, 149, 2261–2269. 10.1007/s00705-004-0367-y [DOI] [PubMed] [Google Scholar]
- Organtini, L. J. , Allison, A. B. , Lukk, T. , Parrish, C. R. , & Hafenstein, S. (2015). Global displacement of canine parvovirus by a host‐adapted variant: Structural comparison between pandemic viruses with distinct host ranges. Journal of Virology, 89, 1909–1912. 10.1128/JVI.02611-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish, C. R. , Aquadro, C. F. , Strassheim, M. L. , Evermann, J. F. , Sgro, J. Y. , & Mohammed, H. O. (1991). Rapid antigenic‐type replacement and DNA sequence evolution of canine parvovirus. Journal of Virology, 65, 6544–6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish, C. R. , O’Connell, P. H. , Evermann, J. F. , & Carmichael, L. E. (1985). Natural variation of canine parvovirus. Science, 230, 1046–1048. 10.1126/science.4059921 [DOI] [PubMed] [Google Scholar]
- Pereira, C. A. D. , Leal, E. S. , & Durigon, E. L. (2007). Selective regimen shift and demographic growth increase associated with the emergence of high‐fitness variants of canine parvovirus. Infection, Genetics and Evolution, 7, 399–409. 10.1016/j.meegid.2006.03.007 [DOI] [PubMed] [Google Scholar]
- Pérez, R. , Calleros, L. , Marandino, A. , Sarute, N. , Iraola, G. , Grecco, S. , … Panzera, Y. (2014). Phylogenetic and genome‐wide deep‐sequencing analyses of canine parvovirus reveal co‐infection with field variants and emergence of a recent recombinant strain. PLoS ONE, 9, e111779 10.1371/journal.pone.0111779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phromnoi, S. , Sirinarumitr, K. , & Sirinarumitr, T. (2010). Sequence analysis of VP2 gene of canine parvovirus isolates in Thailand. Virus Genes, 41, 23–29. 10.1007/s11262-010-0475-6 [DOI] [PubMed] [Google Scholar]
- Purpari, G. , Mira, F. , Di Bella, S. , Di Pietro, S. , Giudice, E. , & Guercio, A. (2018). Investigation on canine parvovirus circulation in dogs from Sicily (Italy) by biomolecular assay. Acta Veterinaria (Beograd), 68(1), 80–94. [Google Scholar]
- Reed, A. P. , Jones, E. V. , & Miller, T. J. (1988). Nucleotide sequence and genome organization of canine parvovirus. Journal of Virology, 62, 266–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelton, L. A. , Parrish, C. R. , Truyen, U. , & Holmes, E. C. (2005). High rate of viral evolution associated with the emergence of carnivore parvovirus. Proceedings of the National Academy of Sciences of the United States of America, 102, 379–384. 10.1073/pnas.0406765102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soma, T. , Taharaguchi, S. , Ohinata, T. , Ishii, H. , & Hara, M. (2013). Analysis of the VP2 protein gene of canine parvovirus strains from affected dogs in Japan. Research in Veterinary Science, 94, 368–371. 10.1016/j.rvsc.2012.09.013 [DOI] [PubMed] [Google Scholar]
- Touihri, L. , Bouzid, I. , Daoud, R. , Desario, C. , El Goulli, A. F. , Decaro, N. , … Bahloul, C. (2009). Molecular characterization of canine parvovirus‐2 variants circulating in Tunisia. Virus Genes, 38, 249–258. 10.1007/s11262-008-0314-1 [DOI] [PubMed] [Google Scholar]
- Truyen, U. (1999). Emergence and recent evolution of canine parvovirus. Veterinary Microbiology, 69, 47–50. 10.1016/S0378-1135(99)00086-3 [DOI] [PubMed] [Google Scholar]
- Truyen, U. (2006). Evolution of canine parvovirus–a need for new vaccines? Veterinary Microbiology, 117, 9–13. 10.1016/j.vetmic.2006.04.003 [DOI] [PubMed] [Google Scholar]
- Tucciarone, C. M. , Franzo, G. , Mazzetto, E. , Legnardi, M. , Caldin, M. , Furlanello, T. , … Drigo, M. (2018). Molecular insight into Italian canine parvovirus heterogeneity and comparison with the worldwide scenario. Infection, Genetics and Evolution, 66, 171–179. 10.1016/j.meegid.2018.09.021 [DOI] [PubMed] [Google Scholar]
- Wang, J. , Lin, P. , Zhao, H. , Cheng, Y. , Jiang, Z. , Zhu, H. , … Cheng, S. (2016). Continuing evolution of canine parvovirus in China: Isolation of novel variants with an Ala5Gly mutation in the VP2 protein. Infection, Genetics and Evolution, 38, 73–78. 10.1016/j.meegid.2015.12.009 [DOI] [PubMed] [Google Scholar]
- Xu, J. , Guo, H. C. , Wei, Y. Q. , Shu, L. , Wang, J. , Li, J. S. , … Sun, S. Q. (2015). Phylogenetic analysis of canine parvovirus isolates from Sichuan and Gansu Provinces of China in 2011. Transboundary and Emerging Diseases, 62, 91–95. 10.1111/tbed.12078 [DOI] [PubMed] [Google Scholar]
- Yi, L. , Tong, M. , Cheng, Y. , Song, W. , & Cheng, S. (2016). Phylogenetic analysis of canine parvovirus VP2 gene in China. Transboundary and Emerging Diseases, 63, e262–e269. 10.1111/tbed.12268 [DOI] [PubMed] [Google Scholar]
- Zhang, R. , Yang, S. , Zhang, W. , Zhang, T. , Xie, Z. , Feng, H. , … Xia, X. (2010). Phylogenetic analysis of the VP2 gene of canine parvoviruses circulating in China. Virus Genes, 40, 397–402. 10.1007/s11262-010-0466-7 [DOI] [PubMed] [Google Scholar]
- Zhang, Z. , Schwartz, S. , Wagner, L. , & Miller, W. (2000). A greedy algorithm for aligning DNA sequences. Journal of Computational Biology, 7, 203–214. 10.1089/10665270050081478 [DOI] [PubMed] [Google Scholar]
- Zhao, H. , Wang, J. , Jiang, Y. , Cheng, Y. , Lin, P. , Zhu, H. , … Cheng, S. (2017). Typing of canine parvovirus strains circulating in North‐East China. Transboundary and Emerging Diseases, 64, 495–503. 10.1111/tbed.12390 [DOI] [PubMed] [Google Scholar]
- Zhou, P. , Zeng, W. , Zhang, X. , & Li, S. (2017). The genetic evolution of canine parvovirus – A new perspective. PLoS ONE, 12, e0175035 10.1371/journal.pone.0175035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang, Q. , Qiu, Y. , Pan, Z. , Wang, S. , Wang, B. , Wu, W. , … Wang, K.‐C. (2019). Genome sequence characterization of canine parvoviruses prevalent in the Sichuan province of China. Transboundary and Emerging Diseases, 66, 897–907. 10.1111/tbed.13100 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


