Abstract
Hold tight: Bismuth complexes including ranitidine bismuth citrate effectively inhibit the nucleoside triphosphate hydrolase and DNA unwinding activities of the SARS coronavirus (SCV) helicase and dramatically reduce SCV replication levels in infected cells. This suggests that bismuth‐based drugs should be further evaluated for the treatment of SCV infections in vivo. ss=single‐stranded DNA, ds=duplex DNA.

Keywords: antiviral agents, bismuth, enzymes, helical structures, SARS coronavirus
Severe acute respiratory syndrome (SARS) is a serious type of pneumonia that was first recognized in late 2002 and was subsequently shown to be caused by the SARS coronavirus (SCV).1 The SCV genome was sequenced within weeks of the initial identification2, 3 and further analysis revealed that SCV is most closely related to the group 2 coronaviruses.4 SCV‐like viruses are known to have a variety of animal reservoirs including palm civets, raccoon dogs, and horseshoe bats.5, 6 SCV is also able to infect cats, which warns of further possible transmission routes for the virus.7 Surveillance and infection‐control measures successfully contained SCV in 2003, but treatment will be the only possibility should the virus re‐emerge before an effective vaccine has been developed. The SCV S protein,8 main proteinase,9, 10 cysteine proteinases,11 nucleoside triphosphate hydrolase (NTPase)/helicase,11, 12 and RNA polymerase13 proteins have all been identified as potential targets for antiviral therapy and have already undergone preliminary characterization.
We previously overexpressed and biochemically characterized the SCV NTPase/helicase protein (nsp13‐pp1ab) and found that it belongs to a distinct class of 5′ to 3′ viral helicases12 that are analogous to the closely related helicase from human coronavirus 229E.14 It has also been shown that the SCV NTPase/helicase has RNA capping activity and is able to unwind both RNA and DNA duplexes.15 Helicases have previously been identified as attractive targets for antiviral drug design.16, 17 We previously identified SCV helicase inhibitors that inhibit the virus in cell culture from a diverse library of over 50 000 compounds.18 Furthermore, we characterized the bananins as a class of compounds that can inhibit both the helicase and the virus in cell cultures.19
A 100‐residue cysteine‐rich metal binding domain (MBD) is located at the N terminus of the SCV helicase protein. The analogous MBD of a closely related arterivirus helicase was shown to be involved in subgenomic mRNA synthesis, genome replication, and virion biogenesis.20 We have previously shown an unusually strong cysteine–bismuth interaction in metallothioneins21 and so we tested the hypothesis that bismuth ions might bind within the MBD, thereby affecting the enzymatic activities of the helicase.
In our initial set of assays, we tested the bismuth compounds for their ability to inhibit the oligonucleotide (oligo‐(dT)24)‐stimulated ATPase (ATP=adenosine 5′‐triphosphate) activity of the SCV NTPase/helicase (Figure 1).12, 22 Phosphate release owing to ATP hydrolysis was measured at various concentrations of each of the potential inhibitors and is expressed as a percentage of the ATPase rate in the absence of inhibitor (see the http://www.wiley-vch.de/contents/jc_2002/2007/z701021_s.pdf). This primary screen revealed the bismuth‐based compounds to be effective inhibitors of the SCV helicase. Bismuth nitrilotriacetate ([Bi(nta)]), bismuth nitrate ([Bi(nit)]), bismuth tricysteine complex ([Bi(cys)3]), and ranitidine bismuth citrate (RBC) appeared to be the most efficacious inhibitors of ATPase activity, all showing significant inhibition at a concentration of 1 μm (Figure 1 a). The similarity in inhibitory potential of these three compounds suggests that the bismuth ion plays the key role in the inhibitory effects. Bismuth ethylenediaminetetraacetate ([Bi(edta)]) was a surprisingly poor inhibitor, indicating that the complexing groups should not bind to the bismuth (Bi3+) too tightly. Owing to the clinical use of RBC, we chose this compound for further studies.
Figure 1.
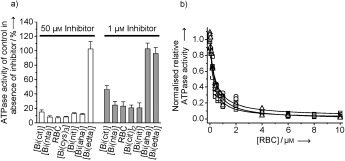
Inhibition of the ATPase activity of the SCV helicase by various bismuth complexes. a) ATPase activity of the SCV helicase in the presence of a 50 μm (left) or 1 μm (right) concentration of various potential inhibitors. ATPase activity is expressed as a percentage of the activity in the absence of inhibitor (representing 100 %). b) Titration of the ATPase activity of the SCV helicase with RBC in the presence of 1 μm U24 (□), the presence of 1 μm (dT)24 (▵), or in the absence of polynucleotide (○). Data have been normalized with respect to the activity in the absence of inhibitor.
To investigate whether RBC specifically inhibited the RNA‐ or DNA‐stimulated or the basal ATPase activity of the helicase, we performed three identical sets of experiments in the presence of oligo‐U24 (as an RNA mimic) in the presence of oligo‐(dT)24 (as a DNA mimic) or in the absence of oligonucleotide. As shown in Figure 1 b, RBC inhibits the DNA‐ and RNA‐stimulated, as well as the nonstimulated, ATPase activity of the helicase to a similar extent. The IC50 values were calculated by fitting the logistic equation for RBC inhibition to each dataset: IC50=248(±75), 350(±82), and 377(±92) nM for U24‐stimulated ATPase, (dT)24‐stimulated ATPase, and nonstimulated ATPase.
After establishing that RBC could effectively inhibit the ATPase activities of the helicase, we next investigated whether it could also inhibit the duplex‐unwinding activity. We used a previously developed radioassay (see the http://www.wiley-vch.de/contents/jc_2002/2007/z701021_s.pdf)12 that measures the inhibition of the helicase's ability to separate the two strands of a small DNA molecule (5T‐DNA). This comprised a short oligonucleotide labeled with 32PO4 at the 5′ end (Figure 2 a, “ss”, the released strand) annealed to a complementary longer strand, generating a partial duplex (Figure 2 a, “ds”) with a 5′ poly‐(dT) overhang. Duplex separation experiments were performed over a range of RBC concentrations. We observed that the duplex was almost fully unwound in the absence of RBC (Figure 2 a, lane A); however, the activity was significantly inhibited in the presence of 0.25 μm RBC (Figure 2 a, lane B), and at higher concentrations of RBC, the unwinding activity was almost totally inhibited (0.5, 1.0, and 2.5 μm RBC for Figure 2 a lanes C, D, and E, respectively). This data set indicated that the helicaseIC50 was in the range of 0.25–0.75 μm. To verify these DNA‐unwinding inhibition results and to obtain a more precise IC50 value, we used a helicase assay based on fluorescence resonant energy transfer (FRET) between the fluorophore Cy3 and the quencher black hole quencher 2 (BHQ‐2) as previously described by using for a different helicase (see the http://www.wiley-vch.de/contents/jc_2002/2007/z701021_s.pdf).19, 23 We altered the partial DNA–duplex substrate molecule to make it compatible with the 5′ to 3′ polarity of the SCV helicase. Conducting the reaction in a fluorimetric cuvette, the SCV helicase was first allowed to equilibrate with the duplex and, where indicated, with RBC at the appropriate concentration by using the same buffer system as the ATPase assays. The reaction was initiated by the addition of adenosine 5′‐triphosphate (ATP; 0.5 mm), and the increase in fluorescence (λ ex=550 nm and λ em=570 nm) owing to release of the Cy3 strand was measured after one minute to determine the extent of reaction. The data were collected and fitted to a logistic equation (Figure 2 b) with the helicaseIC50 value of 0.6 μm in good agreement with that of the radiolabeled assay of helicase activity discussed earlier.
Figure 2.
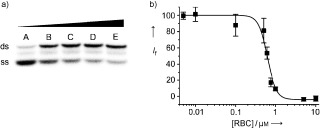
RBC inhibits the DNA unwinding activities of the SCV helicase. a) Titration of the duplex (ds)DNA‐unwinding activity of the SARS helicase with increasing concentrations of RBC observed by radiography of the labeled duplex after acrylamide gel electrophoresis. The lower band, shown the strongest in lane A, represents the (labeled) released single‐stranded (ss) DNA, whilst the upper band, shown the strongest in lane E, represents the duplex DNA. Lane A, no RBC; lanes B to E: 0.25, 0.50, 1.0, and 2.5 μm RBC, respectively. b) Titration of the dsDNA‐unwinding activity of the SCV helicase with increasing concentrations of RBC as observed by a novel FRET assay. The data are expressed as a percentage of the control reaction in the absence of inhibitor. I f=Cy3 relative fluorescence.
The Bi3+ ion is known to have a high affinity for thiolate sulfur.21, 24 We therefore measured the UV absorbance spectrum of the SCV helicase, both in the absence and presence of 1 μm RBC (Figure 3 a). After the addition of RBC, a broad shoulder in the difference spectra was observed between the wavelengths of 300–350 nm, which is characteristic of the formation of Bi—S bonds. This shoulder is very similar in profile to that reported for bismuth binding to cysteine residues of metallothioneins.21 The binding event appeared to be fairly rapid and to be greater than 95 % complete within two minutes after the addition of the RBC. Very slight further increases in the absorption were observed over the following two hours. This was unlike the binding of bismuth to metallothionein or to transferrin, which proceeded over several hours.21, 25
Figure 3.
a) The effect of RBC on the UV absorption spectrum of the SCV helicase. The SCV helicase absorption spectrum was measured before (—) and after (‐ ‐ ‐ ‐) the addition of 1 μm RBC. All spectra are corrected for the buffer solution and the contribution to absorption from RBC itself. b) The titration of the Zn2+ ion into the MBD of the SCV helicase monitored by a PAR/PMPS assay. Inset: Ratio of Zn2+ ions versus MBD. c) The changes in absorbance at approximately 350 nm upon addition of different molar equivalents of Bi3+ ion (ratio of Bi3+ versus MBD).
It is likely that the binding of the Zn2+ ion to the SCV helicase MBD is essential for enzymatic activity.26 We therefore cloned, expressed, and purified the MBD from the SCV to study its biophysical properties in isolation (see the http://www.wiley-vch.de/contents/jc_2002/2007/z701021_s.pdf). The Zn2+‐bound MBD had a retention time of 8.2 min, as determined by gel filtration chromatography (Superdex 75 10/300 GL), which corresponds to a molecular weight of about 42 kDa (see http://www.wiley-vch.de/contents/jc_2002/2007/z701021_s.pdf). This indicates that the Zn2+‐bound MBD is a trimer in solution. The stoichiometry of Zn2+ ion binding to the MBD was measured by using the zinc chelator 4‐(2‐pyridylazo)resorcinol (PAR) together with the thiol‐modifying reagent p‐hydroxymercuriphenylsulfonic acid (PMPS).27, 28 The absorbance of Zn(par)2 at approximately 500 nm increased gradually with the addition of PMPS to a buffered solution of the MBD and PAR (Figure 3 b). The zinc concentration was calculated by using a Zn2+–PAR standard curve. This revealed that the total zinc concentration was about 2.8(±0.2) times that of the MBD, which is indicative of about three Zn2+ ions binding to each MBD monomer (Figure 3 b, inset).
We then performed a similar titration experiment to measure the ratio of Bi3+ ion binding to the MBD. A new absorbance peak appeared at approximately 350 nm in the the UV/Vis spectrum of the MBD, which increased in intensity almost linearly from 0 to approximately 3 mol equivalents upon the gradual addition of Bi3+ ion. No significant increases in absorption were observed upon further addition of the Bi3+ ion, suggesting that the MBD monomer also binds 2.8(±0.3) Bi3+ ions (Figure 3 c), and that the Bi3+ ion effectively competes for the Zn2+ ion binding sites. Taken in combination with the enzymatic data, this suggests that the Bi3+ ion binds strongly at the metal binding domain of the helicase, thereby disrupting and inhibiting both the NTPase and helicase activities. It may be noted that point mutations within the MBD of the helicase from the closely related equine arteritis visus (EAV) led to severe deficiencies in viral replication in infected cell cultures.20, 29
To further investigate the inhibitory properties of RBC, we assessed its anti‐SCV activities against a clinical isolate (SCV strain GZ50) in FRhK‐4 cells. For our initial experiments, we examined cytopathogenic effects by phase‐contrast microscopy 36 h after infection with the virus (see the http://www.wiley-vch.de/contents/jc_2002/2007/z701021_s.pdf). As shown in Figure 4 a, the uninfected cells appeared smooth whilst infected cells showed prominent ridgelike structures along the membranes. Such structures were less prominent after treatment with 100 μm of RBC, but this effect was difficult to quantify by eye. To confirm this result, we used a viral viability assay to measure the effects of RBC treatment. We measured viral reproduction in the presence of RBC by using a TCID50 protocol to determine the number of viable virus particles in the cell‐culture supernatant 36 h post‐SCV infection (see the http://www.wiley-vch.de/contents/jc_2002/2007/z701021_s.pdf).30, 31 RBC significantly inhibited SCV reproduction with the viral titer reduced by approximately 80 % at 25 μm, and concentrations of 500 μm RBC almost completely inhibited SCV infection, replication, and/or release. These data were fitted to the logistic equation to give a 50 % effective concentration (EC50) value of 5.9(±4) μM for viable SCV. Expressing this by using a logarithmic scale for cell viability (Figure 4 b), an 90 % effective concentration (EC90) value of 50(±8) μm was obtained. To measure the cytotoxicity of RBC in cell culture, we used a standard 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide (MTT) assay based on the reduction of the MTT tetrazolium salt to the formazan dye in a reaction catalyzed by mitochondrial succinate dehydrogenase, which is only present in metabolically active cells (see the http://www.wiley-vch.de/contents/jc_2002/2007/z701021_s.pdf).32 The growth rates of RBC‐treated and untreated cells were also compared. The mushroom toxin α‐amanitin was used as a positive control. Fitting data to the logistic equation yielded a 50% cytotoxicity concentration (CC50) of 5.0 mm (Figure 4 c). The EC50 for RBC inhibition of SCV was measured to be 5.9 μm. These results revealed a specificity constant (CC50/EC50) of 847, indicating that RBC acts in a very effective and specific manner in this monkey‐kidney cell‐culture system.
Figure 4.
RBC inhibits SCV infectivity and reproduction. a) Effect of RBC on replication of SCV in FRhK‐4 cells. Where indicated, RBC was added 10 minutes prior to infection and cells were fixed 36 h after infection. Uninfected: uninfected cells. Infected: cells infected in the absence of RBC. Treated: cells infected in the presence of 100 μm RBC. b) Effect of RBC on the infectivity of SCV. Infectivity was measured by measuring the viral titer by back titration of serially diluted culture supernatant 36 h after infection. Data are expressed as a percentage of a control in which no RBC was added to the cell‐culture supernatant. Inset: Viral viability shown by using a logarithmic scale to highlight the differences. c) Cytotoxicity of RBC in cell culture measured by a MTT assay.
In an attempt to delineate the mechanism of RBC‐mediated inhibition of viral replication, the kinetics of SCV infection in the presence of RBC were determined. Quantitative real‐time PCR (Q‐RT‐PCR) was used to monitor the relative quantities of viral RNA (as specified by primers to the SCV spike gene) and cellular RNA (as specified by primers to the housekeeping gene β‐actin) (see the http://www.wiley-vch.de/contents/jc_2002/2007/z701021_s.pdf).30–33 It was found that 6 h after the infection, RBC had little effect on the relative quantities of viral RNA (Figure 5). During this time, even high concentrations of RBC (400 μm) were unable to reduce the viral RNA levels by 50 % relative to a control in the absence of the drug. However, after 12 h, the RBC had a greater effect, with between 50–100 μm RBC causing a 50 % reduction in viral RNA levels. After 24–48 h, a concentration of 12.5–25 μm RBC caused an approximate 50 % reduction in viral RNA levels. These data therefore suggest that RBC plays a greater inhibitory role during the later stages of the replicative cycle. As the SCV helicase acts at later stages of the cycle, this would be consistent with RBC inhibiting SCV helicase functionality within the cell.
Figure 5.
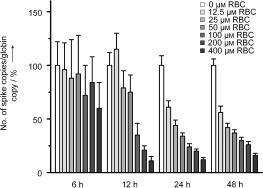
Effects of RBC on the kinetics of SCV infection as measured by Q‐RT‐PCR. SCV infection was measured by primers specific to the SCV spike gene and compared with the housekeeping gene β‐actin as an endogenous control. Data were measured at 6, 12, 24, and 48 h postinfection. Where indicated, RBC was added 1 h prior to infection at the concentration shown.
A distinct benefit of developing bismuth‐based compounds for therapeutic intervention is that many drugs, such as RBC, are already used clinically to treat disorders within the alimentary tract.34 This is particularly relevant in this case as SCV was previously found to be present in the faeces of 97 % of SARS patients, and it is possible that this is an important route of viral transmission.35 Moreover, symptoms of diarrhea were observed in more than 38 % of hospital patients with confirmed SARS infections. Bismuth compounds could be tested against SCV in a prophylactic role, as part of a combination therapy, or in the treatment of infections caused by other nidoviruses.
In summary, we have tested several bismuth complexes as potential inhibitors of the SCV helicase protein. RBC was found to be a particularly strong inhibitor of the ATPase activity of the SCV helicase protein, with an IC50 value of 0.3 μm. Our FRET‐based assays showed that RBC inhibited the DNA duplex unwinding activity with an IC50 value of 0.6 μm. UV/Vis data confirmed that bismuth binds to cysteine residues on the helicase protein. Analogous binding behavior towards the N‐terminal metal binding domain indicates that the bismuth binds to this cysteine‐rich region. In cell culture, RBC effectively inhibited SCV reproduction with an EC50 value of 5.9 μm and exhibited moderately low cytotoxicity with a CC50 value of 5 mm. Q‐RT‐PCR experiments indicate that RBC inhibits a late‐stage process in the SCV replicative cycle, which is consistent with the helicase being the target of these drugs within the cell. Taking into consideration their proposed mechanism of action, high selectivity, and present clinical usage, we suggest that bismuth‐based drugs should be further evaluated for the treatment of SCV infections in vivo.
Experimental Section
Bismuth citrate ([Bi(cit)]), [Bi(nta)], [Bi(cys)3], [Bi(edta)], and bismuth acetohydroxamate ([Bi(aha)]) complexes were prepared by mixing bismuth salts with the appropriate amounts of ligands (e.g. acetohydroxamate (aha)) as described previously.36 Ranitidine bismuth citrate was provided by GlaxoWellcome China Ltd.
The helicase was prepared in 20 mm Tris‐HCl (0.5 mL; pH 6.8; Tris=tris(hydroxymethyl)aminomethane), 200 mm NaCl at a concentration of 0.22 mg mL−1. RBC was then added to a final concentration of 1 μm, then an appropriate delay (minutes) was allowed for binding before measuring the UV/Vis spectrum. The MBD was prepared in 20 mm Tris‐HCl pH 8.5, 200 mm NaCl. For the Zn2+ ion titration experiment, 10 mm PMPS was added gradually to a mixture of 5 μm MBD and 100 μm PAR (0.4 mL, MBD/PAR=1:20) in a UV/Vis cuvette, and the absorbance at 500 nm was recorded. The Zn2+–PAR standard curve was constructed by measuring the UV/Vis absorbance at 500 nm of 100 μm PAR with various dilutions of a zinc atomic absorption standard solution.
For the Bi3+ ion titration experiments, 1 mm [Bi(nta)] was gradually added to 5 μm MBD in 20 mm Tris‐HCl pH 8.5 and 200 mm NaCl solution (total volume=0.4 mL), to a final concentration of 40 μm (8 mol equivalents of MBD). Binding of the Bi3+ ion to MBD was examined by UV/Vis spectroscopy by monitoring the changes in absorption at approximately 350 nm.
Supporting information
Supporting information for this article is available on the WWW under http://www.wiley-vch.de/contents/jc_2002/2007/z701021_s.pdf or from the author.
This work was supported by the Hong Kong Health, Welfare and Food Bureau under the Research Fund for the Control of Infectious Diseases, the RGC of Hong Kong (HKU7040/05P and 7546/06M), and Livzon Pharmaceutical Ltd. SARS=severe acute respiratory syndrome.
Contributor Information
Hongzhe Sun, Email: hsun@hku.hk
Jian‐Dong Huang, Email: jdhuang@hku.hk
References
- 1.
- 1a. Ksiazek T. G., et al., N. Engl. J. Med. 2003, 348, 1953–1966; [DOI] [PubMed] [Google Scholar]
- 1b. Peiris J. S., et al., Lancet 2003, 361, 1319–1325; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1c. Fouchier R. A., Kuiken T., Schutten M., van Amerongen G., van Doornum G. J., van den Hoogen B. G., Peiris M., Lim W., Stohr K., Osterhaus A. D., Nature 2003, 423, 240; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1d. Kuiken T., et al., Lancet 2003, 362, 263–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Marra M. A., et al., Science 2003, 300, 1399–1404. [DOI] [PubMed] [Google Scholar]
- 3. Rota P. A., et al., Science 2003, 300, 1394–1399. [DOI] [PubMed] [Google Scholar]
- 4. Mayor L. R., Fleming K. P., Muller A., Balding D. J., Sternberg M. J., J. Mol. Biol. 2004, 340, 991–1004. [DOI] [PubMed] [Google Scholar]
- 5. Guan Y., et al., Science 2003, 302, 276–278. [DOI] [PubMed] [Google Scholar]
- 6. Lau S. K. P., Woo P. C. Y., Li K. S. M., Huang Y., Tsoi H. W., Wong B. H. L., Wong S. S. Y., Leung S. Y., Chan K. H., Yuen K. Y., Proc. Natl. Acad. Sci. USA 2005, 102, 14040–14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Martina B. E., Haagmans B. L., Kuiken T., Fouchier R. A., Rimmelzwaan G. F., Amerongen G. Van, Peiris J. S., Lim W., Osterhaus A. D., Nature 2003, 425, 915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sui J., et al., Proc. Natl. Acad. Sci. USA 2004, 101, 2536–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Anand K., Ziebuhr J., Wadhwani P., Mesters J. R., Hilgenfeld R., Science 2003, 300, 1763–1767. [DOI] [PubMed] [Google Scholar]
- 10. Yang H., et al., Proc. Natl. Acad. Sci. USA 2003, 100, 13190–13195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thiel V., et al., J. Gen. Virol. 2003, 84, 2305–2315. [DOI] [PubMed] [Google Scholar]
- 12. Tanner J. A., Watt R. M., Chai Y. B., Lu L. Y., Lin M. C., Peiris J. S., Poon L. L., Kung H. F., Huang J. D., J. Biol. Chem. 2003, 278, 39578–39582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Campanacci V., et al., Acta Crystallogr. D 2003, 59, 1628–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Seybert A., Hegyi A., Siddell S. G., Ziebuhr J., RNA 2000, 6, 1056–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ivanov K. A., Thiel V., Dobbe J. C., van der Meer Y., Snijder E. J., Ziebuhr J., J. Virol. 2004, 78, 5619–5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kleymann G., et al., Nat. Med. 2002, 8, 392–398. [DOI] [PubMed] [Google Scholar]
- 17. Crute J. J., Grygon C. A., Hargrave K. D., Simoneau B., Faucher A. M., Bolger G., Kibler P., Liuzzi M., Cordingley M. G., Nat. Med. 2002, 8, 386–391. [DOI] [PubMed] [Google Scholar]
- 18. Kao R. Y., et al., Chem. Biol. 2004, 11, 1293–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tanner J. A., et al., Chem. Biol. 2005, 12, 303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. van Dinten L. C., van Tol H., Gorbalenya A. E., Snijder E. J., J. Virol. 2000, 74, 5213–5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sun H., Li H., Harvey I., Sadler P. J., J. Biol. Chem. 1999, 274, 29094–29101. [DOI] [PubMed] [Google Scholar]
- 22. Porter D. J., J. Biol. Chem. 1998, 273, 7390–7396. [DOI] [PubMed] [Google Scholar]
- 23. Boguszewska‐Chachulska A. M., Krawczyk M., Stankiewicz A., Gozdek A., Haenni A. L., Strokovskaya L., FEBS Lett. 2004, 567, 253–258. [DOI] [PubMed] [Google Scholar]
- 24. Sadler P. J., Sun H., Li H., Chem. Eur. J. 1996, 2, 701–708. [Google Scholar]
- 25. Li H., Sadler P. J., Sun H., J. Biol. Chem. 1996, 271, 9483–9489. [PubMed] [Google Scholar]
- 26. Seybert A., Posthuma C. C., van Dinten L. C., Snijder E. J., Gorbalenya A. E., Ziebuhr J., J. Virol. 2005, 79, 696–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Daniels M. J., Turner‐Cavet J. S., Selkirk R., Sun H., Parkinson J. A., Sadler P. J., Robinson N. J., J. Biol. Chem. 1998, 273, 22957–22961. [DOI] [PubMed] [Google Scholar]
- 28. Jakob U., Eser M., Bardwell J. C., J. Biol. Chem. 2000, 275, 38302–38310. [DOI] [PubMed] [Google Scholar]
- 29. van Marle G., van Dinten L. C., Spaan W. J., Luytjes W., Snijder E. J., J. Virol. 1999, 73, 5274–5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. He M. L., Zheng B. J., Peng Y., Peiris J. S., Poon L. L., Yuen K. Y., Lin M. C., Kung H. F., Guan Y., JAMA J. Am. Med. Assoc. 2003, 290, 2665–2666. [DOI] [PubMed] [Google Scholar]
- 31. Zheng B. J., et al., Antivir. Ther. 2004, 9, 365–374. [PubMed] [Google Scholar]
- 32. Mosmann T., J. Immunol. Methods 1983, 65, 55–63. [DOI] [PubMed] [Google Scholar]
- 33. He M. L., Wu J., Chen Y., Lin M. C., Lau G. K., Kung H. F., Biochem. Biophys. Res. Commun. 2002, 295, 1102–1107. [DOI] [PubMed] [Google Scholar]
- 34.
- 34a. Suerbaum S., Michetti P., N. Engl. J. Med. 2002, 347, 1175–1186; [DOI] [PubMed] [Google Scholar]
- 34b. Sun H., Zhang L., Szeto K. Y., Met. Ions Biol. Syst. 2004, 41, 333–378; [PubMed] [Google Scholar]
- 34c. Ge R. G., Sun H., Acc. Chem. Res. 2007, 40, 267–274. [DOI] [PubMed] [Google Scholar]
- 35. Peiris J. S., et al., Lancet 2003, 361, 1767–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Summers S. P., Abboud K. A., Farrah S. R., Palenik G. J., Inorg. Chem. 1994, 33, 88–92. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information for this article is available on the WWW under http://www.wiley-vch.de/contents/jc_2002/2007/z701021_s.pdf or from the author.


