Abstract
To cite this article: Turchiarelli V, Schinkel J, Molenkamp R, Foschino Barbaro MP, Carpagnano GE, Spanevello A, Lutter R, Bel EH, Sterk PJ. Repeated virus identification in the airways of patients with mild and severe asthma during prospective follow‐up. Allergy 2011; 66: 1099–1106.
Abstract
Background: Respiratory viruses may persist in the airways of asthmatics between episodes of clinical worsening. We hypothesized that patients with clinically stable, severe asthma exhibit increased and more prolonged viral presence in the airways as compared to mild asthmatics and healthy controls.
Methods: Thirty‐five subjects (no cold symptoms >4 weeks) entered a 12‐week prospective study using three groups: clinically stable mild asthma (GINA 2) (n = 12, age 34.1 ± 13.4 year), severe asthma (GINA 4) (n = 12, age 49.3 ± 14.8 year) and healthy controls (n = 11, age 37.9 ± 14.2 year). All subjects underwent spirometry and completed a written questionnaire on asthma symptoms at baseline. Nasal and throat swabs, induced sputum samples, exhaled breath condensate and gelatine‐filtered expired air were analysed at 0, 6 and 12 weeks by a multiplex real‐time PCR assay for 14 respiratory viruses using adequate positive and negative controls.
Results: Thirty‐two of 525 patient assessments (6%) showed a virus‐positive sample. Among the 14 respiratory viruses examined, HRV, adenovirus, respiratory syncytial virus, parainfluenza 3&4, human bocavirus, influenza B and coronavirus were detected. When combining all sampling methods, on average 18% of controls and 30% of mild and severe asthmatics were virus positive, which was not different between the groups (P = 0.34). The longitudinal data showed a changing rather than persistent viral presence over time.
Conclusion: Patients with clinically stable asthma and healthy controls have similar detection rates of respiratory viruses in samples from nasopharynx, sputum and exhaled air. This indicates that viral presence in the airways of stable (severe) asthmatics varies over time rather than being persistent.
Keywords: diagnosis, latent viral infection, noninvasive sampling, persistent viral infection, rhinovirus
Asthma is a chronic respiratory disease characterized by variable airflow limitation, airway hyperresponsiveness and airway inflammation, accompanied with episodic symptoms of shortness of breath, cough, wheezing and chest tightness (1). There is evidence that respiratory infections are a contributor to the pathogenesis of asthma (2) and that they play a role in the inception, exacerbations and chronicity of airways inflammation in asthma.
First, infant human rhinovirus (HRV) infections appear to be the strongest predictor of subsequent childhood wheezing at age 3 (3, 4). Second, respiratory viruses are closely associated with exacerbations of pre‐existing asthma (5, 6, 7). And third, it has been postulated that latent or persistent infection can contribute to the chronicity of airway inflammation (8, 9), potentially driven by epithelial‐derived pro‐inflammatory cytokines and chemokines (10, 11). A recent cross‐sectional study has demonstrated elevated presence of rhinovirus in endobronchial biopsies from patients with stable asthma as compared to control subjects (12). This indicates that rhinovirus can be present in lower airway tissue in asthma, but it is unclear whether such viral presence persists over time. This requires prospective follow‐up studies with repeated viral sampling.
Invasive intrabronchial procedures are less suitable for repeated detection of airways pathogens during prospective monitoring in asthma. There are, however, multiple noninvasive opportunities, such as nasal sampling or induced sputum (IS), which have successfully been used to detect respiratory pathogens (13). In addition, it cannot be excluded that exhaled air or exhaled breath condensate (EBC) contains high molecular compounds that are suitable for real‐time PCR analysis (14). Real‐time multiplex PCR technology is a quantitative and very sensitive technique for virus detection, based on measuring fluorophoric‐labelled oligonucleotides. It can be applied to multiple targets in the same PCR and permits rapid viral identification (15, 16).
Therefore, we hypothesized that patients with clinically stable, severe asthma exhibit increased and more prolonged presence of viruses in the airways as compared to patients with mild asthma and healthy controls. Second, we postulated that such presence can be monitored by longitudinal, noninvasive sampling of the airways, using induced sputum, exhaled breath and standard nasal/throat swabs. Therefore, the primary aim of the study was to perform viral identification in patients with asthma and controls by noninvasive assessment and to associate the presence and types of respiratory virus with clinical severity of asthma. The second aim was to examine whether the assessment of viral presence could be reproduced at 6 and 12 weeks after the initial sampling. Finally, we aimed to compare the viral identification between different sampling techniques, including induced sputum, EBC, gelatine‐filtered expired air and nasal/throat swabs.
Materials and methods
Subjects
A total number of 35 subjects were recruited to participate in this study. All subjects were nonsmokers (18–65 years). The study population consisted of three groups: 12 patients with mild asthma, 12 patients with severe asthma and 11 controls (Table 1). The diagnosis of asthma was based on the GINA Guidelines (1), and asthma severity was defined based on the 2007 severe asthma consensus (17). Patients were selected among outpatients visiting the Academic Medical Centre, Amsterdam, whereas controls were recruited by advertisements on the hospital premises.
Table 1.
Baseline characteristics of the studied subjects
| Controls | Mild asthma | Severe asthma | |
|---|---|---|---|
| Number (N) | 11 | 12 | 12 |
| Age | 37.9 (14.2) | 34.1 (13.4) | 49.3 (14.8) |
| Sex, male/female | 6/5 | 9/3 | 3/9 |
| FEV1pre, %predicted | 107.6 (9.8) | 91.5 (17.8)* | 63.7 (15.1)** |
| FEV1post, %predicted | 113.6 (12.4) | 98.3 (18.2) | 74.6 (13.7) |
Data are expressed as mean and standard deviation (SD). *P = 0.05 and **P = 0.001 as compared to controls.
The mild asthma group was composed of 12 nonsmoking patients with episodic chest symptoms, prebronchodilator FEV1 > 75% predicted, documented reversibility in FEV1 > 200 ml by 400 μg inhaled salbutamol and/or airway hyperresponsiveness (PC20 < 4 mg/ml methacholine bromide) during the past 12 months. The patients were treated with low doses of inhaled corticosteroids (<1000 μg/day inhaled beclomethasone equivalent) and/or long‐acting bronchodilators.
The severe asthma group consisted of 12 nonsmoking patients with episodic chest symptoms, reversibility in FEV1 > 200 ml and/or documented hyperresponsiveness (PC20 methacholine <4 mg/ml) during the past 5 years, use of corticosteroids (≥1000 μg/day inhaled beclomethasone equivalent or oral corticosteroids) and long‐acting bronchodilators for more than 12 months and one severe asthma exacerbation requiring an oral steroid burst during the past 12 months.
The control group was composed of 11 subjects, with a negative history of chest symptoms, prebronchodilator FEV1 > 75%predicted and documented absence of airway hyperresponsiveness (PC20 > 8 mg/ml).
At the time of the study, all patients with asthma were clinically stable, with no loss of control or acute asthma exacerbation as defined by the recent American Thoracic Society/European Respiratory Society (ATS/ERS) recommendations (7) and no evidence of chest infection during the past 6 weeks. The Academic Medical Centre Ethics Committee (Amsterdam, NL, USA) granted ethic approval for the study, and all participants gave written informed consent.
Study design
The study had three groups, prospective design with three visits at 0, 6 and 12 weeks. During a 2‐week screening phase, medical history, asthma control questionnaire (ACQ), physical examination, standard spirometry, postbronchodilator spirometry and/or a methacholine challenge were performed on two separate days.
At baseline, EBC, nasopharyngeal flocked swab, throat flocked swab, induced sputum and gelatine‐filtered expired air were obtained from all subjects. The patients and controls repeated the same procedures at 6 and 12 weeks. The study was performed during the months August until March. All samples were used for the multiplex PCR analysis. Between the visits, all subjects were contacted twice by e‐mail or phone to assess asthma control by ACQ (18) and to inquire about symptoms of intercurrent respiratory tract infections. In case of the latter, the upcoming visit was postponed by 4 weeks.
Induced sputum
Sputum induction was performed according to ERS recommendations (19). In short, patients received pretreatment with 400 μg inhaled salbutamol. FEV1 was measured by standardized spirometry before three episodes of 5‐min inhalation of aerosolized 4.5% hypertonic saline solution generated by ultrasonic nebulizer (KLAVAmed, Bielefeld, Germany). For safety reasons, in severe asthma patients, sputum was induced according to a validated algorithm starting with isotonic saline (20). Prior to sputum production, subjects were asked to rinse their mouth with water, swallow and blow the nose to minimize contamination of sputum with saliva and postnasal drip fluid.
Sputum processing
Whole sputum samples were processed according to a validated protocol (21). An equal volume of dithiotreitol (10 mM DTT in 135 mM Tris buffer, pH 8.0) was added to the sputum (DTT; Sigma Chemical Company, St Louis, MO, USA). The sample was incubated in a shaking bath of 37°C for 15 min with periodical gentle manual mixing to ensure complete homogenization. Cell numbers were counted in a Bürker counting chamber. Slides were stained with Jenner–Giemsa and Romanovsky (Diff‐Quick; Dade Behring, Düdingen, Switzerland). Supernatants were stored at −80°C until further analysis by multiplex viral PCR. Sputum samples that contained <20% nonsquamous cells or <50% viable cells were excluded from analysis.
Exhaled breath condensate
Exhaled breath condensate was collected by Ecoscreen (Erich Jaeger GmbH, Hochberg, Germany) according to the ERS recommendations (22). Patients breathed through a nonrebreathing valve that separates inhaled from exhaled air, while wearing a nose clip. To promote the generation of aerosols from the lower airways, patients were asked to perform three forced expiratory reserve volume (ERV) manoeuvres down to residual volume directly into the mouthpiece. Condensate was frozen at a temperature of −20°C. After 10 min of tidal breathing, the sampling tube was removed from the cooling system and the condensate probe was thawed and poured into a polyethylene storage tube (Expender, Hamburg, Germany). The EBC was stored at −80°C.
Gelatine‐filtered expired air
Patients were asked to perform two forced expirations (ERV manoeuvres) down to residual volume into an expiratory tube containing a gelatinized filter for aerosol capture (Sartorius, Germany). Gelatine filters were dissolved in MagnaPure lysis solution (1 ml) from the total nucleic acid extraction kit (Roche, Penzberg, Germany) and stored at −80°C.
Nasopharyngeal swabs and throat swabs
Nasal and throat swabs were chosen for upper airway assessment (23). For nasopharyngeal collection, a flexible, cotton‐tip, twisted plastic wire‐shaft swab (Copan Italia, Brescia, Italy) was passed through the nostril until resistance at the posterior nasopharynx was encountered. For throat collection, a cotton‐tipped, soft plastic‐shaft swab was passed through the mouth and rubbed over the tonsils and posterior pharynx. The swabs were placed in viral transport media. Samples were kept at −70°C until further processing (23).
Microbiological assessment
All samples from nasal swab, throat swab, induced sputum, gelatine filter and EBC were tested with an internally controlled multiplex PCR assay for 14 respiratory viruses: adenovirus, enterovirus, influenza virus A and B, rhinovirus (HRV), respiratory syncytial virus (RSV), metapneumovirus (MPV), parechovirus, parainfluenza 1–4 (PIV1–4), coronavirus and human bocavirus (hBoV). Multiplex PCR was performed essentially as described before (15, 24). Total nucleic acid was isolated by MagNa Pure LC (Roche). Briefly, 200 μl of sample was isolated by using the total nucleic acid extraction kit (Roche). The purified nucleic acid was eluted in 50 μl of elution buffer. Subsequently, cDNA was generated essentially as described previously (15, 24). The final reverse transcription reaction (50 μl) contained 1500 ng of random hexamers, 1× CMB1 buffer [10 mM Tris–HCl (pH 8.3) 50 mM KCl, 0.1% Triton X‐100], 5 mM MgCl2, 20 U of Superscript II (Invitrogen, Paisley, UK), 120 μM of each dNTP, 4 U of RNAsin and 40 μl of NA eluate. Subsequently, the mixture was incubated at 42°C for 30 min. Multiplex PCRs were performed on a Roche LC480 (Roche). Typical reactions contained 10 μl of 2× Probes Master (Roche), 900 nM of primer (each) 200 nM of probe (each) and 5 μl of cDNA in a total volume of 20 μl. Cycling conditions were as follows: 2 min at 50°C and 10 min at 95°C, followed by 45 cycles each consisting of 15 s at 95°C and 1 min at 60°C. Data were analysed using the LC480 software (Roche). Colour compensation objects were created as described in the LC480 manual. A positive control for each virus was included in the run, as well as a negative control (15, 24).
Statistical analysis
Statistical analysis was performed using the spss software (version 16.0 for Windows; SPSS Inc, Chicago, IL, USA). Normal distributions were examined using the Kolmogorov–Smirnov test. Baseline characteristics between the groups were compared using the unpaired two‐tailed t‐test in combination with corrections for multiple comparisons (Bonferroni). Between‐group differences in the frequency of positive virus identification were analysed using crosstabs and the two‐sided chi‐squared test. The current sample size of 12 subjects per group allowed detection of 2.5‐fold difference in positive viral detection rates between the groups at statistical power of 0.80.
Results
The baseline characteristics of the three groups are listed in Table 1. A total number of 525 samples were analysed by PCR (35 subjects by five techniques at each of the three time points). Overall, 32 of 525 patient assessments (6%) showed a virus‐positive sample (Table 2). Taking all sampling methods together, on average, 2/11 (18%) of healthy controls and 3.6/12 (30%) of mild and severe asthmatics showed any virus positivity at a given visit (1, 2).
Table 2.
Viral identification (number and % of samples) as obtained from any time points (0, 6 and 12 weeks)
| 0 weeks (%) | 6 weeks (%) | 12 weeks (%) | |
|---|---|---|---|
| IS | 4 (11) | 4 (11) | 4 (11) |
| EBC | 2 (5.7) | 1 (2.8) | 2 (5.7) |
| GF | 1 (2.8) | 0 (0) | 1 (2.8) |
| NS | 1 (2.8) | 2 (5.7) | 0 (0) |
| TS | 3 (8.6) | 3 (8.6) | 4 (11) |
IS, induced sputum; EBC, exhaled breath condensate; GF, filtered expired air by gelatinized filter; NS, nasopharyngeal swab; TS, throat swab.
Figure 1.
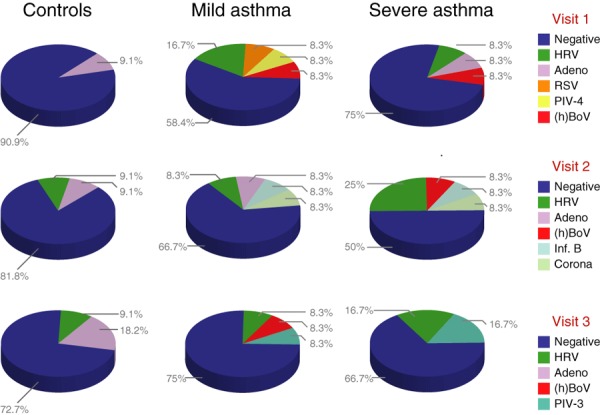
Viral identification rates as obtained by any of the five sampling techniques at baseline (upper panel), 6 weeks (middle panel) and 12 weeks (lower panel) in the healthy controls (left panel), mild asthmatics (middle panel) and severe asthmatics (right panel).
Figure 2.
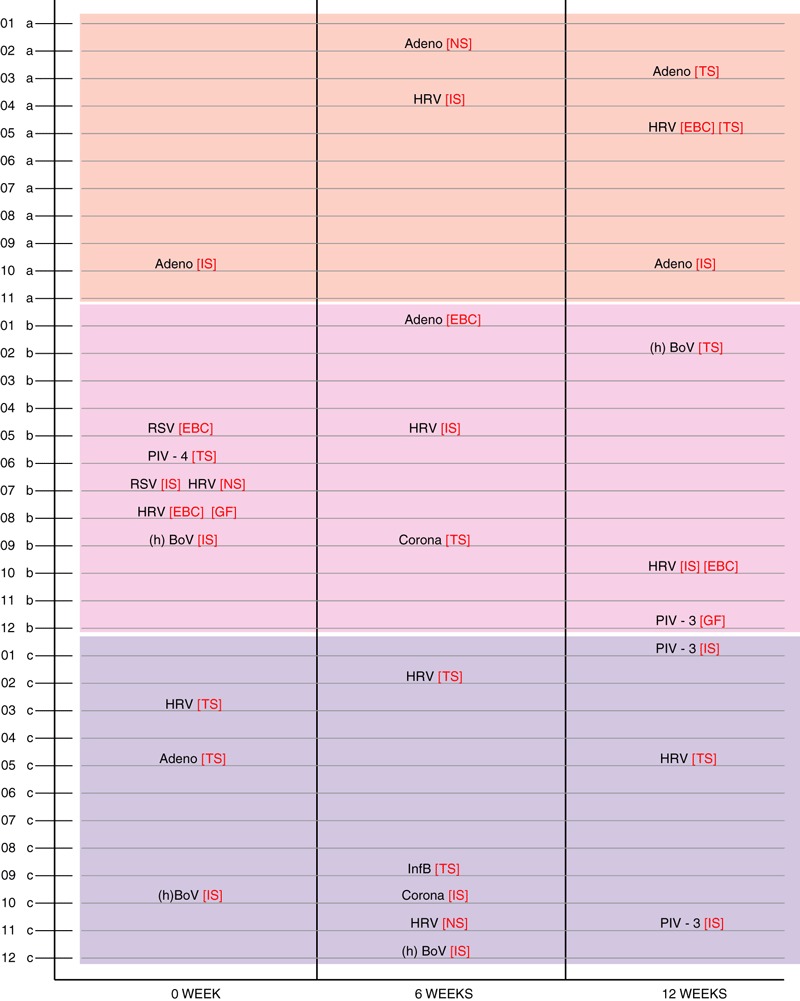
Viral identification in individual subjects in the control (upper panel), mild asthma (middle panel) and severe asthma (lower panel) groups at three different time points (0, 6 and 12 weeks).
Asthmatics vs controls
The microbiological findings for the three visits in mild asthma, severe asthma and controls are shown in Fig. 1. Among the 14 respiratory viruses examined, HRV, adenovirus, RSV, parainfluenza 3&4, hBoV, influenza B and coronavirus were detected. In contrast, all samples were negative for enterovirus, influenza A, MPV, parechovirus and parainfluenza 1&2. Even though the controls showed the lowest viral detection rates at all three visits, there were no between‐group differences in the total number of positive samples (P = 0.34) (Fig. 1).
Comparison between sampling techniques
The total virus identification by the different sampling techniques is shown in Table 2. The highest viral detection rates came from samples obtained from induced sputum and throat swabs, whereas gelatine‐filtered expired air, EBC and nasal swabs were the three sampling techniques yielded the least positive samples. However, the fractions of positive samples were not significantly different between the five sampling methods (P = 0.20). The identified viruses were not consistent between induced sputum and throat swabs in 21 among 22 positive cases.
Repeatability of viral detection over time
As shown in Fig. 1, the virus distribution showed a large variability between 0, 6 and 12 weeks of follow‐up. RSV and parainfluenza‐4 were only detected during one visit, whereas adenovirus, rhinovirus and hBoV were detected in the patient populations at all three visits. However, none of the patients showed evidence of persistent infections by the same virus (Fig. 2).
Discussion
This study shows that repeated, luminal sampling of the airways identifies the presence of respiratory viruses in patients with clinically stable asthma as well as healthy controls. Adenovirus, rhinovirus, RSV, PIV 3&4, influenza B virus, coronavirus, hBoV were identified, whereas enterovirus, influenza A MPV, parechovirus and parainfluenza 1–2 remained undetected. Notably, positive viral identification rates did not significantly differ between patients with mild and severe asthma and healthy controls, nor did infections persist during the 12‐week follow‐up period. These findings suggest that clinically stable asthma is not characterized by elevated or persistent viral presence in the intraluminal airways.
To our knowledge, this is the first study addressing the longitudinal persistence of viral presence in asthma, by repeated sampling of the nasopharynx, induced sputum and exhaled air using a pentaplex PCR for 14 respiratory viruses. Our results extend the cross‐sectional findings by Harju et al. (13) who observed no significant differences in the presence of four respiratory viruses (rhinovirus, enterovirus, RSV and adenovirus) in induced sputum between clinically stable asthmatics and controls. Nevertheless, when examining bronchial tissue, it has been reported that rhinovirus is more frequently detected in clinically stable asthmatics as compared to controls (12). This discrepancy may be attributed to differences in viral presence between airway tissue and intraluminal samples. The latter is supported by a recent cross‐sectional study examining 13 respiratory viruses in bronchoalveolar lavage also showing similar detection rates between asymptomatic asthmatic children and controls (25). Taken together, the present data suggest that respiratory viruses are not more prevalent in the intraluminal airways of asthmatics as compared to controls.
The currently observed overall viral detection rate of 6% is considerably lower than the rates reported by invasive techniques such as bronchoalveolar lavage or bronchial biopsies (12, 25). However, when collectively examining all sampling techniques for each subject, 30% of the asthmatics showed virus positivity at a given visit (1, 2). Hence, different detection rates between studies are likely to be because of technical, temporal and/or biological factors, as invasive methods are sampling different locations of the respiratory tract where viral presence may differ from regions sampled by noninvasive methods (13). Although some studies concluded that nasal washes are slightly more sensitive in viral detection than swabs (26), others have reported that their sensitivity is equal in adults (27). We chose the latter, because nasal swabs are easier to carry out than nasal washes and are causing less discomfort. We observed particularly low detection rate in the gelatine‐filtered expired air and the EBC, which was not unexpected (28). Even though the sampling rates did not differ significantly between the five techniques in the present study, our results suggest that induced sputum may represent a better option in this respect. This fits in with previous studies using induced sputum for viral detection (13, 29) and suggests there is a discrepancy between upper and intrapulmonary airway sampling with regard to the detection of respiratory viruses in asthma and controls.
We made large efforts for adequate selection of subjects and to use optimized methodology. First, the patients were carefully selected based on the latest asthma guidelines (1). On top of this, we added objective criteria to confirm or exclude variable airways obstruction in patients and controls, respectively. Furthermore, the distinction between mild and severe asthma was based on objective and quantitative criteria, including medication usage (17). We were careful to exclude patients who had symptoms of respiratory infections in the past 4 weeks. This timing may not totally exclude the possibility of a recent viral illness, because experimental infections have shown that viruses can occasionally persist in the lower airways for more than 4 weeks (30). Finally, we cannot exclude a medication bias, particularly regarding the dose of inhaled steroids, between the groups. However, this difference was inevitable when selecting mild and severe asthma.
Second, our microbiological findings were obtained by the pentaplex PCR, which is able to yield results in qualitative manner. This pentaplex approach is increasingly applied to routine practice of molecular diagnostics for the respiratory viruses, showing an almost complete concordance between the multiplex PCR and the corresponding single‐target PCR (15). Although crossing point (CP) values could give some indication of high or low viral loads, the assay is essentially qualitative and does not allow statements about replicative properties of the virus in the airways of asthmatic patients and healthy controls. The implicit limitation of our approach is that we did not test for the presence of other nonviral respiratory pathogens that have been linked to the persistence and severity of asthma. This was beyond the objective of the present study. Finally, the current sample size provided a power of 0.80 to detect differences of 2.5‐fold between the groups. We reasoned that clinical or pathophysiological relevance could only be obtained at >2‐fold differences in viral detection rates between the groups. That is why we accepted a 2.5‐fold difference with 12 subjects per group. Rather than expanding the sample size, and thereby increasing the statistical power, we chose to sample each subject at three subsequent time points. Therefore, the present sample size does not suffice to detect smaller differences in virus detection rates between the groups. This should be taken into account when designing future studies.
How can these results be interpreted? Regardless of the sampling techniques used in the present study, it appears that the identification of viruses in the airways of patients with clinically stable asthma does not exceed those in normal controls. Neither was there a difference between severe and mild asthma. This suggests that pre‐existing inflammation and/or impaired innate immune defence do not enhance viral presence in the airways in asthma (31). However, by using 6‐week sampling windows, we cannot exclude the possibility that pre‐existing cytokine profiles or airway inflammation in asthma may still impede viral clearance in asthma (32). Nevertheless, none of the samples showed consistent presence of viruses in individual subjects between 0, 6 and 12 weeks. The variability of virus strains over time extends previous findings in infants (33) and illustrates that series of infections rather than persistent infections occur both in asthmatic and in healthy adults. Taken together, our findings argue against the hypothesis that the maintenance and severity of asthma are driven by persistent virus infections.
The actual viruses that were detected in our study are of clinical interest. Obviously, the viral spectrum is influenced by seasonal variation (34). Nevertheless, the detection of bocavirus was surprising, because this has been reported mainly in paediatric patients with respiratory diseases, but rarely in adults (35, 36). This may suggest a link between host characteristics or medication usage and the presence of bocavirus virus in the airways in asthmatics. This needs to be confirmed by larger prospective studies in asthmatics and controls. Furthermore, we cannot exclude that the presence of atopy with or without exposure to relevant allergens may have affected our results. However, this question was beyond the scope of the present study. Finally, it cannot be excluded that other microbes than respiratory viruses are influencing the maintenance and course of asthma, because a recent study based on culture independent, PCR techniques provided evidence suggestive of differential bacterial presence in bronchial brushings and bronchoalveolar lavage between clinically stable asthmatics and controls (36).
In conclusion, the present study shows that patients with clinically stable asthma and healthy controls have similar detection rates of respiratory viruses in samples from nasopharynx, sputum and exhaled air. Our data indicate that viral presence in the airways of stable asthmatics varies over time rather than being persistent. These findings suggest that the maintenance and chronicity of asthma is not linked to persistent viral presence in the airways.
Authors contributions
VT and PJS conceived, designed the study and drafted the manuscript. VT executed the clinical part of the study. JS, RM and RL executed the laboratory part of the study. MPF, GEC and AS participated in the conception and design of the study. EHB was involved in drafting the manuscript. All authors read and approved the final manuscript prior to submission.
Conflict of interest
The authors declare they have no competing interests.
Edited by: Anthony Frew
References
- 1. Global Initiative for Asthma. From the Global Strategy for Asthma management and prevention, updated 2008. Available at: http://www.ginasthma.org/.
- 2. Dahlberg PE, Busse WW. Is intrinsic asthma synonymous with infection? Clin Exp Allergy 2009;39:1324–1329. [DOI] [PubMed] [Google Scholar]
- 3. Lemanske RF Jr, Jackson DJ, Gangnon RE, Evans MD, Li Z, Shult PA et al. Rhinovirus illnesses during infancy predict subsequent childhood wheezing. J Allergy Clin Immunol 2005;116:571–577. [DOI] [PubMed] [Google Scholar]
- 4. Jackson DJ, Gangnon RE, Evans MD, Roberg KA, Anderson EL, Pappas TE et al. Wheezing rhinovirus illnesses in early life predict asthma development in high‐risk children. Am J Respir Crit Care Med 2008;178:667–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Johnston SL, Pattemore PK, Sanderson G, Smith S, Lampe F, Josephs L et al. Community study of role of viral infections in exacerbations of asthma in 9–11 year old children. BMJ 1995;310:1225–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nicholson KG, Kent J, Ireland DC. Respiratory viruses and exacerbations of asthma in adults. BMJ 1993;307:982–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reddel HK, Taylor DR, Bateman ED, Boulet LP, Boushey HA, Busse WW et al. An official American Thoracic Society/European Respiratory Society statement: asthma control and exacerbations: standardizing endpoints for clinical asthma trials and clinical practice. Am J Respir Crit Care Med 2009;180:59–99. [DOI] [PubMed] [Google Scholar]
- 8. Macek V, Sorli J, Kopriva S, Marin J. Persistent adenoviral infection and chronic airway obstruction in children. Am J Respir Crit Care Med 1994;150:7–10. [DOI] [PubMed] [Google Scholar]
- 9. Hogg JC. Role of latent viral infections in chronic obstructive pulmonary disease and asthma. Am J Respir Crit Care Med 2001;164:S71–S75. [DOI] [PubMed] [Google Scholar]
- 10. Konno S, Grindle KA, Lee WM, Schroth MK, Mosser AG, Brockmann‐Schneider RA et al. Interferon‐gamma enhances rhinovirus‐induced RANTES secretion by airway epithelial cells. Am J Respir Cell Mol Biol 2002;26:594–601. [DOI] [PubMed] [Google Scholar]
- 11. Koetzler R, Zaheer RS, Wiehler S, Holden NS, Giembycz MA, Proud D. Nitric oxide inhibits human rhinovirus‐induced transcriptional activation of CXCL10 in airway epithelial cells. J Allergy Clin Immunol 2009;123:201–208. [DOI] [PubMed] [Google Scholar]
- 12. Wos M, Sanak M, Soja J, Olechnowicz H, Busse WW, Szczeklik A. The presence of rhinovirus in lower airways of patients with bronchial asthma. Am J Respir Crit Care Med 2008;177:1082–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harju TH, Leinonen M, Nokso‐Koivisto J, Korhonen T, Raty R, He Q et al. Pathogenic bacteria and viruses in induced sputum or pharyngeal secretions of adults with stable asthma. Thorax 2006;61:579–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carpagnano GE, Spanevello A, Beghe B, Prato R, Barbaro MP. Microsatellite alterations suggestive of organ‐specific asthma and atopy in exhaled breath condensate. Allergy 2010;65:404–405. [DOI] [PubMed] [Google Scholar]
- 15. Molenkamp R, van der HA, Schinkel J, Beld M. Simultaneous detection of five different DNA targets by real‐time Taqman PCR using the Roche LightCycler480: application in viral molecular diagnostics. J Virol Methods 2007;141:205–211. [DOI] [PubMed] [Google Scholar]
- 16. Lee WM, Grindle K, Pappas T, Marshall DJ, Moser MJ, Beaty EL et al. High‐throughput, sensitive, and accurate multiplex PCR‐microsphere flow cytometry system for large‐scale comprehensive detection of respiratory viruses. J Clin Microbiol 2007;45:2626–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chanez P, Wenzel SE, Anderson GP, Anto JM, Bel EH, Boulet LP et al. Severe asthma in adults: what are the important questions? J Allergy Clin Immunol 2007;119:1337–1348. [DOI] [PubMed] [Google Scholar]
- 18. Juniper EF, Bousquet J, Abetz L, Bateman ED. Identifying ‘well‐controlled’ and ‘not well‐controlled’ asthma using the Asthma Control Questionnaire. Respir Med 2006;100:616–621. [DOI] [PubMed] [Google Scholar]
- 19. Djukanovic R, Sterk PJ, Fahy JV, Hargreave FE. Standardised methodology of sputum induction and processing. Eur Respir J Suppl 2002;37:1s–2s. [DOI] [PubMed] [Google Scholar]
- 20. Ten Brinke A, de Lange C, Zwinderman AH, Rabe KF, Sterk PJ, Bel EH. Sputum induction in severe asthma by a standardized protocol – predictors of excessive bronchoconstriction. Am J Respir Crit Care Med 2001;164:749–753. [DOI] [PubMed] [Google Scholar]
- 21. In‘tVeen JC, de Gouw HW, Smits HH, Sont JK, Hiemstra PS, Sterk PJ et al. Repeatability of cellular and soluble markers of inflammation in induced sputum from patients with asthma. Eur Respir J 1996;9:2441–2447. [DOI] [PubMed] [Google Scholar]
- 22. Horvath I, Hunt J, Barnes PJ, Alving K, Antczak A, Baraldi E et al. Exhaled breath condensate: methodological recommendations and unresolved questions. Eur Respir J 2005;26:523–548. [DOI] [PubMed] [Google Scholar]
- 23. bu‐Diab A, Azzeh M, Ghneim R, Ghneim R, Zoughbi M, Turkuman S et al. Comparison between pernasal flocked swabs and nasopharyngeal aspirates for detection of common respiratory viruses in samples from children. J Clin Microbiol 2008;46:2414–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Beld M, Minnaar R, Weel J, Sol C, Damen M, van der Avoort H et al. Highly sensitive assay for detection of enterovirus in clinical specimens by reverse transcription‐PCR with an armored RNA internal control. J Clin Microbiol 2004;42:3059–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thavagnanam S, Christie SN, Doherty GM, Coyle PV, Shields MD, Heaney LG. Respiratory viral infection in lower airways of asymptomatic children. Acta Paediatr 2010;99:394–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Spyridaki IS, Christodoulou I, de Beer L, Hovland V, Kurowski M, Olszewska‐Ziaber A et al. Comparison of four nasal sampling methods for the detection of viral pathogens by RT‐PCR‐A GA(2)LEN project. J Virol Methods 2009;156:102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gooskens J, Swaan CM, Claas EC, Kroes AC. Rapid molecular detection of influenza outbreaks in nursing homes. J Clin Virol 2008;41:7–12. [DOI] [PubMed] [Google Scholar]
- 28. St George K, Fuschino ME, Mokhiber K, Triner W, Spivack SD. Exhaled breath condensate appears to be an unsuitable specimen type for the detection of influenza viruses with nucleic acid‐based methods. J Virol Methods 2010;163:144–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Simpson JL, Moric I, Wark PAB, Johnston SL, Gibson PG. Use of induced sputum for the diagnosis of influenza and infections in asthma: a comparison of diagnostic techniques. J Clin Virol 2003;26:339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kling S, Donninger H, Williams Z, Vermeulen J, Weinberg E, Latiff K et al. Persistence of rhinovirus RNA after asthma exacerbation in children. Clin Exp Allergy 2005;35:672–678. [DOI] [PubMed] [Google Scholar]
- 31. Wark PA, Johnston SL, Bucchieri F, Powell R, Puddicombe S, Laza‐Stanca V et al. Asthmatic bronchial epithelial cells have a deficient innate immune response to infection with rhinovirus. J Exp Med 2005;201:937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gern JE, Vrtis R, Grindle KA, Swenson C, Busse WW. Relationship of upper and lower airway cytokines to outcom of experimental rhinovirus infection. Am J Respir Crit Care Med 2000;162:2226–2231. [DOI] [PubMed] [Google Scholar]
- 33. Jartti T, Lee WM, Pappas T, Evans M, Lemanske RF, Gern JE. Serial viral infections in infancts with recurrent illnesses. Eur Respir J 2008;32:314–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Arden KE, McErlean P, Nissen MD, Sloots TP, Mackay IM. Frequent detection of human rhinovirues, paramyxoviruses, coronaviruses, and bocavirus during acute respiratory tract infections. J Med Virol 2006;78:1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Maggi F, Andreoli E, Pifferi M, Meschi S, Rocchi J, Bendinelli M. Human bocavirus in Italian patients with respiratory diseases. J Clin Virol 2007;38:321–325. [DOI] [PubMed] [Google Scholar]
- 36. Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C et al. Disordered microbial communities in asthmatic airways. PLoS ONE 2010;5:e8578. [DOI] [PMC free article] [PubMed] [Google Scholar]


