Summary
We investigated bovine coronavirus (BCoV) as an etiological agent in cattle with clinical respiratory and digestive signs using 147 feces and 199 nasal swab samples. A total of 18 test samples (16 feces and 2 nasal swap samples) were detected positive by ELISA and/or RT‐PCR targeting the BCoV N gene. The partial S1 gene regions of BCoVs (An‐4 and An‐11) detected in feces samples from two herd‐mate dairy calves were compared. Virological and serological results indicated that BCoVs are widespread in Turkey and are likely etiological agents in diarrhea cases in calves.
Keywords: bovine, coronavirus, nested RT‐PCR, S1 gene region, sequencing
Bovine coronavirus (BCoV) is a member of the family Coronaviridae and causes severe diarrhea in neonatal calves and winter dysentery in adult cattle (Athanassious et al., 1994; Fukutomi et al., 1999; Cho et al., 2001a; Khalili et al., 2006; Stipp et al., 2009). Bovine coronavirus infection of the upper respiratory tract has also been described in growing calves and adults (Lathrop et al., 2000a, 2000b; Cho et al., 2001b; Hasoksuz et al., 2002b).
Bovine coronavirus contains five major structural proteins: the nucleocapsid (N) protein, the transmembrane (M) protein, the hemagglutinin/esterase (HE) protein, the spike (S) protein, and small membrane E protein (Chouljenko et al., 1998). The BCoV S protein is a very large membrane glycoprotein of 1363 amino acids and is cleaved at amino acid positions 768 and 769 to form two subunits termed S1 and S2 (Yoo and Deregt, 2001). S2 is a transmembrane protein. S1 is responsible for receptor binding, haemagglutination and induction of neutralizing antibodies in the host species (Gallagher and Buchmeier, 2001). The BCoV S1 gene shows a low level of sequence homology while the S2 subunit is highly conserved, as it is in other coronaviruses (Hasoksuz et al., 2002a). Some antigenic and biological variations have been shown in the S1 gene region especially in field strains, although BCoVs belongs to a single serotype.
It has been reported that some BCoVs isolated from respiratory tracts had different antigenic and biological properties from those of BCoV isolated from the enteric tract (Fukutomi et al., 1999; Hasoksuz et al., 1999a, 1999b, 2002a). On the other hand, there are reports indicating that there are no differences between BCoVs that cause enteric and respiratory infections; these have been referred to as enteric coronaviruses (EBCoV) and respiratory coronaviruses (RBCoV), respectively (Reynolds et al., 1985; Tsunemitsu and Saif, 1995). In the most recent studies, questions arose as to whether the same virus strain could be responsible for the simultaneous appearance of enteric and respiratory disease in the same animal and there was a suggestion that the mutation of the virus during infection could occur. This mutation is often in the S1 region of BCoV. Antibodies have an important role in protection against this infection. Thus, the mutations determined in this region might have an influence on the choice of vaccine strain.
In this study, we aimed (i) to report the results of the partial characterization of the gene coding for the hypervariable region of the S1 subunit of the S protein of BCoVs detected in feces samples from two herd‐mates calves with diarrhea, and (ii) to report the epidemiological data of BCoV infection in dairy herds in Turkey.
A total of 147 fecal and 199 nasal swab samples collected from cattle with respiratory and/or enteric infections in 13 herds were used for the virological studies. The feces samples were tested for BCoVs using a commercial ELISA kit (BioX, Jemelle, Belgium) and nested RT‐PCR while the nasal samples were tested only by nested RT‐PCR. Subsequently, samples that were found positive for BCoV were inoculated onto HRT‐18 cells for virus isolation and the S1 subunit of the S gene was sequenced. A total of 569 blood serum samples were also tested for antibodies against BCoVs using standard virus neutralization test (VNT), in a 1/5 serum dilution (Table 1).
Table 1.
The number of test samples and the results of test for virological and serological studies
| Herds | Province | Number of the materials | Results | |||||
|---|---|---|---|---|---|---|---|---|
| Serum | Nasal Swab | Feces | Feces (ELISA) | Feces (Nested RT‐PCR) | Nasal Swab (Nested RT‐PCR) | Serum VNT+ (%) | ||
| I | Kars | 100 | 22 | 22 | 3 | 3 | – | 43 (43.0) |
| II | Eskişehir | 168 | 29 | 12 | 1 | 3 | 2 | 121 (72.0) |
| III | Van | 30 | 15 | 15 | 2 | 2 | – | 21 (70.0) |
| IV | Aksaray | 90 | 49 | 64 | 5 | 5 | – | 61 (67.7) |
| V | Bursa | 124 | 21 | 8 | – | – | – | 59 (47.5) |
| VI | Aydın | 45 | 35 | – | – | – | – | 35 (77.7) |
| VII | Tokat | – | – | 5 | – | – | – | |
| VIII | Mersin | – | 12 | 7 | – | – | – | |
| IX | Samsun | – | – | 1 | – | – | – | |
| X | Ankara‐1 | – | – | 12 | 2 | 2 | – | |
| XI | Ankara‐2 | – | – | 1 | 1 | 1 | – | |
| XII | Kırklareli‐1 | – | 4 | – | – | – | – | |
| XIII | Kırklareli‐2 | 12 | 12 | – | – | – | – | – |
| Total | 569 | 199 | 147 | 14 (9.5%) | 16 (10.8%) | 2 (1.0%) | 340 (59.7) | |
VNT, virus neutralization test.
Bovine coronavirus genomic RNA was extracted from nasal swab and fecal samples using an acid guanidinium thiocyanate‐phenol‐chloroform extraction method as described elsewhere (Chomczynski and Sacchi, 1987). The RNA pellet was dissolved in 20 μl of sterile distilled water and used for cDNA synthesis. Complementary DNA synthesis was performed using the ReverAid cDNA Synthesis kit as described in its manual using random hexamere primers (MBI, Fermentas, Vilnius, Lithuania). The detection of the RNA was performed using the nested RT‐PCR as described elsewhere (Cho et al., 2001a). The list of primers used for nested PCR for the diagnosis of BCoV and for the sequencing of the hypervariable S1 region of the S protein are given in Table 2.
Table 2.
Primers used for diagnostic (N protein) and sequence analysis (hypervariable S1 region of S glycoprotein) purposes
| Procedure | Target gene | Primers (5′→3′)* | Location | Product (bp) |
|---|---|---|---|---|
| RT‐PCR | N | GCAATCCAGTAGTAGAGCGT (F) CTTAGTGGCATCCTTGCCAA (R) | 21–40 731–750 | 730 |
| Nested RT‐PCR | N | GCCGATCAGTCCGACCAATG (F) AGAATGTCAGCCGGGGTAG (R) | 79–98 467–485 | 407 |
| Sequence analysis | S‐1 | GTT TCT GTT AGC AGG TTT AA (F) ATA TTA CAC CTA TCC CCT TG (R) | 1329–13481797–1816 | 488 |
Asterisk indicates direction of the primers as F, forward; R, reverse.
The spike gene encoding the S1 subunit of the S protein of two Turkish BCoV isolates (An‐4 and An‐11) was partially sequenced. Briefly, a DNA fragment (488 bp in length) obtained from RT‐PCR performed with S1 sequence primers (Table 2) was excised from the gel and purified using a commercial gel extraction kit (GeneMark, Tainan, Taiwan) according to the manufacturer’s protocol. The purified PCR products were sequenced using the Quick Start DTSC kit (Beckman Coulter, Brea, Ca, USA) in the CEQ 8000 facility (Beckman Coulter) of the department. The sequences were then submitted to GenBank for assignation of accession numbers. The phylogenetic relationship between the EBCoVs detected in this study and the BCoVs of enteric and/or respiratory origin detected in different countries was examined using neighbor joining analysis with bootstrap analysis (1000 replicates) (Felsenstein, 1985; Saitou and Nei, 1987) and Kimura 2‐parameter correction was conducted using the mega software package v4.0 (Tamura et al., 2007).
Out of 147 fecal samples tested for the presence of BCoV, 14 (9.5%) and 16 (10.8%) were found to be positive by ELISA and nested RT‐PCR, respectively. Fourteen fecal samples were positive by both ELISA and nested PCR, while two samples were positive by nested RT‐PCR only. In addition, the nested RT‐PCR revealed two (1.0%) out of 199 swab samples positive with the expected size (407 bp in length) of amplicon (Table 1). It is noted that both nasal and fecal samples from two calves (Code: An‐8, An‐11) were positive for BCoV RNA. BCoV was isolated from 18 samples including two nasal swab and 16 fecal samples. All samples, which produced cytopathic effect characterized by syncytium formation on HRT‐18 cells after second or third passages were confirmed as positive for BCoV by single round RT‐PCR targeting the N gene (730 bp in size). Virological results and individual data of animals are shown in Table 1. The S1 gene region of BCoV could be amplified from five of the 18 feces samples and from the BCoV Mebus strain using primers (S1NS and S1NA) described by Brandao et al. (2006) (Fig. 2). The sequencing of the S1 coding regions of An‐4 and An‐11 isolates identified 450 and 487 nucleotides respectively (GenBank accession numbers GQ259978 and GQ259979, respectively). The S1 sequences of the isolates showed 88.2% identity. The nucleotide similarity between An‐4 and the Mebus strain was 88.8% and 92.5% between An‐11 and the Mebus strain. Comparison of amino acid sequences, deduced from the S1 nucleotide sequence, showed some amino acid changes between BCoVs from two herd–mate calves and also between the Mebus strain and BCoVs detected in this study (Fig. 1). Phylogenetic analysis using the Neighbor Joining method revealed two local isolates (An‐4 and An‐11) were clustered in different groups (Fig. 2). In addition to the virological data, VNT indicated that BCoV was widespread in Turkey (Table 1), which concurs with previous studies reported in Turkey (Alkan, 1998; Alkan et al., 2003; Hasoksuz et al., 2005).
Figure 2.
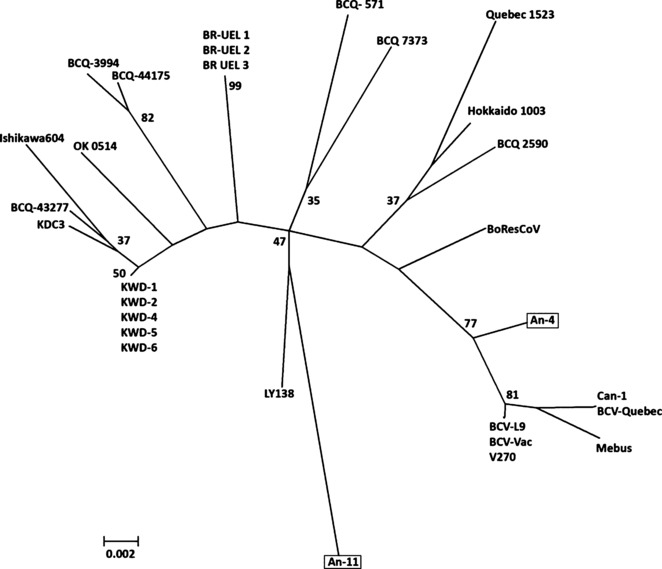
Phylogenetic analysis (Neighbor Joining method) of Turkish bovine coronavirus (BCoV) isolates (An‐4 and An‐11) in a stretch of nucleotides of the gene coding for the hypervariable region of the S1 subunit of S protein of BCoVs of respiratoric (n = 6), BCQ 43277 (AF239308), BCQ 3994 (AF339836), BCQ 44175 (AF239309), OK 0514 (AF058944), BResCoV (M80844), Hokkaido/10/03 (AB277108); enteric (n = 9), Mebus (U00735), LY 138 (AF058942), KDC3 (DQ389634), BCQ 571 (U06093), BCQ 7373 (AF239306), Quebec 1523 (AH010062), V270 (EF193075), BCV‐Quebec (D00662), Ishikawa/6/04 (AB277125); winter dysentery (n = 6) KWD 1 (AY935637), KWD 2 (AY935638), KWD 4 (AY935640), KWD 5 (AY935641), KWD 6 (AY935642), BCQ 2590 (AF239317); and unknown origins (n = 6) Canada1 (AF313395), BR‐UEL1 (DQ479421), BR‐UEL2 (DQ479421), BR‐UEL3 (DQ479421), BCV‐L9 (M64667), BCV‐Vac (M64668). Numbers at the nodes indicate bootstrapping values. Bar represents nucleotide substitutions per position.
Figure 1.

Comparison of the predicted amino acid sequences of hypervariable S1 region of the S glycoprotein of Turkish (An‐4 and An‐11) and other bovine coronavirus.
Data showed that BCoV was often detected in feces samples from calves with diarrhea (Table 1). In contrast to this, however, one of the BCoV isolates (Ka‐55) was recovered from a 3 year‐old cow with diarrhea. It is known that BCoV is associated with acute diarrhea of adult cattle, referred to as winter dysentery (Athanassious et al., 1994), and also the same field strain can cause different clinical infections in cattle in different age groups (calves, heifers and adults) (Park et al., 2006; Decaro et al., 2008). In the present study, BCoV was detected from both nasal swab (n = 2) and feces samples (n = 16), consistent with previous reported by several studies (Fukutomi et al., 1999; Lathrop et al., 2000a, 2000b; Cho et al., 2001b; Hasoksuz et al., 2002b; Park et al., 2006; Kanno et al., 2009).Genetic and antigenic variation related to tissue tropism has been the subject of many studies (Chouljenko et al., 2001; Gelinas et al., 2001; Hasoksuz et al., 1999a, 2002a; Tsunemitsu and Saif, 1995). In this study, BCoVs were detected from both nasal and fecal samples in two calves (An‐8 and An‐11) in the same herd (Herd No II) (Table 3). Nucleotide sequence analysis could be performed only in two BCoV isolates recovered from fecal samples from the animals (An‐4 and An‐11).
Table 3.
The individual data of animals detected as positive for bovine coronavirus infection
| Herd No | Sample Code | Material | ELISA | RT‐PCR | VI | Neutralizing antibodies | Age at sampling (month) | Clinical remarks | |
|---|---|---|---|---|---|---|---|---|---|
| N | S1 | ||||||||
| I | Ka‐55 | Feces | + | + | − | + | ≥1/160 | 3 years | Diarrhea |
| Ka‐71 | Feces | + | + | + | + | − | 2 | Diarrhea | |
| Ka‐84 | Feces | + | + | − | + | ≥1/160 | 2 | – | |
| II | An‐4 | Feces | + | + | +* | + | ≥1/160 | 1 | – |
| An‐8 | Feces | − | + | − | + | 1/80 | 1 | Diarrhea | |
| Nasal swab | NT | + | − | + | |||||
| An‐11 | Feces | − | + | +* | + | 1/80 | 1 | Diarrhea | |
| Nasal swab | NT | + | − | + | |||||
| III | V‐7 | Feces | + | + | + | + | − | 1 | Diarrhea |
| V‐27 | Feces | + | + | − | + | − | 1 | Diarrhea | |
| IV | Ko‐422/05 | Feces | + | + | − | + | NT | 0–3 | Diarrhea |
| Ko‐412/05 | Feces | + | + | − | + | NT | 0–3 | Diarrhea | |
| Ko‐419/05 | Feces | + | + | − | + | NT | 0–3 | Diarrhea | |
| Ko‐432/05 | Feces | + | + | − | + | NT | 0–3 | Diarrhea | |
| Ko‐433/05 | Feces | + | + | − | + | NT | 0–3 | Diarrhea | |
| X | Ba‐162548 | Feces | + | + | − | + | NT | 0–3 | Diarrhea |
| Ba‐080506 | Feces | + | + | + | + | NT | 0–3 | Diarrhea | |
| XI | A‐1 | Feces | + | + | − | + | NT | 0–3 | Diarrhea |
Asterisk indicates amplicons sequenced.
NT, not tested; N, N gene; S1, S1 gene; VI, virus isolation.
In conclusion, data obtained from this study showed that BCoV is common in Turkish cattle herds and is a causative agent of respiratory and digestive system infections in calves and adult cattle. The partial characterizations of the S1 gene region of two EBCoV from diarrheic calves were reported. An investigation to determine the biological properties of field isolates recovered in this study and to sequence their HE gene regions is in progress. Further studies involving more BCoVs isolates from cattle with different clinical signs will also be required to compare field strains from different origins in Turkey, in particular their molecular, antigenic and biologic characterizations.
Acknowledgements
This research was supported by a grant of Turkish Scientific and Technical Research Council (Project No: TOVAG 104 0 490).
References
- Alkan, F. , 1998: Buzağı ishallerinde rotavirus ve coronavirusların rolü. Ankara Üniv. Vet. Fak. Derg. 45, 29–37. [Google Scholar]
- Alkan, F. , Bilge–Dağalp S., Can‐Şahna K., and Özgünlük I., 2003: Sığırlarda coronavirus enfeksiyonunun epidemiyolojisi. Ankara Üniv. Vet. Fak. Derg. 50, 59–64. [Google Scholar]
- Athanassious, R. , Marsolais G., Assaf R., Dea S., Descoteaux J.P., Dulude S., and Montpetit C., 1994: Detection of coronavirus and type a rotavirus in neonatal calf diarrhea and winter dysentery of cattle in Quebec: evaluation of threeddiagnostic methods. Can. Vet. J. 35, 163–169. [PMC free article] [PubMed] [Google Scholar]
- Brandao, P.E. , Gregori F., Richtzenhain L.J., Rosales C.A.R., Villarreal L.Y.B., and Jerez J.A., 2006: Molecular analysis of Brazilian strains of bovine coronavirus (BCoV) reveals a deletion within the hypervariable region of the S1 subunit of the spike glycoprotein also found in human coronavirus OC43. Arch. Virol. 151, 1735–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, K.O. , Hasoksuz M., Nielsen P.R., Chang K.O., Lathrop S., and Saif L.J., 2001a: Cross‐protection studies between respiratory and calf diarrhea and winter dysentery coronavirus strains in calves and RT‐PCR and nested PCR for their detection. Arch. Virol. 146, 2401–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, K.O. , Hoet A., Lorech S.C., Wittum T.E., and Saif L.J., 2001b: Evaluation of concurrent shedding of bovine coronavirus via the respiratory tract and enteric route in feedlot cattle. Am. J. Vet. Res. 62, 1436–1441. [DOI] [PubMed] [Google Scholar]
- Chomczynski, P. , and Sacchi N., 1987: Single‐step method of RNA isolation by acid guanidinium thiocyanate‐phenol‐chloroform extraction. Anal. Biochem. 162, 156–159. [DOI] [PubMed] [Google Scholar]
- Chouljenko, V.N. , Kousoulas K.G., Lin X., and Storz J., 1998: Nucleotide and predicted amino acid sequences of all genes encoded by the 3′ genomic portion (9.5 Kb) of respiratorybovine coronaviruses and comparisons among respiratory and enteric coronaviruses. Virus Gene 17, 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouljenko, V.N. , Lin X.Q., Storz J., Kousoulas K.G., and Gorbalenya A.E., 2001: Comparison of genomic and predicted amino acid sequences of respiratory and enteric bovine coronaviruses isolated from the same animal with fatal shipping pneumonia. J. Gen. Virol. 82, 2927–2933. [DOI] [PubMed] [Google Scholar]
- Decaro, N. , Mari V., Desario C., Campolo M., Elia G., Martella V., Greco G., Cirone F., Colaianni M.L., Cordioli P., and Buonavoglia C., 2008: Severe outbreak of bovine coronavirus infection in dairy cattle during the warmer season. Vet. Microbiol. 126, 30–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J. , 1985: Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. [DOI] [PubMed] [Google Scholar]
- Fukutomi, T. , Tsunemitsu H., and Akashi A., 1999: Detection of bovine coronaviruses from adult cows with epizootic diarrhea and their antigenic and biological diversities. Arch. Virol. 144, 997–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher, T.M. , and Buchmeier M.J., 2001: Coronavirus spike proteins in viral entry and pathogenesis. Virology 279, 371–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelinas, A.M. , Boutin M., Sasseville A.M., and Dea D., 2001: Bovine coronaviruses associated with enteric and respiratory disease in Canadian dairy cattle display different reactivities to anti‐HE monoclonal antibodies and distinct amino acid changes in their HE, S and NS4.9 protein. Virus Res. 76, 43–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasoksuz, M. , Lathrop S., Gadfield K., and Saif L.J., 1999a: Isolation of bovine respiratory coronaviruses from feedlot cattle and comparison of their biologic and antigenic properties with bovine enteric coronaviruses. Am. J. Vet. Res. 60, 1227–1233. [PubMed] [Google Scholar]
- Hasoksuz, M. , Lathrop S., Al‐dubaib M.A., Lewis P., and Saif L.J., 1999b: Antigenic variation among bovine enteric coronaviruses (BECV) and bovine respiratory coronaviruses (BRCV) detected using monoclonal antibodies. Arch. Virol. 144, 2441–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasoksuz, M. , Sreevatsan S., Cho K.O., Hoet A.E., and Saif L.J., 2002a: Molecular analysis of the S1 subunit of the spike glycoprotein of respiratory and enteric bovine coronavirus isolates. Virus Res. 84, 101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasoksuz, M. , Hoet A.E., Loerch S.C., Wittum T.L., Nielsen P.R., and Saif L.J., 2002b: Detection of respiratory and enteric shedding of bovine coronaviruses in cattle in an Ohio feedlot. J. Vet. Diagn. Invest. 14, 308–313. [DOI] [PubMed] [Google Scholar]
- Hasoksuz, M. , Kayar A., Dodurka T., and Ilgaz A., 2005: Detection of respiratory and enteric shedding of bovine coronaviruses in cattle in northwestern Turkey. Acta Vet. Hungarica 53, 137–146. [DOI] [PubMed] [Google Scholar]
- Kanno, T. , Kamiyoshi T., Ishihare R., Hatama S., and Uchida I., 2009: Phylogenetic studies of bovine coronaviruses isolated in Japan. J. Vet. Med. Sci. 71, 83–86. [DOI] [PubMed] [Google Scholar]
- Khalili, M. , Morshedi A., Keyvanfar H., and Hemmatzadeh F., 2006: Detection of bovine coronavirus by RT‐PCR in a field study. Veterinarski Arhiv. 76, 291–296. [Google Scholar]
- Lathrop, S.L. , Wittum T.E., Brock K.V., Loerch S.C., Perino L.J., Bingham H.R., Mc Collum F.T., and Saif L.J., 2000a: Association between infection of the respiratory tract attributable to bovine coronavirus and health and growth performance of cattle in feedlots. Am. J. Vet. Res. 61, 1062–1066. [DOI] [PubMed] [Google Scholar]
- Lathrop, S.L. , Wittum T.E., Loerch S.C., Perino L.J., and Saif L.J., 2000b: Antibody titers against bovine coronavirus and shedding of the virus via the respiratory tract in feedlot cattle. Am. J. Vet. Res. 61, 1057–1061. [DOI] [PubMed] [Google Scholar]
- Park, S.J. , Jeong C., Yoon S.S., Choy H.E., Saif L.J., Park S.H., Kim Y.J., Jeong J.H., Park S.I., Kim H.H., Lee B.J., Cho H.S., Kim S.K., Kang M., and Cho K.O., 2006: Detection and characterization of bovine coronaviruses in fecal specimens of adult cattle with diarrhea during the warmer seasons. J. Clin. Microbiol. 44, 3178–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds, D.J. , Debney T.G., Hall G.A., Thomas L.H., and Parsons K.R., 1985: Studies on the relationship between coronaviruses from the intestinal and respiratory tracts of calves. Arch. Virol. 85, 71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou, N. , and Nei M., 1987: The neighbor‐joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Stipp, D.T. , Barry A.F., Alfieri A.F., Takiuchi E., Amude A.M., and Alfieri A.A., 2009: Frequency of BCoV detection by a semi‐nested PCR assay in faeces of calves from Brazilian cattle herds. Trop. Anim. Health Prod. 41, 1563–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K. , Dudley J., Nei M., and Kumar S., 2007: MEGA 4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599. [DOI] [PubMed] [Google Scholar]
- Tsunemitsu, H. , and Saif L.J., 1995: Antigenic and biological comparisons of bovine coronaviruses derived from neonatal calf diarrhea and winter dysentery of adult cattle. Arch. Virol. 140, 1303–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, D. , and Deregt D., 2001: A single amino acid change within antigenic domain II of the spike protein of bovine coronavirus confers resistance to virus neutralization. Clin. Diagn. Lab. Immunol. 8, 297–302. [DOI] [PMC free article] [PubMed] [Google Scholar]


