CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest.
To the Editor,
Chronic rhinosinusitis (CRS) is an inflammatory disorder of the paranasal sinuses occurring with and without nasal polyps (CRSsNP and CRSwNP). Although not objectively demonstrated, an initial viral insult is commonly described by patients prior to the development of CRS. If viruses were demonstrated to play a role in CRS, novel prophylactic and/or therapeutic targets might be uncovered.
Findings in previous studies investigating CRS and viruses are variable.1, 2, 3, 4 Possible reasons include small sample sizes, unvalidated collection methods, seasonal limitation, heterogenous CRS cohorts, and limited viral species screening. No studies to date have investigated disease severity in relation to viral presence.
We aimed to investigate the sinonasal virome of patients with CRS in relation to disease phenotype, to compare it to healthy controls, and to explore any association between more severe disease and viral presence. Cytobrush samples were taken from the sinonasal passages, and DNA/RNA extracts underwent PCR for a number of viral species and strains. The Herpesviridae were excluded due to their near‐ubiquity in adult sinuses. Methodology details appear in the Data S1.
A total of 288 patients were recruited: 71 controls, 133 CRSsNP, and 84 CRSwNP (Table S1). Of the 288, 45 patients were virus‐positive: 5 control, 27 CRSsNP, and 13 CRSwNP (Figure 1). The rate of viral positivity was significantly higher in the CRSsNP group (P < 0.05).
Figure 1.

Detection of virus by PCR in controls and in patients with chronic rhinosinusitis (CRS). Number of patients where virus was detected and not detected in controls, CRSsNP and CRSwNP. Comparison of positivity of virus detected, *P < 0.05, chi‐square test
Objective disease severity scores (Lund‐Mackay [LMS] and Lund‐Kennedy [LKS]) revealed significantly worse disease in the CRSsNP virus‐positive cohort compared with the CRSsNP virus‐negative cohort (P < 0.05, Figure 2). No significant differences were observed in the control or CRSwNP cohorts. Subjective scores (Sino‐Nasal Outcome Test 22 [SNOT‐22] and Adelaide Disease Severity Score [ADSS]) revealed no difference between patients with or without virus in any of the groups (Figure 2). PCR cycle thresholds also revealed no difference between virus‐positive or virus‐negative individuals (Table S2). Viral species detected did not vary significantly from the previously published studies; these were largely rhinovirus and coronavirus (Tables S2 and S3 and Figure S1). Peak viral detection occurred in spring and winter; there was no significant difference in detection when analyzed by season (Figure S2).
Figure 2.
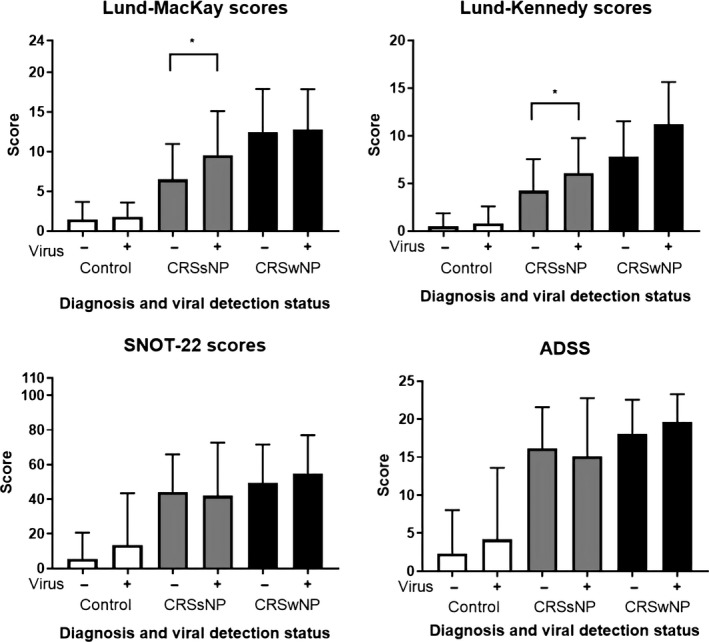
Objective (top) and subjective (bottom) disease scores in virus‐negative and virus‐positive patients. Absolute improvement units grouped and compared by diagnosis (control, CRSsNP, or CRSwNP), * P < 0.05, Kruskal‐Wallis tests
This study identified common respiratory viruses as more prevalent in patients with CRSsNP than in controls. It is the first study to demonstrate their significant association with more severe radiological and endoscopic disease in virus‐positive CRSsNP patients but not virus‐positive patients with CRSwNP.
The lack of any significant difference in subjective symptom scores in any of the groups is not unexpected. The absence of correlation between subjective and objective measures of disease severity has been well documented.5 Although the inclusion of nonrhinologic questions in the SNOT‐22 score is a possible explanation, no difference was observed when using the more specific ADSS. Another possible explanation may be the timing of sampling. As most viruses tested in the assay are shed from the nasopharynx up to 3 weeks after symptom resolution, it is possible that sampling occurred either during this time or early in the infection prior to symptom development.
The viruses identified largely were consistent with those seen in previous CRS studies, with the exception that this study did not identify metapneumovirus. The main viruses observed across all cohorts were rhinovirus and coronavirus, with influenza featuring strongly in the CRS group. However, it seems likely there is no one virus with a particular contribution to CRS not also seen in the general population. The size of the virus‐positive cohort limited subanalysis of viral species/strains with regard to disease severity. As such, we were unable to determine whether one particular viral species is associated with worse disease.
Seasonality is an important concern when sampling for respiratory viruses. These are known to be most prevalent in winter and the early part of spring, an observation supported by this study. Importantly, we also showed strong viral positivity in summer and autumn. This highlights a clear shortfall of previous CRS virome studies that limited sampling to winter and spring.
The mechanisms underlying the viral contribution to CRS are unknown. Similarly unknown is whether the higher rates of viral infection here observed are a cause or a consequence of CRS. CRS has been well established as a bacterial disease, encompassing bacterial overgrowth, superantigen and biofilm formation, and disruption of the microbiome.6 The link between the bacterial and viral hypotheses of CRS aetiopathogenesis may lie in the ability of viruses to prime the airways for bacterial infection. Viruses damage the epithelial barrier by increasing mucus production, reducing ciliary presence, and reducing tight junction expression. Viruses bind directly to bacteria and upregulate host cell surface molecules to facilitate bacterial‐host adherence. Viruses hamper the innate immune system with effects on neutrophil and macrophage recruitment and impairment of natural killer, antigen presenting and T cell activity, leaving the mucosa at risk for bacterial invasion. Viruses alter temperature and variably exhaust or increase the availability of micronutrients, which can allow bacteria in planktonic and biofilm form to proliferate (References in Data S1). The general population, however, is subject to a near‐constant onslaught from respiratory viruses but only 16% of individuals develop CRS. The link between these two entities may be related to the interferons (IFN) and their signaling pathways. These cytokines are expressed by almost all cells in response to pathogen invasion and play key roles in innate antiviral immunity. Deficient IFN responses to viral infection have been consistently demonstrated in asthma, a similar disease process to CRS, as well as in CRS itself.7, 8, 9 We hypothesize this lack of early antiviral activity in patients with CRS may result in more frequent, severe, or persistent viral infections, paving the way for the bacterial invasion so critical for disease development. Further research, however, is required to clarify this hypothesis.
Our results confirm a long‐held suspicion that viruses are more common in CRS than in the general population. Viral presence is associated with more severe sinus disease measured by LMS and LKS. This has the potential to lead to exciting new developments in viral prophylaxis and antiviral therapy in the prevention and possible treatment of CRS.
Supporting information
ACKNOWLEDGMENTS
The authors thank Professor Eric Gowans for his assistance in establishing the methodology and Dr. Ahmed Bassiouni for his advice on statistical analysis. The authors also acknowledge the contributions of Drs. Lisa Cherian, Stephanie Fong, Claire Frauenfelder, Michael Gouzos, Glenn Jenkins, Stephen Kao, Giri Krishnan, Mian Ooi, Sathish Paramasivan, Neil Tan, Rowan Valentine, Mr. Andrew Hayes, and Mr. Aden Lau in the collection of samples, and Drs. Clare Cooksley and Mahnaz Ramezanpour in the processing of samples.
REFERENCES
- 1. Cho GS, Moon BJ, Lee BJ, et al. High rates of detection of respiratory viruses in the nasal washes and mucosae of patients with chronic rhinosinusitis. J Clin Microbiol. 2013;51(3):979‐984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jang YJ, Kwon HJ, Park HW, Lee BJ. Detection of rhinovirus in turbinate epithelial cells of chronic sinusitis. Am J Rhinol. 2006;20(6):634‐636. [DOI] [PubMed] [Google Scholar]
- 3. Wood AJ, Antoszewska H, Fraser J, Douglas RG. Is chronic rhinosinusitis caused by persistent respiratory virus infection? Int Forum Allergy Rhinol. 2011;1(2):95‐100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rowan NR, Lee S, Sahu N, et al. The role of viruses in the clinical presentation of chronic rhinosinusitis. Am J Rhinol Allergy. 2015;29(6):197‐200 [DOI] [PubMed] [Google Scholar]
- 5. Ryan WR, Ramachandra T, Hwang PH. Correlations between symptoms, nasal endoscopy, and in‐office computed tomography in post‐surgical chronic rhinosinusitis patients. Laryngoscope. 2011;121(3):674‐678. [DOI] [PubMed] [Google Scholar]
- 6. Brook I. The role of bacteria in chronic rhinosinusitis. Otolaryngol Clin North Am. 2005;38(6):1171‐1192. [DOI] [PubMed] [Google Scholar]
- 7. Sykes A, Macintyre J, Edwards MR, et al. Rhinovirus‐induced interferon production is not deficient in well controlled asthma. Thorax. 2014;69(3):240‐246. [DOI] [PubMed] [Google Scholar]
- 8. Wark PA, Johnston SL, Bucchieri F, et al. Asthmatic bronchial epithelial cells have a deficient innate immune response to infection with rhinovirus. J Exp Med. 2005;201(6):937‐947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim JH, Kim YS, Cho GS, et al. Human rhinovirus‐induced proinflammatory cytokine and interferon‐beta responses in nasal epithelial cells from chronic rhinosinusitis patients. Allergy Asthma Immunol Res. 2015;7(5):489‐496. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


