Abstract
As an emerging disease, the porcine epidemic diarrhoea virus has caused substantial economic losses to the pork industry in Mexico, leading to piglet mortality rates of up to 100%. For detection, sequencing and genetic characterization of the virus, 68 samples of one‐week‐old piglets from pork farms in 17 states of Mexico were analysed. In total, 53 samples were positive by real‐time RT‐PCR, confirming the presence of the virus in 15 states. Twenty‐eight samples from 10 states were amplified by endpoint RT‐PCR, and 20 sequences of the spike gene were obtained. A phylogenetic analysis based on the spike gene demonstrated that all Mexican strains are in Group II and are classified as non‐Indel‐S emerging variants. Three strains showed amino acid insertions: PEDv/MEX/GTO/LI‐DMZC15/2015 and PEDv/MEX/QRO/LI‐DMZC45/2016 showed one amino acid insertion (424Y425 and 447D448, respectively), and PEDv/MEX/QRO/LI‐DMZC49/2019 showed one and two amino acid insertions (422C423 and 537SQ538), with the second insertion in the COE region. These results provide evidence of the prevalence of emerging, non‐Indel‐S strains of the virus are currently circulating in Mexico during 2016–2018, when three of which have amino acid insertions: PEDv/MEX/GTO/IN‐DMZC15/2015 and PEDv/MEX/QRO/IN‐DMZC45/2016 have one amino acid insertion each (424Y425 and 447D448, respectively), and PEDv/MEX/QRO/IN‐DMZC49/2019 has one (422C423) and two amino acid insertions (537SQ538), the latter being in the COE region, which could generate new antigenic variants.
Keywords: emerging strains, Mexico, porcine epidemic diarrhoea virus, spike gene
1. INTRODUCTION
The porcine epidemic diarrhoea virus (PEDv) was first identified in 1971. This virus remained in Europe and Asia until 2013 and was first reported in the United States in April 2016 (Jarvis et al., 2016). Its presence was officially recognized in Mexico before the World Organization for Animal Health (OIE) in May 2014 (OIE, 2014). PEDv belongs to the Alphacoronavirus genus of the Coronaviridae family. It is a single‐stranded RNA virus, and the spike protein is a type I membrane glycoprotein with signal peptides; it plays an important role in the induction of neutralizing antibodies, receptor binding and viral entry, and it is the protein with the most variable sequences in the genome of coronaviruses (Weiss & Leibowitz, 2011). The strains isolated during the initial outbreaks in the United States were classified as highly pathogenic. In 2014, a strain with low pathogenicity was found in Ohio, USA; it was known as Indel‐S because it has three nucleotide deletions and one insertion in the S gene, similar to the classic CV777 strain (Wang, Byrum, & Zhang, 2014). Thus, the circulation of two great strain groups was established: the Indel‐S type and the non‐Indel‐S type (Lee, 2015). Indel‐S‐type strains have been found in several countries where the disease is endemic; however, they have been described as highly pathogenic strains (Stadler et al., 2015). The circulation of Indel‐S and non‐Indel‐S strains was recently described in Mexico, reporting PEDv prevalence levels of 30% (OIE, 2014) and 88.8% (Lara‐Romero et al., 2017). Previous reports on porcine epidemic diarrhoea in other countries suggest that the high genetic diversity of the circulating strains could produce emerging strains not yet identified in Mexico. Therefore, the aim of this study was to report the genetic variability of PEDv in samples from farms that showed high‐mortality diarrhoeic outbreaks in piglets.
2. MATERIALS AND METHODS
2.1. Samples
In total, 68 (47 intestine and 21 faeces) samples were obtained from one‐week‐old piglets from pig farms in 17 states in Mexico from 2016 to 2018. In these farms, disease was observed to affect 80% of the farm population, causing severe watery diarrhoea, vomiting and dehydration, which led to 80%–95% mortality in piglets that were a few days old.
2.2. RT‐PCR
RNA was purified from the supernatant with the QIAmp Viral RNA Mini Extraction Kit (Qiagen). Nucleic acids were analysed for the viral antigen using a commercial Real‐Time RT‐PCR Kit (Real‐Time RT‐PCR PEDV/SDCV; Pockit, GeneReach) following the manufacturer's directions. An endpoint RT‐PCR targeting the S gene was performed on positive samples using the Superscript II RT Commercial Kit (Invitrogen, Thermo Fisher Scientific); the primer used was SRF/PEDv/All‐R3 primer (5′–CCTTCTTGCACTGACATTACCAC–3′). For the PCR, the SRF/PEDv/S‐F1 (5′–GTGTCATCACCGAAAAGTTGGC–3′) and SRF/PEDv/S‐R2 (5′‐GTGGGCAATAAAGAACAATGACAGC–3′) primers were used. The amplification was performed with a LongAmp™ Hot Start Taq 2X Master Mix Kit (New England BioLabs).
2.3. Sequencing
cDNA of the genes of interest obtained in the amplification step was sequenced using the Ion Personal Genome Machine (PGM) platform, following the supplier's directions for DNA sequencing. Briefly, 100 ng of cDNA was used to build a library with the Ion Xpress™ Plus Fragment Library Kit (Life Technologies). The sequencing reaction was carried out on an Ion 314 Chip v2 and with the Ion PGM™ Hi‐Q™ Sequencing Kit (Life Technologies). cDNA was physically fragmented with a Bioruptor® Sonication System (Diagenode) until fragments of 200 base pairs (bp) were obtained. Adapters were ligated to these fragments, the ends were repaired, and DNA was purified. DNA quantity, quality and integrity were evaluated using the High Sensitivity DNA Kit (Agilent). A library template was obtained using the Ion PGM™ Hi‐Q™ OT2 Kit (Life Technologies) by emulsion PCR in the Ion OneTouch™ 2 System. Readings were filtered with the FastQC plug‐in v3.4.1.1, selecting only those with a Q score ≥ 20 (6,074,916 reads); the depth was higher than 300×.
2.4. Analysis of sequences
The sequences obtained were edited with BioEdit Sequence Alignment Editor v.7.2.6, keeping only the ORF coding for each protein. The sequences were aligned by the Clustal W method with the spike protein from Indel‐S‐ and non‐Indel‐S‐type emerging and classical strains using the USA/Colorado/2013 (KF272920) strain as a reference. A phylogenetic tree was then built by the neighbour‐joining method with 1,000 bootstrap replications using the MEGA 7 software.
3. RESULTS AND DISCUSSION
In this study, of the 68 processed samples from 17 states, 53 (77.94%) were positive by real‐time RT‐PCR; these positive samples were collected in 15 states in Mexico: Mexico City, Estado de México, Guanajuato, Hidalgo, Jalisco, Michoacán, Morelos, Nuevo León, Oaxaca, Puebla, Querétaro, San Luis Potosí, Sonora, Tlaxcala and Veracruz (Figure 1). The PEDv S gene was amplified by endpoint RT‐PCR, yielding 28 positive samples (52.83%) from 10 of the 17 states sampled. This study reports the presence of the PEDv S gene in the states of Hidalgo, Oaxaca and San Luis Potosí for the first time (Lara‐Romero et al., 2017; OIE, 2014). The coronavirus S protein is a type I membrane glycoprotein that interacts with cell receptors during virus entry (Weiss & Leibowitz, 2011). Various genotypes of the S protein gene have been described based on their genetic traits and the period when the strains emerged: GIa (classical), GIb (emergent, Indel‐S), GIIa (emergent, non‐Indel‐S from the Americas) and GIIb (emergent, non‐Indel‐S from Asia) (Jarvis et al., 2016; Lee, 2015). Previous studies in Mexico have identified circulating strains with a GII genotype (from the Americas) as well as strains with a GI genotype (Indel‐S) (Lara‐Romero et al., 2017). The 20 strains isolated in this study (Table 1) are in the GIIa genotype branch and were identified as emerging, non‐Indel‐S variants (Figure 2); the strains PEDv/MEX/JAL/LI‐DMZC11/2015 and PEDv/MEX/MICH/LI‐DMZC19/2015 are located in a cluster related to emerging strains from Asia, and all other strains are in clusters along with strains from the Americas (Jarvis et al., 2016; Lee, 2015), indicating that a wide range of strains are circulating in Mexico. The similarity rates for the S gene among classical and emerging virus strains ranged from 96% to 98%. Within the cluster of US strains, the similarity rate is even higher (99.3%–99.6%) than among emerging strains from China (98.1%–99.5%) (Lin, Saif, Marthaler, & Wang, 2016). Our results indicate a similarity rate of 99.3%–99.9% among Mexican strains with respect to emerging, non‐Indel‐S strains. The strains reported in this study showed a similarity rate of 95%–95.9% with emerging Indel‐S strains and of 92.7%–93.2% with classical reference strains.
Figure 1.
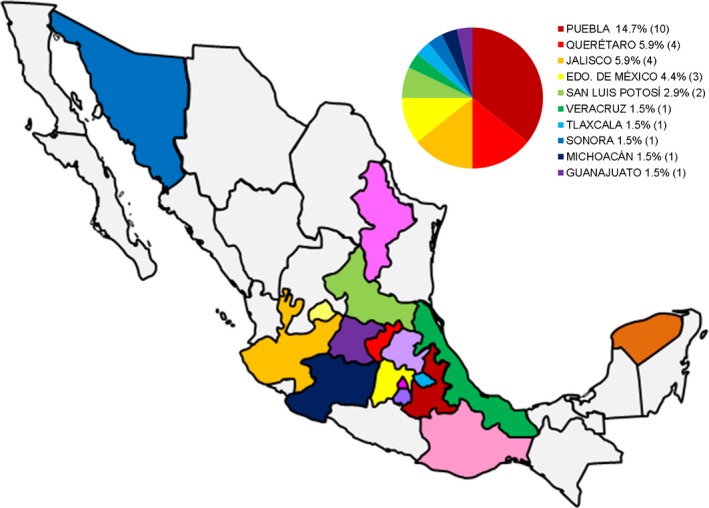
Sampled states in Mexico. All sampled states are shown on the map in different colours. The chart shows those states where positive samples were collected, as revealed by endpoint RT‐PCR targeting the PEDv S gene [Colour figure can be viewed at http://wileyonlinelibrary.com]
Table 1.
Information regarding strains detected in this study
| Strain name | Specimen | Collection date | Location | GenBank accession number |
|---|---|---|---|---|
| PEDv/MEX/PUE/LI‐DMZC0/2015 | Intestine | 19 March 2015 | Puebla | MN091345 |
| PEDv/MEX/TLAX/LI‐DMZC3/2014 | Intestine | 6 February 2014 | Tlaxcala | MN091346 |
| PEDv/MEX/PUE/LI‐DMZC5/2014 | Intestine | 15 March 2014 | Puebla | MN091347 |
| PEDv/MEX/EDOMEX/LI‐DMZC6/2014 | Intestine | 15 March 2014 | Edo. de México | MN091348 |
| PEDv/MEX/SLP/LI‐DMZC9/2015 | Intestine | 26 March 2015 | San Luis Potosí | MN091349 |
| PEDv/MEX/JAL/LI‐DMZC10/2015 | Intestine | 26 March 2015 | Jalisco | MN091350 |
| PEDv/MEX/JAL/LI‐DMZC11/2015 | Intestine | 26 March 2015 | Jalisco | MN091351 |
| PEDv/MEX/PUE/LI‐DMZC12/2015 | Intestine | 15 April 2015 | Puebla | MN091352 |
| PEDv/MEX/PUE/LI‐DMZC14−2/2015 | Intestine | 6 March 2015 | Puebla | MN091353 |
| PEDv/MEX/GTO/LI‐DMZC15/2015 | Intestine | 7 May 2015 | Guanajuato | MN091354 |
| PEDv/MEX/PUE/LI‐DMZC16/2015 | Intestine | 29 May 2015 | Puebla | MN091355 |
| PEDv/MEX/MICH/LI‐DMZC19/2015 | Intestine | 10 August 2015 | Michoacán | MN091356 |
| PEDv/MEX/PUE/LI‐DMZC20/2015 | Faeces | 7 August 2015 | Puebla | MN091357 |
| PEDv/MEX/VER/LI‐DMZC22/2015 | Intestine | 3 November 2015 | Veracruz | MN091358 |
| PEDv/MEX/SON/LI‐DMZC23/2015 | Faeces | 14 September 2015 | Sonora | MN091359 |
| PEDv/MEX/JAL/LI‐DMZC25/2015 | Intestine | 8 October 2015 | Jalisco | MN091360 |
| PEDv/MEX/PUE/LI‐DMZC44/2016 | Intestine | 27 January 2016 | Puebla | MN091361 |
| PEDv/MEX/PQRO/LI‐DMZC45/2016 | Intestine | 21 January 2016 | Querétaro | MN091362 |
| PEDv/MEX/PUE/LI‐DMZC48/2016 | Intestine | 3 March 2016 | Puebla | MN091363 |
| PEDv/MEX/QRO/LI‐DMZC49/2016 | Intestine | 5 April 2016 | Querétaro | MN091364 |
Figure 2.
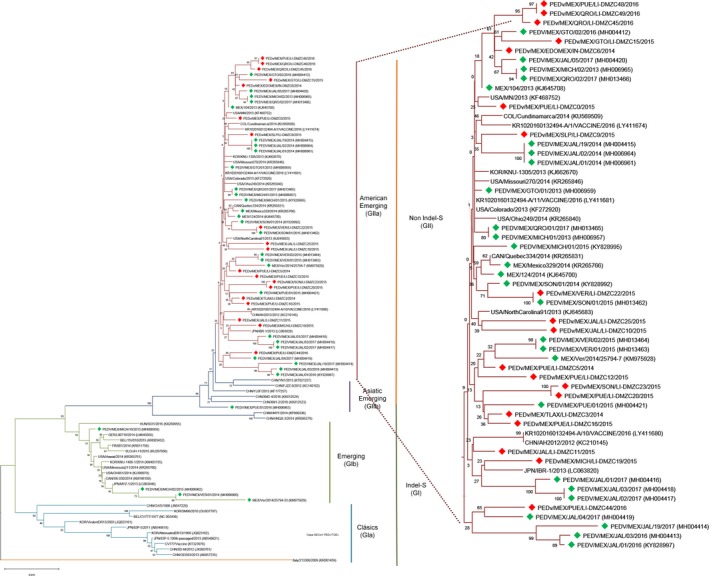
Phylogenetic analysis of the complete sequence of the S gene, along with reference strains. The strains herein reported are marked in red, while previously described Mexican strains are marked in green. The tree was built with the neighbor‐joining method, using the maximum likelihood for nucleotide distance and 1,000 bootstrap replications, with MEGA7 [Colour figure can be viewed at http://wileyonlinelibrary.com]
The neutralizing epitopes on the S protein are SS2 (amino acid position 748–755), SS6 (amino acid position 764–771) (Sun et al., 2008) and 2C10 (amino acid position 1368–1374) (Cruz, Kim, & Shin, 2006). In this study, the reference strain USA/Colorado/2013 was chosen for comparison because it showed a higher similarity rate with the GIIa genotype (Figure 3). Sequencing analysis revealed the positions of the epitopes 751YSNIGVCK758, 767SQSGQVKI774 and 1371GPRLQPY1377 (SS2, SS6 and 2C10, respectively). No changes were observed in the epitopes SS2 and 2C10 in any of the 20 strains analysed, while a strain from Puebla isolated in 2014 (PEDv/MEX/PUE/LI‐DMZC5/2014) showed an L/S767F substitution on the SS6 epitope, a position previously described with different mutations in other strains (Hao, Xue, He, Wang, & Cao, 2014).
Figure 3.

Identification of amino acid substitution sites in the S protein from the PEDv/LIDMZC strain with respect to the reference sequence USA/Colorado/2013 (KF272920). Substitutions in the regions of the COE and SS6 epitopes are highlighted in a red square [Colour figure can be viewed at http://wileyonlinelibrary.com]
The 2‐amino acid insertion 537SQ538 was identified herein in the COE region in the strain PEDv/MEX/QRO/LI‐DMZC49/2016; this insertion could give the S protein a different biological activity. Recently, 11 substitutions were found in 7 different Mexican strains (Lara‐Romero et al., 2017); one of these substitutions (H524R) was found in a strain from Sonora. In this work, amino acid substitutions were found in strains from Puebla, Veracruz and Querétaro. These data suggest that antigenic variations among strain groups are given by changes in 8–11 amino acids on this neutralizing epitope (Lin et al., 2016). The substitution of three serine residues (A522S, A554S and G599S) on the COE epitope and two serine residues on the SS6 epitope (L767S and D769S) was found in all the strains studied herein.
4. CONCLUSION
The presence of PEDv in 15 states in Mexico was demonstrated in this study, and a frequency of 77.9% was observed. Three strains were found with amino acid insertions: PEDv/MEX/GTO/IN‐DMZC15/2015 and PEDv/MEX/QRO/IN‐DMZC45/2016 with one amino acid insertion each (424Y425 and 447D448, respectively) and PEDv/MEX/QRO/IN‐DMZC49/2019 with one (422C423) and two amino acid insertions (537SQ538), the latter being in the COE region. It was demonstrated that the circulating strains are emerging, non‐Indel‐S strains (GIIa genogroup).
CONFLICT OF INTEREST
The authors declare no financial or commercial conflict of interest.
ETHICAL APPROVAL
The manuscript does not contain clinical studies or patient data; the samples used in this study were voluntarily provided by veterinarians and pig farmers.
ACKNOWLEDGEMENTS
Saúl Reveles is a doctoral student of the Posgrado en Ciencias de la Producción y de la Salud Animal, Universidad Nacional Autónoma de México (UNAM). This research was supported by SAGARPA (Register No: 38445‐1635‐30‐VI‐14) and the International Pig Veterinary Society Foundation 2014. We thank the ‘Departamento de Medicina y Zootecnia de Cerdos' and ‘Unidad de Investigación' from the FMVZ‐UNAM.
Reveles‐Félix S, Carreón‐Nápoles R, Mendoza‐Elvira S, et al. Emerging strains of porcine epidemic diarrhoea virus (PEDv) in Mexico. Transbound Emerg Dis. 2020;67:1035–1041. 10.1111/tbed.13426
REFERENCES
- Cruz, D. J. M. , Kim, C. , & Shin, H. (2006). Phage‐displayed peptides having antigenic similarities with porcine epidemic diarrhea virus (PEDV) neutralizing epitopes. Virology, 354(1), 28–34. 10.1016/j.virol.2006.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, J. , Xue, C. , He, L. , Wang, Y. , & Cao, Y. (2014). Bioinformatics insight into the spike glycoprotein gene of field porcine epidemic diarrhea strains during 2011–2013 in Guangdong, China. Virus Genes, 49(1), 58–67. 10.1007/s11262-014-1055-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis, M. C. , Lam, H. C. , Zhang, Y. , Wang, L. , Hesse, R. A. , Hause, B. M. , … Marthaler, D. (2016). Genomic and evolutionary inferences between American and global strains of porcine epidemic diarrhea virus. Preventive Veterinary Medicine, 123, 175–184. 10.1016/j.prevetmed.2015.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara‐Romero, R. , Gómez‐Núñez, L. , Cerriteño‐Sánchez, J. L. , Márquez‐Valdelamar, L. , Mendoza‐Elvira, S. , Ramírez‐Mendoza, H. , & Rivera‐Benítez, J. F. (2017). Molecular characterization of the spike gene of the porcine epidemic diarrhea virus in Mexico, 2013–2016. Virus Genes, 54(2), 215–224. 10.1007/s11262-017-1528-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C. (2015). Porcine epidemic diarrhea virus: An emerging and re‐emerging epizootic swine virus. Virology Journal, 12, 193 10.1186/s12985-015-0421-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C.‐M. , Saif, L. J. , Marthaler, D. , & Wang, Q. (2016). Evolution, antigenicity and pathogenicity of global porcine epidemic diarrhea virus strains. Virus Research, 226, 20–39. 10.1016/j.virusres.2016.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- OIE (2014). Infección por el Virus de la Diarrea Epidémica Porcina, México. Ficha Técnica de la OIE, p. 4 Retrieved from http://www.oie.int/es/nuestra-experiencia-cientifica/informaciones-especificas-y-recomendaciones/porcine-epidemic-diarrhoea/ [Google Scholar]
- Stadler, J. , Zoels, S. , Fux, R. , Hanke, D. , Pohlmann, A. , Blome, S. , … Ladinig, A. (2015). Emergence of porcine epidemic diarrhea virus in southern Germany. BMC Veterinary Research, 11(1), 1–8. 10.1186/s12917-015-0454-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, D. , Feng, L. I. , Shi, H. , Chen, J. , Cui, X. , Chen, H. , … Tong, G. (2008). Identification of two novel B cell epitopes on porcine epidemic diarrhea virus spike protein. Veterinary Microbiology, 131(1‐2), 73–81. 10.1016/j.vetmic.2008.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Byrum, B. , & Zhang, Y. (2014). New variant of porcine epidemic diarrhea virus, United States, 2014. Emerging Infectious Diseases, 20(5), 917–919. 10.3201/eid2005.140195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, S. R. , & Leibowitz, J. L. (2011). Coronavirus pathogenesis. Advances in Virus Research, 81, 85–164. 10.1016/B978-0-12-385885-6.00009-2 [DOI] [PMC free article] [PubMed] [Google Scholar]


