Abstract
This study evaluated removal of live emerging waterborne pathogens by pilot‐scale conventional treatment with alum coagulation, flocculation, sedimentation, and filtration. The microbes tested were Cryptosporidium parvum oocysts, Encephalitozoon intestinalis spores, enteropathogenic Escherichia coli O157:H7, Aeromonas hydrophila, and bacteriophage MS2. The study showed the effects of filter run time, alternative loading rates, alternative filter media, and pH on pathogen removal. Results indicated that turbidity breakthrough was accompanied by breakthrough of all pathogens tested in this study. Results also suggest that the breakthrough of A. hydrophila and E. coli O157:H7 occurred more rapidly than that of turbidity. In general, filtration rate or alternative filter media configurations had no apparent effect on pathogen removal.
Keywords: Pathogens, Filtration, Cryptosporidium, Filters, Surface Water, Bacteriophages, Filter Media, pH, Giardia
Emerging infectious diseases are defined as those whose documented incidence in humans has increased during the past 20 years or threatens to rise in the near future (Institute of Medicine, 1992). A variety of viral, bacterial, and protozoan agents have been implicated as potentially important emerging waterborne agents. Although little is known about the fate, ecology, and distribution of these microorganisms in the aquatic environment, some of these microorganisms have already been identified as the cause of waterborne disease outbreaks (Mullens, 1996; CDC, 1993; CDC, 1991a; CDC, 1991b; Kaplan et al, 1982a; Kaplan et al, 1982b). Beyond these documented cases, other emerging pathogens have been identified in natural water sources and therefore have the potential to cause waterborne disease outbreaks (Moe, 1997; Avery & Undeen, 1987; Clark et al, 1982).

Emerging viral pathogens include the caliciviruses, such as Norwalk and Norwalk‐like viruses, and other small round structured viruses such as astroviruses and coronaviruses. Enteric adenoviruses are also considered to be emerging viral pathogens. Several bacteria, including Escherichia coli O157:H7, Aeromonas hydrophila, Mycobacterium avium, and Helicobacter pylori, have also been identified as emerging waterborne pathogens. Emerging protozoan parasites include five genera of microsporidian protozoa that are known to cause disease in humans: Nosema, Pleistophora, Enterocytozoon, Encephalitozoon, and Microsporidium (Garcia & Bruckner, 1997). Other recent waterborne outbreaks have been attributed to the protozoa Cyclospora cayetanensis and Toxoplasma gondii (Mullens, 1996; CDC, 1991b).
Regulatory response to pathogen emergence. Giardia and Cryptosporidium were the primary emerging protozoan pathogens of concern in the 1980s and 1990s. The emergence of Giardia as a waterborne pathogen in the late 1970s and early 1980s helped reinforce the need for a multiple‐barrier approach to water treatment. To address this need, the US Environmental Protection Agency (USEPA) issued the Surface Water Treatment Rule (SWTR) in 1989 (USEPA, 1989). This rule required filtration for most utilities that treat surface water and established minimum levels of disinfection for all utilities that treat surface water. In addition, the emergence of Cryptosporidium in the late 1980s and 1990s forced the USEPA to issue and propose a series of Enhanced SWTRs (USEPA, 2002; USEPA, 2000; USEPA, 1998; USEPA, 1994). These regulatory actions trace how a pathogen can emerge to substantially alter regulatory policies and treatment practices. Given the significant effect these new rules have had on the drinking water industry, it is appropriate to ask whether some other organism could emerge to trigger more changes in both policy and practice.
The observed or suspected presence of pathogens in water systems implies that they entered the systems by direct contamination of the distribution system or by surviving the conventional, multiple‐barrier approach to drinking water treatment. The potential for the latter of these two possible routes has not yet been addressed for the emerging pathogens noted previously. This article provides data that begin to fill this information gap.
Objective. The objective of this research was to evaluate the removal of several emerging waterborne pathogens by using pilot‐scale conventional treatment with alum coagulation, flocculation, sedimentation, and filtration. The study focused on the effect of filter run time, alternative filter loading rates, alternative filter media, and coagulation pH on pathogen removal.
Experimental Procedures
Although many studies have focused on the effects of particle size on particle removal in clean bed filters, little research has been conducted to determine the breakthrough rates of different‐sized particles.
Selected microorganisms. Six pilot‐scale challenge experiments were performed to investigate the effects of various filtration conditions on the removal of live waterborne pathogens by alum coagulation, flocculation, sedimentation, and rapid‐rate granular media filtration. The microbes tested were Cryptosporidium parvum oocysts, Encephalitozoon intestinalis spores, enteropathogenic E. coli O157:H7, A. hydrophila, and bacteriophage MS2. Test microorganisms were pooled to produce a microbial “cocktail” that was used to seed raw waters for all pilot‐scale experiments.
Table 1 summarizes the morphological characteristics of these pathogens; however, the characteristics presented here may have only an indirect influence on pathogen removal. Once coagulation and flocculation have been completed, these pathogens would likely be associated with floc particles having significantly different sizes and shapes from those shown in the table. It is the characteristics of these floc particles that have a direct influence on pathogen removal through sedimentation and granular media filtration. Nevertheless, the pathogen characteristics listed in Table 1 play a role in determining the physical and chemical characteristics of the floc particles. Floc characteristics will also depend on other factors including the type and dosage of coagulant being used, the pH of coagulation, and the characteristics of other particles such as suspended silts and clays.
Table 1.
Typical morphological characteristics of selected microorganisms
| Pathogen | Shape | Size—μm |
|---|---|---|
| Cryptosporidium parvum oocysts | Round | 3–7 |
| Encephalitozoon intestinalis spores | Tear drop | (1.3–2.1) × (2.5–3.3) |
| Aeromonas hydrophila | Rod | (0.3–1.0) × (1.0–3.5) |
| Escherichia coli O157:H7 | Rod | (1.0–1.5) × (2.0–6.0) |
| Bacteriophage MS2 | Icosahedral | 0.026–0.027 |
Methods common to all pilot experiments. Continuous‐flow pilot‐plant experiments were run at the University of Wisconsin's pilot‐scale water treatment facility in Madison. The pilot plant received raw water from Lake Mendota in Madison and included two parallel treatment trains (Figures 1–3) operated at a flow rate of 1 gpm (0.06 L/s) per treatment train. Raw water was pumped to a constant‐head reservoir and flowed by gravity through two rapid‐mix tanks, three flocculation chambers, and one sedimentation basin on each train. For the duration of each run, pathogen cocktail and alum were fed at a constant dosage to the first and second rapid‐mix chambers, respectively.
Figure 1.
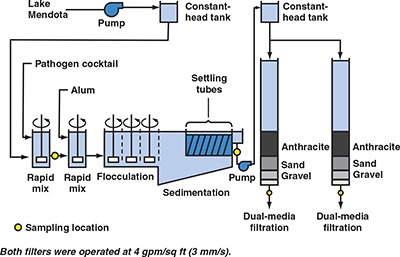
Flow chart for evaluating effect of filter run time on removal of emerging waterborne pathogens
Figure 3.
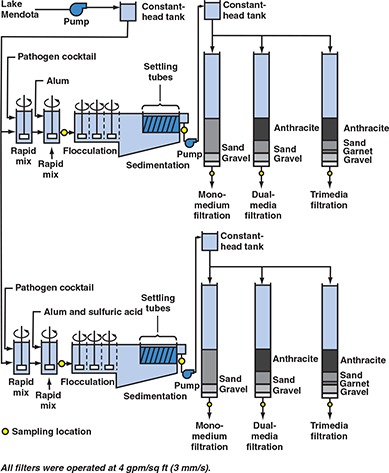
Flow chart for evaluating effect of alternative filter media configuration on removal of emerging waterborne pathogens
For each treatment train, settled water was pumped to a constant‐head tank and then flowed by gravity to a rack of four granular media filter columns. Pumping of settled water was provided because of space limitations in the laboratory, but similar designs have been used in previous studies on Cryptosporidium oocyst and Giardia cyst removal (Patania et al, 1995). Because pumping breaks up floc particles that are not removed by sedimentation, these experiments may represent a worst‐case analysis of pathogen removal in conventional treatment of Lake Mendota water. A detailed description of the treatment plant operating conditions is provided elsewhere (Harrington et al, 2001).
Experiments were conducted to evaluate hydraulic residence time distributions and the influence of settled water pumping on turbidity removal (Harrington et al, 2001). The hydraulic residence time distribution showed that filter effluent samples could be collected no earlier than 5 h after initiation of pathogen feed to the raw water supply. The other tests indicated that settled water pumping had no effect on turbidity removal through filtration provided that the sedimentation basin was cleaned out one day prior to a challenge experiment.
The day before a pathogen challenge run, the rapid‐mix tanks, flocculation chambers, and sedimentation basins were drained, and solids were cleaned out and flushed to the sanitary sewer. The alum dosage for each pilot‐scale test was determined by a series of jar‐test experiments using pathogen‐seeded water approximately 12 h before the pilot‐scale experiment. The selected alum dosage was based on optimal turbidity removal. After the cleaning, the pilot‐scale system was brought back on line without pathogen feed and allowed to run overnight with the selected alum dosage.
On the following day, feeding of the pathogen cocktail was initiated once the pilot plant was determined to be operating in a stable fashion. Table 2 shows average pathogen concentrations in the seeded Lake Mendota water. These concentrations were measured at a location between the two rapid‐mix basins on each train (see yellow circles on Figures 1–3). The cocktail was continuously fed to each pilot train at a flow rate of 5 mL/min from a 20 L (5.3 gal) high‐density polyethylene container. The container was placed in a cooler with ice packs to help maintain the stability of the pathogen cocktail. A mixing system was designed to help maintain a uniform suspension of pathogens within the container. Details of the pathogen feed system and the preparation methods for the stock pathogen suspension are provided elsewhere (Harrington et al, 2001).
Table 2.
Average pathogen concentrations in pathogen‐seeded Lake Mendota water
| Pathogen | Average Concentration |
|---|---|
| Cryptosporidium parvum oocysts | 1×105 oocysts/L |
| Encephalitozoon intestinalis spores | 2×105 spores/L |
| Aeromonas hydrophila | 2×107cfu/L |
| Escherichia coli O157:H7 | 6×107cfu/L |
| Bacteriophage MS2 | 5×107 pfu/L |
Five hours after the pathogen feed was initiated, the filter effluent was presumed to be at steady state with respect to the pathogen feed. During this 5 h period, pathogen samples were collected at a location between the two rapid‐mix tanks (yellow circles on Figures 1–3) to evaluate the stability of the pathogen feed during this part of the run. These samples were collected 15 min, 2 h, and 4 h after initiation of the pathogen feed.
After the 5 h period, all filters were backwashed and simultaneously placed back on line after backwashing was completed. To track turbidity during the ripening phase, filter effluent turbidities were monitored every 5 min for 3 h after the filters were placed back on line. Head loss across each filter was recorded every 30 min during the ripening phase. Once ripening was completed, turbidity and head loss were monitored every 2–7 h until the experiment was terminated.
Methods specific to evaluating filter run time. The pilot plant was used on two separate occasions to evaluate the effect of filter run time on the performance of dual‐media filters operated at 4 gpm/sq ft (2.7 mm/s). A flow chart for these tests is shown in Figure 1. Duplicate filters were run on a single train during each test to characterize experimental repeatability. The dual‐media filters consisted of 18 in. (46 cm) of crushed anthracite over 12 in. (31 cm) of sand. Table 3 lists the effective size and uniformity coefficient of each filter medium.
Table 3.
Characteristics of filter media used in the pilot‐scale experiments
| Filter Medium | Effective Size mm (in.) | Uniformity Coefficient |
|---|---|---|
| Anthracite | 1.0 (0.04) | 1.3 |
| Sand | 0.5 (0.02) | 1.4 |
| Garnet | 0.3 (0.012) | 1.4 |
In these two experiments, feeding of the pathogen cocktail was initiated and the pilot plant was operated for 5 h, at which point steady‐state conditions were assumed. Both filters were then backwashed and placed back into service. Forty‐five minutes after backwashing, pathogen samples were collected from the spiked raw water, the settled water, and both filter effluents to evaluate removal during the ripening stage (see yellow circles on Figure 1). Additional sets of pathogen samples were collected from the same locations at 5, 20, and 35 h after backwashing. The 35 h time was selected because routine operation of the pilot plant showed that terminal head loss was typically achieved at 40±5 h.
Methods specific to evaluating alternative filter loading rates. On two separate occasions, the pilot plant was used to evaluate the effect of alternative loading rates on the performance of ripened granular media filters (Figure 2). The loading rates studied were 2, 4, 6, and 8 gpm/sq ft (1.4, 2.7, 4.1, and 5.4 mm/s). All eight filters were dual‐media with 18 in. (46 cm) of crushed anthracite over 12 in. (31 cm) of sand; see Table 3 for effective size and uniformity coefficient. Duplicate treatment trains were operated during each test to assess experimental repeatability.
Figure 2.
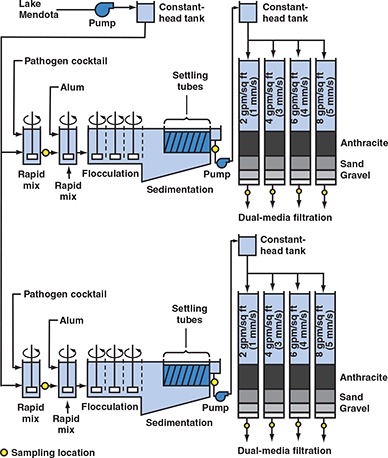
Flow chart for evaluating effect of filtration rate on removal of emerging waterborne pathogens
Once filter effluent turbidities indicated that ripening was complete, samples were collected for pathogen analysis. These samples were collected at all of the locations designated by yellow circles on Figure 2. To demonstrate the stability of the pathogen feed, additional samples were collected from the location between the two rapid‐mix tanks on each train. These samples were collected at 2 h intervals throughout the experiment.
Methods specific to evaluating alternative filter media. Pilot‐scale tests were also performed to evaluate alternative filter media configurations. As shown in Figure 3, these tests compared the performance of monomedium filters, dual‐media filters, and trimedia filters. The monomedium filter contained 30 in. (76 cm) of sand, whereas the dual‐media filter contained 18 in. (46 cm) of anthracite over 12 in. (31 cm) of sand. The trimedia filter contained 3 in. (8 cm) of garnet under 12 in. (31 cm) of sand and 15 in. (38 cm) of anthracite. Table 3 lists the effective size and uniformity coefficients for all of the filter media. All filters were operated at a loading rate of 4 gpm/sq ft (2.7 mm/s).
Once filter effluent turbidities indicated that ripening was complete, samples were collected for pathogen analysis. These samples were collected at all of the locations designated by yellow circles on Figure 3. To demonstrate the stability of the pathogen feed, additional samples were collected from the location between the two rapid‐mix tanks on each train. These samples were collected at 2 h intervals throughout the experiment.
Methods specific to evaluating coagulation pH. This evaluation was combined with the evaluation of alternative filter media (Figure 3). For this experiment, alum coagulation was performed at ambient pH in one treatment train and at a lower pH in the other treatment train. The lower pH was achieved with the addition of sulfuric acid. All other procedures for this test were as previously described.
Control reactions. For each trial, a separate batch reactor containing the microbial cocktail diluted into untreated source water was maintained to monitor any changes in the stability of test microorganisms over the course of each experiment. For each set of samples collected (pretreatment, postsedimentation, and postfiltration), a sample was collected from the control reactor to monitor changes in microbial densities associated with growth or decay over the course of the experiment.
Analytical Methods
Turbidity. Turbidity was measured with a turbidimeter 1 in accordance with method 2130B (Standard Methods, 1996). The turbidimeter was calibrated several times throughout the project with primary standard formazin suspensions (hydrazine sulfate and hexamethylene tetramine) 2 and before every experiment with secondary standards. 3
pH. Sample pH was measured with a glass‐bulb, liquidjunction electrode 4 with manual temperature compensation. The pH meter was calibrated before every experiment with pH 4, 7, and 10 buffers.
Absorbance. Absorbance of ultraviolet light was measured at 254 nm (UV254) with a spectrophotometer 5 in accordance with method 5910B (Standard Methods, 1996).
Enumeration of microorganisms. This section provides a general overview of the methods used for microbial enumeration; methods used for each microbe are detailed elsewhere (Harrington et al, 2001). Relative removals of C. parvum oocysts and E. intestinalis spores were determined by immunolabeling samples with specific antibodies conjugated to fluorescent antibodies and subjecting samples to enumerative analysis by flow cytometry. Samples were supplemented with precise numbers of an internal standard (fluorescent beads), and the absolute numbers of oocysts or spores were then corrected for original sample volume and expressed per millilitre. In cases in which high turbidity or interfering materials rendered flow cytometry analysis ineffective or inadequately sensitive, oocysts and spores were enumerated manually by examining immunolabeled samples under epifluorescence conditions in a fluorescence microscope. 6 Reductions of oocysts and spores were then computed as the log‐transformed ratio of pretreatment and posttreatment microbial densities.
Bacterial concentrations were quantified using a spreadplate technique with selective agar specific for each test microorganism. Serially diluted samples in volumes of 200 μL were inoculated onto selective agar and were distributed uniformly over the agar surface with sterile stainless‐steel rods. For E. coli O157:H7 and A. hydrophila, plastic petri dishes were prepared with 15 mL of MacConkey sorbitol and ampicillin dextrin agar, respectively. Samples were incubated for 22 to 24 h at 35°C and enumerative results were expressed as colony‐forming units per millilitre. To increase the precision of the results and to determine analytical error, plates were prepared in triplicate for each dilution.
Bacteriophage MS2 concentrations were determined according to infectivity assay using the double‐agar‐layer method in 100 mm (3.94 in.) plastic petri dishes. Samples (100 μL) of each sample dilution were added to 5 mL molten top agar, supplemented with 100 μL log‐phase E. coli F‐amp, and plated over nutrient bottom agar containing streptomycin and ampicillin. Samples were incubated at 37°C for 16 to 24 h, with sample dilutions plated in triplicate to characterize analytical error. Virus concentrations were expressed as mean plaque‐forming units per millilitre.
Results
Effect of filter run time on pathogen removal. The alum dose for the experiment described here was set to 50 mg/L; raw water temperatures during the experiment were 14.3±0.3°C. Raw water pH averaged 7.59±0.11 during the experiment, and alum addition lowered the pH to an average of 7.46±0.06 in settled waters. Treatment reduced UV254 by an average of 9%, from 0.074±0.003 cm–1 to 0.067±0.007 cm–1. These standard deviations showed that both pH and UV254 were stable throughout the experiment, suggesting that raw water quality and alum dosage varied to an insignificant degree during the experiment. The variation in pH and UV254 between the two filters was <0.07 pH units and 0.001 cm–1, respectively. Therefore, the chemical quality of the water through filtration was reproducible.
Filter effluent turbidities for the entire experiment are shown in Figure 4, which indicates that turbidity breakthrough was evident after approximately 20 h of filter run time. Although breakthrough appears to have begun at around 16 h of run time, the turbidity values at 20 h of run time were not significantly different from values during the mature phase. From 3 h of filter run time to 20 h of filter run time, filter effluent turbidity averaged 0.16±0.05 ntu. After this period, filter effluent turbidities increased steadily from an average of 0.20 ntu at 20 h to an average of 1.7 ntu at 35 h of filter run time.
Figure 4.
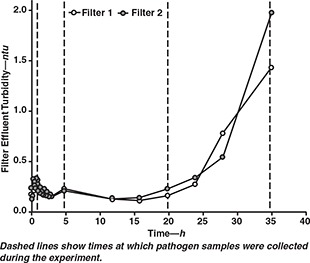
Effect of filter run time on filter effluent turbidity
Figure 4 also shows that the two filters produced similar turbidity results, with filter 1 producing an average turbidity that was 0.02 ntu lower than the average turbidity produced by filter 2. A statistical analysis of the data from the two filters showed that the experimental uncertainty was 0.03 ntu (see Harrington et al, 2001, for details of uncertainty calculations). Therefore, turbidity differences between the two filters were within experimental error and not significant.
Pathogen samples were collected from both filters at all four of the run times shown in Figure 4. Figure 5 shows average pathogen removals for all four of these sampling times. The removals shown in Figure 5 reflect the performance of the entire treatment train from seeded raw water to filtered water. The experimental uncertainty for pathogen removal was ±0.2 log for C. parvum oocysts, adenoviruses, and polioviruses. This uncertainty was ±0.1 log for the remaining microorganisms.
Figure 5.
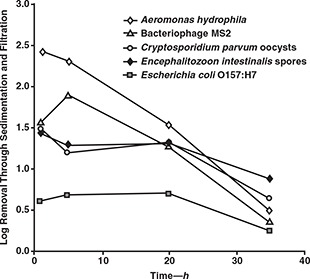
Effect of filter run time on pathogen removal
With the exception of bacteriophage MS2, differences in removal between the 45 min and 5 h sampling times did not exceed the experimental uncertainty for any of the microorganisms. Therefore, pathogen removal during the ripening phase was similar to pathogen removal in the mature filters. Between 5 and 20 h, removals of E. intestinalis spores and C. parvum oocysts remained stable, and this stability was consistent with that observed for filter effluent turbidity. However, removals of A. hydrophila and bacteriophage MS2 declined significantly between these two sample times, suggesting that breakthrough of these microbes occurred earlier than that of turbidity. Beyond 20 h, significant breakthrough was observed for all microorganisms and was consistent with the extensive breakthrough of turbidity. Results showed that E. coli O157:H7 was poorly removed throughout the run, implying that breakthrough of this pathogen took place prior to the 45 min sampling time.
A second experiment with all of the test pathogens also showed early breakthrough of A. hydrophila and E. coli O157:H7. Two additional tests run with only A. hydrophila and E. coli O157:H7 showed that the latter microorganism achieved breakthrough in less than 1 h of filter run time (results for one of these tests are shown in Figure 6).
Figure 6.
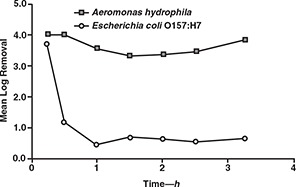
Removal of Aeromonas hydrophila and Escherichia coli O157:H7 during the ripening phase of a filter run
The reasons for the rapid breakthrough of A. hydrophila and E. coli O157:H7 are unclear, but it is possible that this was associated with the size of the bacteria. Filtration theory suggests that 1‐μm particles are the most difficult ones to remove through clean bed, rapid‐rate granular media filters (Yao et al, 1971). This size is comparable to the two bacteria that were studied. However, A. hydrophila was more readily removed than any of the test microbes during the initial 5 h of the filter run. This size also corresponds to that of E. intestinalis spores, which were more poorly removed than most of the other microbes but did not break through as quickly as the two bacteria.
In addition, it is not known whether the bacteria were associated with floc particles when entering the filter. Although many studies have focused on the effects of particle size on particle removal in clean bed filters, little research has been conducted to determine the breakthrough rates of different‐sized particles. Previous research has concluded that particles in the size range of 3 to 7 μm were associated with early breakthrough, but that research did not evaluate particles smaller than 1 μm in size (Moran et al, 1993). It is also possible that surface charge played a role, because measurements showed that A. hydrophila was less negatively charged than either E. coli O157:H7 or E. intestinalis (Harrington et al, 2001). However, the surface charge of particles entering the filter was not measured. Therefore, additional research is needed to evaluate the reasons for the rapid breakthrough of A. hydrophila and E. coli O157:H7.
Effect of filter‐loading rate on pathogen removal. Figure 2 shows the setup for this experiment. As noted previously, pathogen samples were collected after 3 h of filter run time, when the ripening phase was complete. The alum dose was set to 80 mg/L, and raw water temperature during the experiment was 15.5°C. Raw water pH averaged 7.79±0.01 during the experiment, and alum addition lowered the pH to an average of 7.35±0.03 in the settled water. Treatment reduced UV254 by 36% from 0.068 to 0.044 cm–1. The variation in pH and UV254 between the two treatment trains was <0.03 pH units and 0.004 cm–1, respectively. Throughout the experiment, raw and settled water turbidities averaged 3.7±0.2 and 1.7±0.3 ntu, respectively.
Figures 7 and 8 show the filter effluent turbidities and the log removals achieved for the various microorganisms, respectively. These figures indicate that filtration rate had no observable effect on effluent turbidity or pathogen removal across the range from 2 to 8 gpm/sq ft (1.4 to 5.4 mm/s). Although loading rate did not appear to affect effluent quality, higher loading rates were associated with greater head loss.
Figure 7.
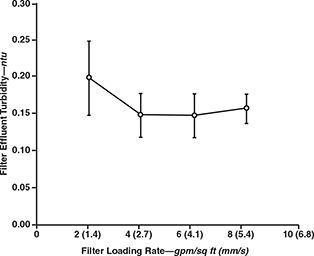
Effect of filter loading rate on filter effluent turbidity
Figure 8.
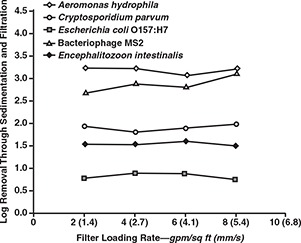
Effect of filter loading rate on pathogen removal
The results were similar to those observed for Cryptosporidium oocysts and Giardia cysts in pilot studies at Contra Costa, Calif., and Seattle, Wash. (Patania et al, 1995). However, the results observed in this study apply to filters that recently completed the ripening phase. Further studies are needed to determine the effects of loading rate on time to pathogen breakthrough.
Effect of alternative filter media on pathogen removal. The experiment described in this section compared monomedium sand filtration, dual‐media anthracite/sand filtration, and trimedia anthracite/sand/garnet filtration (see Figure 3). Raw water temperature and pH were 12 ±0°C and 7.41±0.10, respectively. The alum dose for this experiment was set to 70 mg/L. In one treatment train, alum was fed with sulfuric acid to produce pH 5.72±0.15 in the settled water. In the train having no sulfuric acid addition, alum addition lowered the pH to an average of 6.99±0.06 in the settled water. The treatment train achieved 27% removal of UV254‐absorbing substances, lowering UV254 from 0.063 to 0.046 cm–1. All filters were loaded at 4 gpm/sq ft (2.7 mm/s).
Raw water turbidity averaged 8.6±1.5 ntu through the pilot run. There was no statistically significant difference among the filters for turbidity control. At the time of pathogen sample collection on the pH 7.0 train, filter effluent turbidities varied from 0.10 ntu for the trimedia filter to 0.12 ntu for the dual‐media filter. Turbidities varied from 0.08 ntu for the dual‐media filter to 0.09 ntu for both the sand and trimedia filters on the pH 5.7 train.
Figures 9 and 10 show the pathogen removals achieved at pH 7.0 and pH 5.7, respectively. With the exception of bacteriophage MS2, there was no statistically significant difference among the three filters for microbe removal on the conventional coagulation train. The sand filter was observed to achieve the highest removal of bacteriophage MS2. Similarly, there was no difference among the three filters on the enhanced coagulation train except for removal of C. parvum oocysts. For both exceptions, the sand filter was observed to achieve the highest performance.
Figure 9.
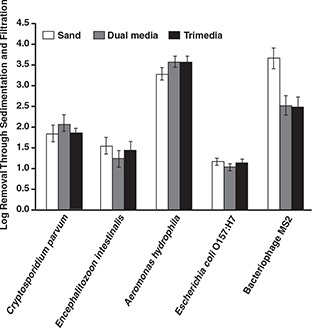
Effect of alternative filter media on pathogen removal at pH 7.0
Figure 10.
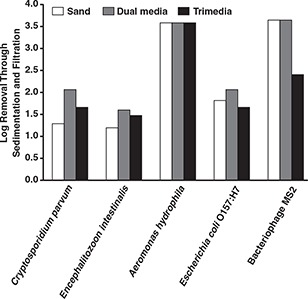
Effect of alternative filter media on pathogen removal at pH 5.7
Effect of coagulation pH on pathogen removal. This experiment was conducted as part of the evaluation of trimedia, dual‐media, and sand filtration. Experimental conditions were described in the previous section. To summarize, an alum dose of 70 mg/L was used for this study, and coagulation was carried out at pH 7.0 in one treatment train and at pH 5.7 in the other treatment train. Figure 3 shows the flow schematic for this experiment.
A small but statistically significant difference was observed between the two pH values for turbidity control. The filters at pH 5.7 achieved effluent turbidities of 0.08 to 0.09 ntu, whereas the filters at pH 7.0 had slightly higher effluent turbidities ranging from 0.11 to 0.12 ntu.
Pathogen removals through sedimentation and dual‐media filtration are shown in Figure 11. As shown in this figure, higher removals of E. coli O157:H7 and bacteriophage MS2 were achieved with a coagulation pH of 5.7.
Figure 11.
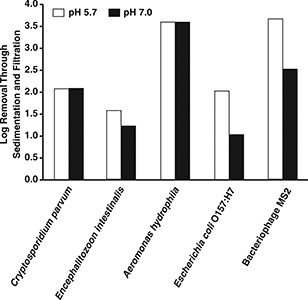
Effect of pH on removal of pathogens through sedimentation and dual‐media filtration
Results suggest a more rapid breakthrough of A. hydrophila and E. coli O157:H7, a finding that signals a public health concern because protection of water consumers from these pathogens may depend solely on disinfection.
In both cases, the difference exceeded 1.0 log and was statistically significant. There was no statistically significant effect of coagulation pH on the other microorganisms. The same conclusions were reached for removals through sedimentation and trimedia filtration.
With monomedium sand filtration, the pH 5.7 treatment train achieved a significantly greater removal of C. parvum oocysts than did the pH 7.0 treatment train (Figure 12). In this case, oocyst removals through sedimentation and sand filtration were 2.6 log for pH 5.7 and 1.8 log for pH 7.0. There was no statistically significant effect of coagulation pH on the removal of the other microorganisms.
Figure 12.
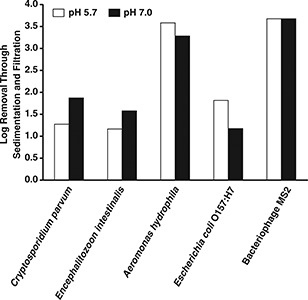
Effect of pH on removal of pathogens through sedimentation and sand filtration
Conclusions
Turbidity breakthrough at the end of a filter run was accompanied by the breakthrough of all pathogens tested in this study. However, the results suggest a more rapid breakthrough of A. hydrophila and E. coli O157:H7. Although the reasons for this are unclear, this finding signals a public health concern because protection of water consumers from these pathogens may depend solely on disinfection. Even if these pathogens are not resistant to disinfection, there may be little protection from these microorganisms in the event of a disinfectant feed system failure.
Filtration rate and alternative filter media configurations had no apparent effect on pathogen removal for the pilot‐scale experiments. It is important to stress that pathogen samples were collected from ripened dual‐media filters. There is a strong likelihood that the onset of pathogen breakthrough would occur at different times for each filtration rate. Additional studies are needed to quantify the effects of filtration rate and filter type on pathogen breakthrough.
In a limited number of cases, a pretreatment and filtration pH of 5.7 achieved better pathogen removal than a pretreatment and filtration pH of 7.0. This enhanced removal was observed for C. parvum oocysts through sand filtration and for E. coli O157:H7 and bacteriophage MS2 through both dual‐media and trimedia filtration. There was no observable effect of pH on any of the other pathogens tested.
Acknowledgment
University of Wisconsin grants were used to purchase pilot‐plant equipment for the pilot‐scale phase of the project. The F.B. Leopold Company (Zelionople, Pa.) provided crushed anthracite filter medium; Best Sand (Chardon, Ohio) and the Northern Gravel Company (Muscatine, Iowa) donated filter sand and support gravel, respectively. The Oak Creek (Wis.) Water and Sewer Utility contributed garnet sand, silica sand, and crushed anthracite for trimedia filtration studies. Installation and startup of the pilot‐plant facilities were achieved with the substantial involvement of Jim Buchholtz, Chris Bone, and Andrew Harris. Andrew Harris and Kristine Hahn provided assistance with the pilot‐scale experiments. David A. Battigelli provided laboratory support and consultation throughout the project; Brad Argue, Rebecca Hoffman, Elizabeth Weisshaar, and David Polchert provided additional laboratory support. Secretarial support was provided by Nanette Kelsey in the University of Wisconsin's Department of Civil and Environmental Engineering. The authors also thank Kathryn Martin, AWWA Research Foundation project manager, and the project advisory committee—Gene Koontz, Rick Sakaji, and Stanley States—for their support and suggestions throughout the project.
Biography
Gregory W. Harrington is an associate professor in the Department of Civil and Environmental Engineering at the University of Wisconsin in Madison. He holds a BS in chemical engineering from Stanford University in Stanford, Calif., and MS and PhD degrees in environmental engineering from the University of North Carolina at Chapel Hill. A recipient of the 1999 CAREER Award from the National Science Foundation and of a 1998 AWWA Publications Award, Harrington has 18 years of experience in drinking water research and professional practice. Irene Xagoraraki is a postdoctoral research associate in the Department of Civil and Environmental Engineering, University of Wisconsin, 1415 Engineering Dr., Madison, WI 53706; e‐mail ixagorar@facstaff.wisc.edu. Prapakorn Assavasilavasukul is a research assistant in the same department. Jon H. Standridge is managing microbiologist at the Wisconsin State Laboratory of Hygiene, University of Wisconsin, Madison.

Footnotes
Model 2100N, Hach Co., Loveland, Colo.
Sigma, St. Louis, Mo.
Gelex, Hach Co., Loveland, Colo.
Fisher Scientific, Pittsburgh, Pa.
Model DR/4000U, Hach Co., Loveland, Colo.
Olympus BH‐X, Olympus America Inc., Melville, N.Y.
References
- Avery, S.W. & Undeen, A.H. , 1987. Isolation of Microsporidia and Other Pathogens From Concentrated Ditch Water. Jour. Amer. Mosquito Control Assoc., 3:1:54. [PubMed] [Google Scholar]
- CDC (Centers for Disease Control) , 1993. Preventing Foodborne Illness: Escherichia coli O157:H7. Centers for Disease Control and Prevention, Atlanta. [Google Scholar]
- CDC , 1991a. Waterborne‐Disease Outbreaks, 1989–1990. Morbidity & Mortality Monthly Rept., 40:1. [PubMed] [Google Scholar]
- CDC , 1991b. Outbreaks of Diarrheal Illness Associated With Cyanobacteria (Blue–Green Algae)‐Like Bodies— Chicago and Nepal. Morbidity & Mortality Monthly Rept., 40:325. [PubMed] [Google Scholar]
- Clark, J.A. ; Burger, G.A. ; & Sabatinos, L.E. , 1982. Characterization of Indicator Bacteria in Municipal Raw Water, Drinking Water, and New Main Water Samples. Can. Jour. Microbiol., 28:1002. [DOI] [PubMed] [Google Scholar]
- Garcia, L.S. & Bruckner, D.A. , 1997. (3rd ed.). Diagnostic Medical Parasitology. ASM Press, Washington. [Google Scholar]
- Harrington, G.W. et al, 2001. Removal of Emerging Waterborne Pathogens. AWWA Res. Fdn., Denver. [Google Scholar]
- Institute of Medicine , 1992. Emerging Infections: Microbial Threats to Health in the United States. Natl. Acad. Press, Washington. [PubMed] [Google Scholar]
- Kaplan, J.E. et al, 1982a. Frequency of a Norwalk‐like Pattern of Illness in Outbreaks of Acute Gastroenteritis. Amer. Jour. Public Health, 72:12:1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, J.E. et al, 1982b. Epidemiology of Norwalk Gastroenteritis and the Role of Norwalk Virus in Outbreaks of Acute Nonbacterial Gastroenteritis. Annals Intern. Med., 96:1:756. [DOI] [PubMed] [Google Scholar]
- Moe, C.L. , 1997. Waterborne Transmission of Infectious Agents. Manual of Environmental Microbiology (Hurst C.J. et al, editors). ASM Press, Washington. [Google Scholar]
- Moran, D.C. et al, 1993. Particle Behavior in Deep‐bed Filtration: Part 1—Ripening and Breakthrough. Jour. AWWA, 85:12:69. [Google Scholar]
- Mullens, A. , 1996. “I Think We Have a Problem in Victoria”: MDs Respond Quickly to Toxoplasmosis Outbreak in BC. Can. Med. Assn. Jour., 154:11:1721. [PMC free article] [PubMed] [Google Scholar]
- Patania, N.L. et al, 1995. Optimization of Filtration for Cyst Removal. AWWA Res. Fdn., Denver. [Google Scholar]
- Standard Methods for the Examination of Water and Wastewater, 1996. (19th ed.). APHA, AWWA, and WEF, Washington. [Google Scholar]
- USEPA (US Environmental Protection Agency) , 2002. 40 CFR Parts 9, 141, and 142; National Primary Drinking Water Regulations. Long‐term 1 Enhanced Surface Water Treatment; Final Rule. Fed. Reg., 67:9:1811. [PubMed] [Google Scholar]
- USEPA , 2000. Stage 2 Microbial and Disinfection By‐products Federal Advisory Committee Agreement in Principle. Fed. Reg., 65:251:83015. [Google Scholar]
- USEPA , 1998. 40 CFR Parts 9, 141, and 142; National Primary Drinking Water Regulations. Interim Enhanced Surface Water Treatment; Final Rule. Fed. Reg., 63:241:69478. [Google Scholar]
- USEPA , 1994. 40 CFR Parts 141 and 142; National Primary Drinking Water Regulations. Enhanced Surface Water Treatment Requirements; Proposed Rule. Fed. Reg., 59:145:38832. [Google Scholar]
- USEPA , 1989. 40 CFR Parts 141 and 142; National Primary Drinking Water Regulations. Filtration, Disinfection, Turbidity, Giardia lamblia, Viruses, Legionella, and Heterotrophic Bacteria; Final Rule. Fed. Reg., 54:124:27486. [Google Scholar]
- Yao, K.M. ; Habibian, M.T. ; & O'Melia, C.R. , 1971. Water and Waste Water Filtration: Concepts and Applications. Envir. Sci. & Technol., 5:11:1105. [Google Scholar]


