Abstract
Background and objective
Noroviruses have been recognised as a significant cause of neonatal enteritis in calves in many countries, but there has been no investigation of their occurrence in Australian cattle. This study aimed to establish whether bovine noroviruses could be detected in faecal samples from Australian dairy cattle. It also sought to determine whether bovine coronaviruses, also associated with neonatal enteritis in calves, could be detected in the same faecal samples.
Methods
A selection of faecal samples that were negative for rotaviruses from dairy farms located in three geographically distinct regions of Victoria were pooled and tested by reverse transcription‐PCR for the presence of noroviruses (genogroup III), neboviruses and bovine coronaviruses.
Results and conclusion
Genetically distinct genogroup III noroviruses were detected in two sample pools from different geographic regions and bovine coronavirus was detected in a third pool of samples. This is the first report of bovine norovirus infection in Australian cattle and suggests that future work is required to determine the significance of these agents as a cause of bovine enteric disease in Australia.
Keywords: bovine coronavirus, bovine norovirus, Caliciviridae, Coronaviridae, cattle, diarrhoea, enteritis
Abbreviations
- GIII.1
genogroup III, genotype 1
- ORF
open reading frame
- RT
reverse transcription
- VLP
virus‐like particle
Studies of viral causes of neonatal enteritis in Australia have focussed on rotaviruses, but several other enteric viruses, including caliciviruses and bovine coronavirus, have been identified in cattle in other countries. Bovine caliciviruses were first detected in the 1970s and 1980s in faeces from diarrhoeic calves in the UK1, 2 and Germany.3 However, it was not until later that these viruses were confirmed to belong to the family Calicivirdae, based on their genomic sequences, and were found to belong to two separate genera, Norovirus (within genogroup III)4, 5 and Nebovirus. 6 The genogroup III noroviruses were further divided into genotypes,7 with this division based on phylogenetic relatedness. The prototype strain for bovine genogroup III genotype 1 noroviruses is Jena virus, (GIII/Bo/DE/1980/GIII.1/Jena) and for bovine genogroup III genotype 2 noroviruses the prototype strain is Newbury agent‐2 (GIII/Bo/UK/1976/GIII.2/Newbury2). In humans, noroviruses are one of the most important aetiological agents of gastroenteritis,8 but our understanding of their significance as a cause of diarrhoea in cattle is limited.
Since their initial detection, a number of studies have used reverse transcription (RT)‐PCR‐based molecular detection methods to ascertain the presence and prevalence of these agents in cattle in a limited number of countries. In addition to the UK and Germany, bovine noroviruses have been detected in the Netherlands,9, 10 the USA,11, 12 New Zealand,13 South Korea,14 Norway,15 France,16 Turkey17 and Tunisia18 and neboviruses have been detected in South Korea,19 France,16 Tunisia18 and the USA.12
Bovine coronaviruses are also associated with diarrhoea in cattle. These viruses belong to the order Nidovirales, family Coronaviridae, subfamily Coronavirinae, genus Betacoronavirus and are enveloped, positive sense RNA viruses.20 In Australia, bovine coronaviruses have been detected in association with diarrhoea21 and respiratory disease in cattle.22
As no bovine caliciviruses from either genus have been detected in Australia, the aim of this study was to search for Australian bovine noroviruses and neboviruses in faeces from diarrhoeic calves and, if they were detected, to compare them with strains characterised in other countries.
Materials and methods
In 2006, faecal samples were collected from calves with diarrhoea on dairy farms in three Victorian regions: South Gippsland, Northern Victoria and the Western District. Table 1 summarises the farms sampled in each of these regions. Faecal samples were classified based on the severity of diarrhoea, on a scale from 1 to 3. For this study, samples that were scored as most severe (3) but were negative for rotavirus, based on polyacrylamide gel electrophoresis of phenol/chloroform extracted RNA (data not shown), were selected for analysis. These samples were pooled by region, as detailed in Table 2.
Table 1.
Faecal samples collected in 2006 from calves with diarrhoea on dairy farms in three regions of Victoria, Australia: South Gippsland, Northern Victoria and the Western District
| Region | Farm | Herd size | Samples | Rotavirus positive | Date of collection |
|---|---|---|---|---|---|
| South Gippsland | 1 | 130 | 3 | 0 | 27/07/2006 |
| 1a | − | 3 | 0 | 27/07/2006 | |
| 2 | − | 18 | 1 | 27/07/2006 | |
| 3 | − | 15 | 1 | 27/07/2006 | |
| 4 | − | 9 | 2 | 27/07/2006 | |
| 5 | − | 12 | 0 | 27/07/2006 | |
| 6 | − | 13 | 1 | 27/07/2006 | |
| Northern Victoria | A | − | 18 | 0 | 24/08/2006 |
| B | − | 14 | 0 | 24/08/2006 | |
| C | − | 9 | 0 | 24/08/2006 | |
| D | − | 27 | 0 | 24/08/2006 | |
| E | − | 16 | 1 | 24/08/2006 | |
| F | − | 18 | 0 | 24/08/2006 | |
| G | − | 10 | 0 | 24/08/2006 | |
| P | − | 9 | 3 | 24/08/2006 | |
| Q | − | 19 | 0 | 24/08/2006 | |
| X | − | 12 | 0 | 24/08/2006 | |
| Western District | A | 240 | 16 | 0 | 26/06/2006 |
| B | 300 | 4 | 0 | 26/06/2006 | |
| C | 350 | 7 | 0 | 26/06/2006 | |
| D | 500 | 21 | 1 | 26/06/2006 | |
| E | 400 | 18 | 5 | 26/06/2006 | |
| F | 400 | 16 | 1 | 26/06/2006 | |
| G | 200 | 10 | 6 | 26/06/2006 | |
| H | 600 | 13 | 1 | 26/06/2006 | |
| L | − | 1 | 1 | 26/06/2006 |
Table 2.
Pooled faecal samples from dairy farms across three regions of Victoria, Australia, with a diarrhoeal severity score of 3, excluding rotavirus positive samples
| Region | Pool | Farm (no. of samples) |
|---|---|---|
| South Gippsland | SG2 | 2 (5) |
| SG3 | 3 (4) | |
| SG5 | 5 (7) | |
| SG6 | 6 (4) | |
| Northern Victoria | NVD | D (7) |
| NVF | F (7) | |
| NV‐Mix | B (2), C (1), G (2), P (1), Q (1), X (1) | |
| Western District | WD‐Mix | B (2), D (1), F (1), G (1), H (1) |
Pooled faecal samples were diluted 1 in 5 and homogenised in phosphate‐buffered saline. Samples were centrifuged at 2700g for 10 min to remove larger particulate matter before the nucleic acid was extracted with the QIAamp viral RNA mini kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. For RT, 5 μL of the extracted nucleic acid was mixed with 100 ng of random oligonucleotide hexamers and incubated at 80°C for 5 min before being placed on ice. After the addition of 1 × first strand buffer (Life Technologies, Carlsbad, CA, USA), 1.5 mmol/L dNTP, 10 mmol/L DTT, 20 U RNaseOUT (Life Technologies) and 100 U SuperScript III (Life Technologies), reactions were incubated at 50°C for 50 min, then at 70°C for 15 min.
PCR reactions used the following primer sets: CBECU‐F/R,23 designed to hybridise with the conserved YGDD polymerase motif and the open reading frame (ORF)1/2 junction of bovine noroviruses (genogroup III); NBU‐F/R,23 designed to hybridise to neboviruses in the same region as CBECU‐F/R; p289/p290,24 designed to hybridise to the polymerase region of the genome of human caliciviruses, but believed to be broadly reactive; and BCoV‐fwd and BCoV‐rev,25 designed to target the gene encoding the nucleocapsid protein of the Nebraska strain of bovine coronavirus, with modification of the forward primer (BCoV‐fwd alt: 5′ CTAACAAGCAGGCTGATGTTAATACC) that allowed detection of equine coronavirus. A positive control was not available for the bovine caliciviruses, but a coronavirus positive control was included (an 87‐bp BCoV‐fwd alt/BCoV‐rev equine coronavirus amplicon cloned into pGEM‐T; Promega, Madison, WI, USA). Negative controls (sterile water) were included in all reactions. Reactions for the calicivirus assays included 1 × GoTaq Flexi buffer (Promega), 2.5 mmol/L MgCl2, 0.2 mmol/L dNTPs, 0.4 μmol/L of each of the forward and reverse primers and 1 U GoTaq polymerase (Promega). Reactions for the coronavirus assays were identical except that 2.0 mmol/L of MgCl2 was used. The same incubation conditions were used for all reactions: 1 cycle at 94°C for 1 min; 30 cycles of 94°C for 30 s, 45°C for 30 s and 68°C for 40 s; and 1 cycle of 68°C for 7 min. For PCR product visualisation, 5 μL of each reaction was electrophoresed through a 2% (w/v) agarose gel containing SYBR Safe DNA gel stain (Invitrogen, Carlsbad, CA, USA) in 0.5 × TBE buffer (1 × TBE is 89 mmol/L Tris, 89 mmol/L boric acid, 2 mmol/L EDTA, pH 8.3) and an image of the gel was captured with a Molecular Imager ChemiDoc XRS+ imaging system (Bio Rad, Hercules, CA, USA) using transillumination with ultraviolet light.
Amplified DNA from the RT‐PCR assay identified as positive on the gel was purified using QIAquick Gel Extraction kits (Qiagen) prior to sequencing using the BigDye version 3.1 cycle sequencing kit (Life Technologies). Bioinformatic analyses of the resulting sequences were performed using the program Geneious (Biomatters: http://www.geneious.com).
Results
A product of the expected size was amplified from the pooled samples SG5 and WD‐mix with CBECU‐F/R primer set, which targeted the genogroup III noroviruses, and from the SG6 sample with the BCoV‐fwd alt/BCoV‐rev primer set (Table 3). DNA sequencing was used to confirm the similarity of the products with the sequences of genogroup III noroviruses and the BCoVs, respectively.
Table 3.
Amplification of enteric viruses from pooled faecal samples from dairy farms across three regions of Victoria, Australia
| Region | Pool | Primer set (expected size) | |||
|---|---|---|---|---|---|
| CBECU‐F/R (532 bp) | NBU‐F/R (549 bp) | p289/p290 (~319–331 bp) | BCoV‐F/R (87 bp) | ||
| South Gippsland | SG2 | – | – | – | – |
| SG3 | – | – | – | – | |
| SG5 | + | – | – | – | |
| SG6 | – | – | – | + | |
| Northern Victoria | NVD | – | – | – | – |
| NVF | – | – | – | – | |
| NV‐mix | – | – | – | – | |
| Western District | WD‐mix | + | – | – | – |
bp, base pairs.
A comparison of the 271 nucleotides (equivalent to nucleotides 4777–5047 of GIII/Bo/UK/1976/GIII.2/Newbury2, AF097917) for which high‐quality sequence obtained from the norovirus products amplified from the SG5 and WD‐mix pools detected 11 nucleotide differences (Figure 1) and resulted in a nucleotide sequence identity of 95.9%. None of the nucleotide changes in this 271 nucleotide region equated to amino acid differences between SG5 and WD‐mix. However, SG5 and WD‐mix showed a single amino acid difference from Newbury2 and 11 amino acid differences from Jena across the 90 deduced amino acids. A search of GenBank using BLAST revealed that the virus with the highest nucleotide sequence identity with the SG5 product was GIII/Bo/NOR/2006/GIII.2/340_1235 (FM242185), with 94.8% nucleotide identity, and the most similar viruses to that detected in the WD‐mix were GIII/Bo/NOR/2006/GIII.2/216_0114 (FM242188) and GIII/Bo/NOR/2006/GIII.2/340_1235 (FM242185), both with 95.2% nucleotide sequence identity. These strains were detected in Norway in 2006 and belong to genotype GIII.2, with the prototype strain being GIII/Bo/UK/1976/GIII.2/Newbury2.
Figure 1.
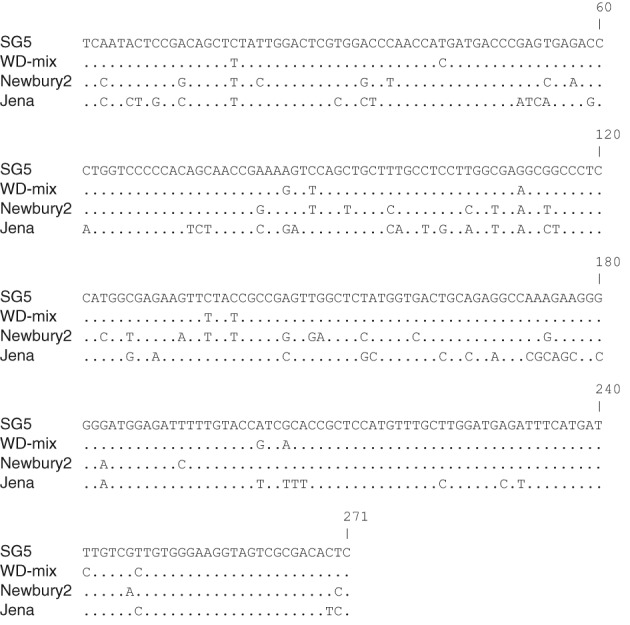
Nucleotide alignment between bovine norovirus sequences from Jena virus (GIII/Bo/DE/1980/GIII.1/Jena, GenBank accession AJ011099), Newbury agent‐2 (GIII/Bo/UK/1976/GIII.2/Newbury2, GenBank accession AF097917) and pools SG5 and WD‐mix. Dots represent identical residues when compared with SG5 as a reference. Alignment spans the nucleotides equivalent to 4777–5047 of Bo/Newbury2/1976/UK.
Genotyping into norovirus GIII.2 was confirmed by phylogenetic analysis with high bootstrap support (Figure 2), with the two Australian viruses grouping most closely with each other and the Norwegian viruses.
Figure 2.
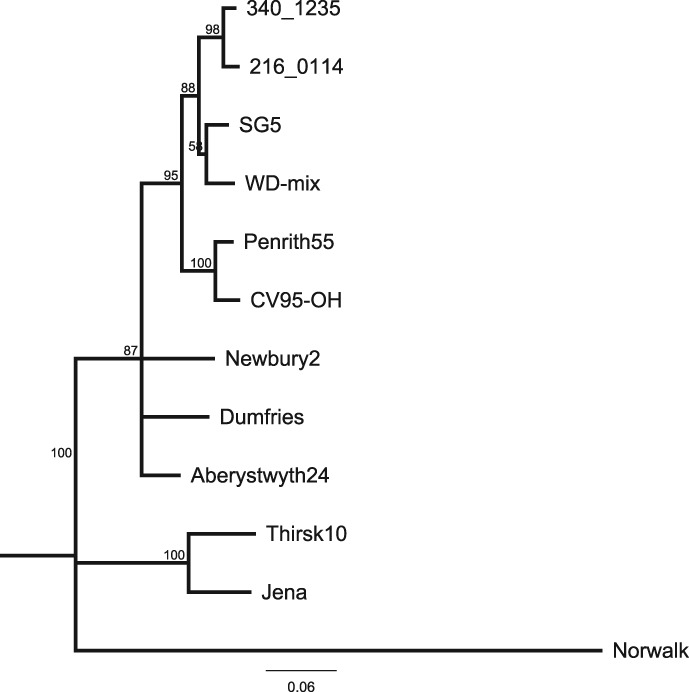
Neighbour joining phylogenetic tree with 1000 bootstrap replicates showing the relationship of two Australian GIII noroviruses to other bovine noroviruses. The human GI.1 Norwalk (GI/Hu/USA/1968/GI.1/Norwalk, M87661) virus was used as an outgroup. Reference GIII bovine noroviruses include the GIII.1 viruses: Jena (GIII/Bo/DE/1980/GIII.1/Jena, AJ011099) and Thirsk10 (GIII/Bo/UK/2000/GIII.1/Thirsk10, AY126468); and the GIII.2 viruses: Newbury2 (GIII/Bo/UK/1976/GIII.2/Newbury2, AF097917), Dumfries (GIII/Bo/UK/1994/GIII.2/Dumfries, AY126474), Penrith55 (GIII/Bo/UK/2000/GIII.2/Penrith55, AY126476), CV95‐OH (GIII/Bo/USA/2002/GIII.2/CV95‐OH, AF542083), Aberystwyth24 (GIII/Bo/UK/2000/GIII.2/Aberystwyth24, AY126475), 340_1235 (GIII/Bo/NOR/2006/GIII.2/340_1235, FM242185) and 216_0114 (GIII/Bo/NOR/2006/GIII.2/216_0114, FM242188). The tree is based on the alignment of 271 base pairs from the polymerase region of the genome. Bootstrap values are expressed as percentages and are shown at the branch points. Scale bar represents substitutions per site.
Discussion
The prevalence of bovine noroviruses has been determined in several countries, but the data are not always comparable because the different studies have had different designs. These differences include the detection method and the primer pairs used, the samples collected (diarrhoeal, non‐diarrhoeal or both), pooled or individual samples and inclusion or exclusion of samples in which other pathogens have been detected. Reported detection rates for the bovine noroviruses range from as low as 8.6% of diarrhoeic faecal samples in Turkey17 and 9.3% of faecal samples from a study in South Korea14 to as high as 49.6% in a study from Norway15 and 53.6% in a study from New Zealand.13 In the current study, bovine noroviruses were detected in two of the eight pooled samples from two different geographical regions.
Because of the pooled nature of the samples tested, it was not possible to determine the prevalence of these viruses. However, given that the bovine noroviruses were detected in two geographically distinct regions of Victoria (South Gippsland and the Western District) and that the sequences obtained were different from each other at the nucleotide level, suggests that the GIII bovine noroviruses may be common in Australian cattle.
Both noroviruses detected in this study were from the GIII.2 genotype. This is consistent with the observation that in more recent years the GIII.2 genotype has predominated over the GIII.1 genotype,14, 15, 16, 17, 18 although in a New Zealand study only GIII.1 bovine noroviruses were detected.13
As a primary focus of this study was to detect and describe bovine caliciviruses in Australian cattle for the first time, faecal samples were pooled. However, to reduce the dilution effect of pooling too many samples, some samples were excluded from the study. Samples were excluded that had previously tested positive for rotaviruses or that scored less than 3 on the diarrhoea severity score. The decision to assay the more clinically severe diarrhoeal samples was supported by the observation that bovine noroviruses have been found to be more commonly associated with watery faeces.12 Although samples testing positive for rotavirus were excluded from the pools in our study, mixed infection with bovine noroviruses and bovine rotaviruses have been described previously,12 so these results likely underestimate the frequency of bovine noroviruses in our sample set.
In addition, there are a number of ways in which the detection of viruses in these samples could have been increased. These include assaying individual samples (rather than pooled samples), additional primer sets (to encompass more of the genetic diversity of noroviruses) and increasing the number of cycles in the PCR screening step. The faecal samples in this study were collected in 2006 and stored at −70°C. The viral extractions were performed 7 years later, in 2013. The length of time spent in storage may have affected the integrity of the viral RNA in the samples, which could explain why only short sequences were recovered.
The bovine norovirus sequences in this study were obtained from the relatively highly conserved polymerase‐encoding region of the genome, as the conservation of this area makes the initial amplification more likely. However, in the future it would be beneficial to also determine the sequence of the capsid‐encoding region, because this region provides important information about the phylogenetic grouping of these viruses. The capsid sequence could also be used to establish whether the isolates in this study had undergone recombination, which in the caliciviruses most commonly occurs between the polymerase and capsid‐encoding regions of the genome.26
To date, there have been no reports of bovine noroviruses associated with disease in humans. However, there has been speculation about the possibility of zoonotic transmission.27, 28, 29 Veterinarians in the Netherlands, particularly those with exposure to cattle, were more frequently found to have IgG antibodies against recombinant bovine norovirus virus‐like particles (VLP) than the general population, who are less likely to come into contact with cattle.27 In addition, a study looking at the acquisition of antibodies to different norovirus genogroups in children in India found antibodies against bovine noroviruses, using recombinant VLPs.30 However, cross‐reactivity could not be ruled out in these studies. In fact, cross‐reactive epitopes have been found in bovine and human norovirus capsids.31, 32
Given that bovine noroviruses have not been reported in humans and human noroviruses have only been detected very infrequently in cattle,33 if zoonotic transmission is possible, it is likely to be an uncommon occurrence. Recombination, including between genogroups, has been reported for noroviruses and other caliciviruses.34, 35, 36, 37, 38 Norovirus recombination most commonly occurs at the junction of the first two ORFs.26, 39, 40 This region of the genome is highly conserved and it is thought that the RNA secondary structure in this region is involved in the recombination process.39 As the majority of noroviruses cannot be cultured, the precise mechanisms of recombination have not been extensively studied and it is not known how much conservation of the RNA sequence or secondary structure conservation is required to allow two strains to recombine.
Although the primary aim of this study was to investigate bovine noroviruses and neboviruses in Australian samples of calf diarrhoea, as neither of these agents has been detected in this country, a secondary aim was to investigate other viral agents of diarrhoea. The samples were previously screened for rotavirus and only samples that were negative for rotavirus were included in further screening. Bovine coronaviruses, which are known to be associated with diarrhoea in Australian calves,21 were also investigated. Although one pooled sample was found to be positive for bovine coronavirus by RT‐PCR and confirmed by sequencing, the length of the amplicon was insufficient to provide any meaningful sequence comparison with other bovine coronavirus sequences. However, it does support the association of this virus with diarrhoea in Australian cattle and suggests it is a potential aetiological agent. Further sequence information from this virus in the future would be informative.
This study is the first report of bovine noroviruses in Australia. Bovine norovirus was detected in faecal samples from two geographically distinct regions and sequence analysis found that these viruses both clustered with the GIII.2 bovine noroviruses. Although this report establishes that bovine noroviruses are present in Australian dairy calves, further investigations are required to fully understand the role of bovine norovirus in diarrhoeal disease in Australian cattle.
Conflicts of interest and sources of funding
The authors declare no conflicts of interest for the work presented here.
Funding for this study was provided by a Special Virology Fund.
Acknowledgment
We thank Cynthia Brown, Faculty of Veterinary and Agricultural Sciences, The University of Melbourne, for excellent technical assistance.
References
- 1. Bridger JC, Hall GA, Brown JF. Characterization of a calici‐like virus (Newbury agent) found in association with astrovirus in bovine diarrhea. Infect Immun 1984;43:133–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Woode GN, Bridger JC. Isolation of small viruses resembling astroviruses and caliciviruses from acute enteritis of calves. J Med Microbiol 1978;11:441–452. [DOI] [PubMed] [Google Scholar]
- 3. Gunther H, Otto P, Heilmann P. [Diarrhea in young calves. 6. Determination of the pathogenicity of a bovine coronavirus and an unidentified icosahedral virus]. Arch Exp Veterinarmed 1984;38:781–792. [PubMed] [Google Scholar]
- 4. Dastjerdi AM, Green J, Gallimore CI et al. The bovine Newbury agent‐2 is genetically more closely related to human SRSVs than to animal caliciviruses. Virology 1999;254:1–5. [DOI] [PubMed] [Google Scholar]
- 5. Liu BL, Lambden PR, Gunther H et al. Molecular characterization of a bovine enteric calicivirus: relationship to the Norwalk‐like viruses. J Virol 1999;73:819–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oliver SL, Asobayire E, Dastjerdi AM et al. Genomic characterization of the unclassified bovine enteric virus Newbury agent‐1 (Newbury1) endorses a new genus in the family Caliciviridae. Virology 2006;350:240–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clarke IN, Estes MK, Green KY et al. Caliciviridae In: King AMQ, Adams MJ, Carstens EB. et al., editors. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, 2011. [Google Scholar]
- 8. Green KY. Caliciviridae: the noroviruses In: Knipe DM, Howley PM, Griffin DE, et al., editors. Field’s virology. 5th edn Lippincott Williams & Wilkins, 2007. [Google Scholar]
- 9. van der Poel WH, Vinje J, van der Heide R et al. Norwalk‐like calicivirus genes in farm animals. Emerg Infect Dis 2000;6:36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van der Poel WH, van der Heide R, Verschoor F et al. Epidemiology of Norwalk‐like virus infections in cattle in the Netherlands. Vet Microbiol 2003;92:297–309. [DOI] [PubMed] [Google Scholar]
- 11. Wise AG, Monroe SS, Hanson LE et al. Molecular characterization of noroviruses detected in diarrheic stools of Michigan and Wisconsin dairy calves: circulation of two distinct subgroups. Virus Res 2004;100:165–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cho YI, Han JI, Wang C et al. Case–control study of microbiological etiology associated with calf diarrhea. Vet Microbiol 2013;166:375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wolf S, Williamson WM, Hewitt J et al. Sensitive multiplex real‐time reverse transcription‐PCR assay for the detection of human and animal noroviruses in clinical and environmental samples. Appl Environ Microbiol 2007;73:5464–5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Park SI, Jeong C, Kim HH et al. Molecular epidemiology of bovine noroviruses in South Korea. Vet Microbiol 2007;124:125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jor E, Myrmel M, Jonassen CM. SYBR Green based real‐time RT‐PCR assay for detection and genotype prediction of bovine noroviruses and assessment of clinical significance in Norway. J Virol Methods 2010;169:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kaplon J, Guenau E, Asdrubal P et al. Possible novel nebovirus genotype in cattle, France. Emerg Infect Dis 2011;17:1120–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yilmaz H, Turan N, Altan E et al. First report on the phylogeny of bovine norovirus in Turkey. Arch Virol 2011;156:143–147. [DOI] [PubMed] [Google Scholar]
- 18. Hassine‐Zaafrane M, Kaplon J, Sdiri‐Loulizi K et al. Molecular prevalence of bovine noroviruses and neboviruses detected in central‐eastern Tunisia. Arch Virol 2012;157:1599–1604. [DOI] [PubMed] [Google Scholar]
- 19. Park SI, Jeong C, Park SJ et al. Molecular detection and characterization of unclassified bovine enteric caliciviruses in South Korea. Vet Microbiol 2008;130:371–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. de Groot RJ, Cowley JA, Enjuanes L et al. Nidovirales In: King AMQ, Adams MJ, Carstens EB. et al., editors. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, 2011. [Google Scholar]
- 21. Izzo MM, Kirkland PD, Gu X et al. Comparison of three diagnostic techniques for detection of rotavirus and coronavirus in calf faeces in Australia. Aust Vet J 2012;90:122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hick PM, Read AJ, Lugton I et al. Coronavirus infection in intensively managed cattle with respiratory disease. Aust Vet J 2012;90:381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Smiley JR, Hoet AE, Traven M et al. Reverse transcription‐PCR assays for detection of bovine enteric caliciviruses (BEC) and analysis of the genetic relationships among BEC and human caliciviruses. J Clin Microbiol 2003;41:3089–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jiang X, Huang PW, Zhong WM et al. Design and evaluation of a primer pair that detects both Norwalk‐ and Sapporo‐like caliciviruses by RT‐PCR. J Virol Methods 1999;83:145–154. [DOI] [PubMed] [Google Scholar]
- 25. Cho YI, Kim WI, Liu S et al. Development of a panel of multiplex real‐time polymerase chain reaction assays for simultaneous detection of major agents causing calf diarrhea in feces. J Vet Diagn Invest 2010;22:509–517. [DOI] [PubMed] [Google Scholar]
- 26. Symes SJ, Job N, Ficorilli N et al. Novel assay to quantify recombination in a calicivirus. Vet Microbiol 2015;177:25–31. [DOI] [PubMed] [Google Scholar]
- 27. Widdowson MA, Rockx B, Schepp R et al. Detection of serum antibodies to bovine norovirus in veterinarians and the general population in the Netherlands. J Med Virol 2005;76:119–128. [DOI] [PubMed] [Google Scholar]
- 28. Palmer S, Brown D, Morgan D. Early qualitative risk assessment of the emerging zoonotic potential of animal diseases. BMJ 2005;331:1256–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oliver SL, Dastjerdi AM, Wong S et al. Molecular characterization of bovine enteric caliciviruses: a distinct third genogroup of noroviruses (Norwalk‐like viruses) unlikely to be of risk to humans. J Virol 2003;77:2789–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Menon VK, George S, Shanti AA et al. Exposure to human and bovine noroviruses in a birth cohort in southern India from 2002 to 2006. J Clin Microbiol 2013;51:2391–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Batten CA, Clarke IN, Kempster SL et al. Characterization of a cross‐reactive linear epitope in human genogroup I and bovine genogroup III norovirus capsid proteins. Virology 2006;356:179–187. [DOI] [PubMed] [Google Scholar]
- 32. Oliver SL, Batten CA, Deng Y et al. Genotype 1 and genotype 2 bovine noroviruses are antigenically distinct but share a cross‐reactive epitope with human noroviruses. J Clin Microbiol 2006;44:992–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mattison K, Shukla A, Cook A et al. Human noroviruses in swine and cattle. Emerg Infect Dis 2007;13:1184–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hansman GS, Takeda N, Oka T et al. Intergenogroup recombination in sapoviruses. Emerg Infect Dis 2005;11:1916–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nayak MK, Balasubramanian G, Sahoo GC et al. Detection of a novel intergenogroup recombinant Norovirus from Kolkata, India. Virology 2008;377:117–123. [DOI] [PubMed] [Google Scholar]
- 36. Phan TG, Kaneshi K, Ueda Y et al. Genetic heterogeneity, evolution, and recombination in noroviruses. J Med Virol 2007;79:1388–1400. [DOI] [PubMed] [Google Scholar]
- 37. Jiang X, Espul C, Zhong WM et al. Characterization of a novel human calicivirus that may be a naturally occurring recombinant. Arch Virol 1999;144:2377–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Oliver SL, Brown DW, Green J et al. A chimeric bovine enteric calicivirus: evidence for genomic recombination in genogroup III of the norovirus genus of the Caliciviridae . Virology 2004;326:231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bull RA, Hansman GS, Clancy LE et al. Norovirus recombination in ORF1/ORF2 overlap. Emerg Infect Dis 2005;11:1079–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Symes SJ, Gunesekere IC, Marshall JA et al. Norovirus mixed infection in an oyster‐associated outbreak: an opportunity for recombination. Arch Virol 2007;152:1075–1086. [DOI] [PubMed] [Google Scholar]


