Abstract
Janibacter hoylei has previously been isolated only from an air sample in the upper atmosphere and clinical significance of J. hoylei was not yet established. Herein, we report a case of bacteremia caused by J. hoylei. An 8‐week‐old previously healthy male infant presented to the emergency room with fever. Blood culture yielded growth of Gram‐positive bacilli and this microorganism could not be identified with conventional phenotypic methods. The isolate was identified by 16S rRNA gene sequencing, and the patient was successfully treated with vancomycin. To our knowledge, this is the first report of the recovery of J. hoylei in humans. This case shows that J. hoylei can be a potential pathogen in young children.
Keywords: Janibacter hoylei, bacteremia, identification
Genus Janibacter is an unusual coryneform bacteria that belongs to the family Intrasporangiaceae. Strains of the genus Janibacter have been isolated from various environmental sources 1. Among Janibacter spp., Janibacter hoylei was first isolated from air samples in the upper atmosphere, and there have been no isolates recovered from other environmental samples 2. Due to its rarity, the etiology and clinical significance of J. hoylei are not well known. We present a case of bacteremia in a pediatric patient in which J. hoylei was isolated from blood culture. To the best of our knowledge, this is the first case of bacteremia caused by J. hoylei.
Case Presentation
An 8‐week‐old male infant presented at Chung‐Ang University Hospital with a chief complaint of fever up to 39 °C. The baby was born by Cesarean section at 39 weeks of gestation and had no medical history. The baby presented with irritability, reduced appetite, and increasing body temperature starting 1 day prior.
The infant had a temperature of 38.6 °C in the emergency room and appeared visibly ill. On physical examination, his lungs were clear without wheezes or rhonchi. A complete blood count showed a white blood cell (WBC) count of 7340 × 109/L (normal, 3000–9000 × 109/L), with 64% lymphocytes and 22% segmented neutrophils. High sensitive C‐reactive protein (CRP), erythrocyte sedimentation rate (ESR), electrolytes, and urinary analysis were normal. To avoid contamination, the collection sites were disinfected with chlorhexidine. While chest radiograph revealed no active lung lesion, moderate air distension was seen on supine abdominal X‐ray. Blood culture, urinary culture, and cerebrospinal fluid (CSF) culture were obtained, and empirical therapy with cefotaxime and vancomycin was administered intravenously. To rule out viral infection, virus assays were performed using antibody detection of Mycoplasma pneumonia (serum), enterovirus PCR (stool and CSF), and multiplex real‐time PCR for viral pathogens (nasopharyngeal swab) including influenza A/B virus, respiratory syncytial viruses A/B, adenovirus, human metapneumovirus, coronavirus 229E/NL63/OC43, parainfluenza viruses 1/2/3/4, Rhinovirus A/B/C, enterovirus, and bocavirus 1/2/3/4 (Anyplex II RV16, Seegene, South Korea). However, except for blood cultures, all tests showed negative results.
Blood culture was positive for growth after 72 h of incubation in a pediatric culture bottle (BacT/Alert PF Pediatric FAN, bioMérieux Inc., Marcy‐l'Etoile, France) by the BacT/Alert 3D blood culture system (bioMérieux Inc.). Direct Gram stain revealed that the isolates were Gram‐positive, not Gram‐variable, coccoidal to rod forms. They were 0.4–0.7 mm in diameter, and occurred in singles, pairs, or irregular clumps. To isolate and identify this microorganism, subcultures of the positive blood culture bottle were performed and yielded growth on a blood agar plate under aerobic conditions. Colonies were creamy, yellowish, circular, and convex, measuring 1 mm in size. The isolate was non‐motile, catalase‐positive, oxidase‐positive, coagulase‐negative, and indole‐negative (Fig. 1).
Figure 1.
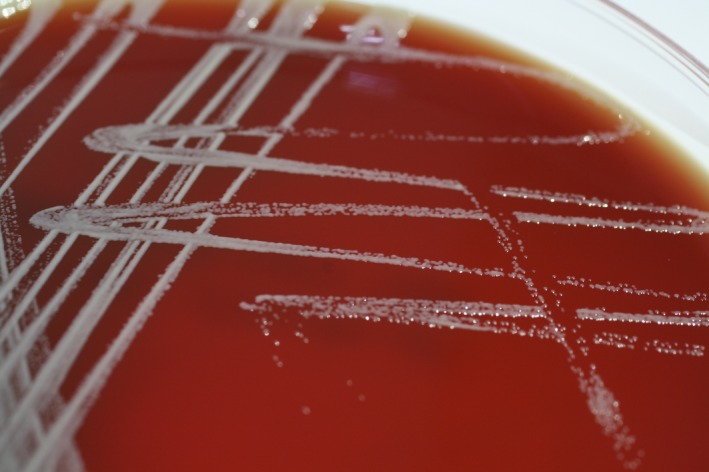
Colonies of Janibacter hoylei on blood agar plates showed creamy, circular, and convex shapes with 1 mm in size.
The Vitek 2 system (bioMérieux Inc.) was used for bacterial identification, and the result was Corynebacterium pseudodiphtheriticum (95% probability) with low discrimination confidence. Additionally, matrix‐assisted laser desorption ionization‐time of flight mass spectrometry (MALDI‐TOF MS) using a VITEK MS system (bioMérieux Inc.) was employed, but the result was indeterminate. The isolate was characterized by 16S rRNA gene sequencing, and a sequence containing 1432 nucleotides was compared for similarity with the GenBank database. There was 99.86% homology of Janibacter hoylei (GenBank accession number NR_104794), followed by 98.81% of Janibacter anopheles (GenBank accession number NR_043218), 98.6% of Janibacter limosus (GenBank accession number NR_026362), and 98.47% of Janibacter cremeus (GenBank accession number NR_114380). For antimicrobial susceptibility tests, we followed the European Committee on Antimicrobial Susceptibility Testing (EUCAST) standardized disk diffusion method 3. The isolate with 0.5 McFarland standard was inoculated onto Mueller–Hinton agar with 5% blood and incubated at 35 °C. Zone diameters were read after 48 hours. Due to the lack of a zone diameter breakpoint for Janibacter spp., we used the zone diameter breakpoint for Corynebacterium spp. according to EUCAST clinical breakpoint tables. Disks of benzylpenicillin, ciprofloxacin, gentamicin, vancomycin, clindamycin, and tetracycline were used, and isolates were shown to be “susceptible” to all agents except benzylpenicillin. Zone diameters were as follows: benzylpenicillin, 12 mm; ciprofloxacin, 34 mm; gentamicin, 46 mm; vancomycin, 47 mm; clindamycin, 29 mm; and tetracycline, 50 mm.
During the hospital stay, the inflammatory markers including CRP and ESR were tested; however, no significant increases were seen. Before antimicrobial susceptibility was reported, the patient's symptoms improved with continuous treatment of cefotaxime with vancomycin, and the baby was discharged without any complications on the fifth day after admission. After discharge, the baby was finally diagnosed with J. hoylei bacteremia.
Conclusions
Since the genus Janibacter, a member of the family Intrasporangiaceae, was first described by Martin et al., a total of nine species have been identified including J. limosus, Janibacter terrae, Janibacter melonis, Janibacter anophelis, Janibacter corallicola, J. hoylei, Janibacter alkaliphilus, J. cremeus, and Janibacter indicus 1. Gram stains for these strains are Gram‐variable or Gram‐positive with coccoidal to rod forms in singles, pairs, or irregular clumps 4. Although the natural habitat is not fully understood, members of the genus Janibacter have been recovered from various environments including environmentally polluted samples, melon, the midgut of mosquitoes, coral, sea sediment, and an air sample 1, 2, 5, 6, 7, 8, 9. Among Janibacter spp., J. hoylei was first isolated from cryotubes used to collect air samples in the upper atmosphere at an altitude of 40–41.4 km and was named after Sir Fred Hoylei, the renowned English astronomer, in 2009 2. The draft genome sequence of this strain was announced 10, but J. hoylei has been previously isolated. To the best of our knowledge, this is the first case of J. hoylei bacteremia and shows that J. hoylei can be isolated from not only high altitude environmental samples but also ground samples.
To date, three literature publications and a total of six cases of bacteremia caused by the genus Janibacter have been reported. In 2005, two cases were simultaneously reported. The pathogen was identified in one case as J. melonis, and the other isolate was not identified 11, 12. Bacteremia caused by J. terrae has also been reported, with a total of four cases described in 2015 13. With the exception of one case caused by J. melonis in a healthy horse‐riding instructor, all five cases had underlying conditions, and three were diagnosed with hematologic malignancy. Therefore, Janibacter spp. may be considered an opportunistic pathogen in immunocompromised patients. However, the previously healthy patient case and our patient suggest that these strains can have pathogenic potential for bacteremia in relatively healthy patients without any other risk factors.
Because symptoms of infection were mild, isolation of J. hoylei from blood culture could be an incidental finding from contamination. However, clinical improvement was clear after treatment with vancomycin, and this finding makes the possibility of a contamination very unlikely. While the previously reported patients infected with other Janibacter spp. had a history of possible direct contact such as insect bite or catheter insertion 11, 13, our case was an 8‐week‐old infant without any medical or travel history and no known contact with Janibacter spp. The exact cause of such bacteremia is unclear; however, we can speculate with a skin colonization of his mother and later ingestion of this strain through breastfeeding.
Conventional biochemical assays do not include associated database entries, because Janibacter spp. have been rarely isolated in the clinical setting. For the same reason, there is no database of genus Janibacter on MALDI‐TOF analyzers. Although these methods could be helpful in identification of Janibacter spp., the only reliable and exact method for identification of Janibacter spp. is sequencing of 16S rRNA. In addition, because 16S rRNA sequencing cannot be applied routinely in microorganism identification, there might be a substantial number of missed or misidentified cases of Janibacter spp. infection.
There are no comprehensive data of antimicrobial susceptibility patterns in Janibacter spp., and only two literature reports have conducted antimicrobial susceptibility testing. Loubinoux et al. reported that unidentified Janibacter isolate was susceptible to penicillin, aminoglycosides, fluoroquinolones, and glycopeptides 12. In a recent report, Fernández‐Natal et al. applied the Clinical Laboratory Standards Institute method for antimicrobial susceptibility of coryneform organisms to J. terrae isolates and showed susceptibility to imipenem, vancomycin, linezolid, daptomycin, tetracycline, ciprofloxacin, erythromycin, gentamicin, and cotrimoxazole 13. In our case, J. hoylei showed susceptibility to ciprofloxacin, gentamicin, vancomycin, clindamycin, and tetracycline. While some differences can be shown in antimicrobial susceptibilities between Janibacter spp., the genus Janibacter is generally susceptible to vancomycin 4. Antibiotic therapy including vancomycin should be adequate treatment for Janibacter spp. infection even if antimicrobial susceptibility testing is not performed.
In conclusion, our case report shows that J. hoylei can be isolated not only from air samples but also from clinical specimens. In addition, Janibacter spp. have pathogenic potential in healthy subjects as well as in immunocompromised patients. Therefore, clinical microbiologists should recognize that these species can be isolated in clinical specimens regardless of host immune status and can only be identified by molecular methods.
Lim YK, Kweon OJ, Kim HR, Kim TH, Lee M‐K. First case of bacteremia caused by Janibacter hoylei . APMIS 2017; 125: 665–668.
References
- 1. Zhang G, Ren H, Wang S, Chen X, Yang Y, Zhang Y, et al. Janibacter indicus sp. nov., isolated from hydrothermal sediment of the Indian Ocean. Int J Syst Evol Microbiol 2014;64:2353–7. [DOI] [PubMed] [Google Scholar]
- 2. Shivaji S, Chaturvedi P, Begum Z, Pindi PK, Manorama R, Padmanaban DA, et al. Janibacter hoylei sp. nov., Bacillus isronensis sp. nov. and Bacillus aryabhattai sp. nov., isolated from cryotubes used for collecting air from the upper atmosphere. Int J Syst Evol Microbiol 2009;59:2977–86. [DOI] [PubMed] [Google Scholar]
- 3. The European Committee on Antimicrobial Susceptibility Testing . Breakpoint tables for interpretation of MICs and zone diameters. Version 5.0, 2015. http://www.eucast.org.
- 4. Jorgensen JH, Carroll KC, Funke G, Landry ML, Richter SS, Warnock DW. Manual of Clinical Microbiology, 11th Edn Washington, DC: ASM Press, 2015: 414–36. [Google Scholar]
- 5. Martin K, Schumann P, Rainey FA, Schuetze B, Groth I. Janibacter limosus gen. nov., sp. nov., a new actinomycete with meso‐diaminopimelic acid in the cell wall. Int J Syst Bacteriol 1997;47:529–34. [DOI] [PubMed] [Google Scholar]
- 6. Yoon JH, Lee HB, Yeo SH, Choi JE. Janibacter melonis sp. nov., isolated from abnormally spoiled oriental melon in Korea. Int J Syst Evol Microbiol 2004;54:1975–80. [DOI] [PubMed] [Google Scholar]
- 7. Kampfer P, Terenius O, Lindh JM, Faye I. Janibacter anophelis sp. nov., isolated from the midgut of Anopheles arabiensis . Int J Syst Evol Microbiol 2006;56:389–92. [DOI] [PubMed] [Google Scholar]
- 8. Kageyama A, Takahashi Y, Yasumoto‐Hirose M, Kasai H, Shizuri Y, Omura S. Janibacter corallicola sp. nov., isolated from coral in Palau. J Gen Appl Microbiol 2007;53:185–9. [DOI] [PubMed] [Google Scholar]
- 9. Li J, Long LJ, Yang LL, Xu Y, Wang FZ, Li QX, et al. Janibacter alkaliphilus sp. nov., isolated from coral Anthogorgia sp. Antonie Van Leeuwenhoek 2012;102:157–62. [DOI] [PubMed] [Google Scholar]
- 10. Pawar SP, Dhotre DP, Shetty SA, Chowdhury SP, Chaudhari BL, Shouche YS. Genome sequence of Janibacter hoylei MTCC8307, isolated from the stratospheric air. J Bacteriol 2012;194:6629–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Elsayed S, Zhang K. Bacteremia caused by Janibacter melonis . J Clin Microbiol 2005;43:3537–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Loubinoux J, Rio B, Mihaila L, Fois E, Le Fleche A, Grimont PA, et al. Bacteremia caused by an undescribed species of Janibacter . J Clin Microbiol 2005;43:3564–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fernandez‐Natal MI, Saez‐Nieto JA, Medina‐Pascual MJ, Valdezate‐Ramos S, Guerra‐Laso JM, Rodriguez‐Pollan RH, et al. First report of bacteremia by Janibacter terrae in humans. Infection 2015;43:103–6. [DOI] [PubMed] [Google Scholar]


