Abstract
Background
A detailed laboratory investigation identified bovine coronavirus (BCoV) as the aetiological agent in an outbreak of respiratory disease at a semi‐intensive beef cattle feedlot in south‐east Australia. The outbreak caused 30% morbidity in the resident population and also affected two cohorts of cattle that were newly introduced to the property.
Methods
At slaughter, pulmonary consolidation and inflammatory lesions in the trachea were identified in 15 of 49 animals. Pasteurella multocida or Histophilus somni was cultured from 3 of 7 animals with lesions. Histopathological examination revealed multifocal non‐suppurative bronchointerstitial pneumonia with formation of epithelial syncytial cells, sometimes associated with suppurative bronchopneumonia.
Results
BCoV was detected in nasal swabs and pulmonary lesions using real‐time reverse transcriptase–polymerase chain reaction (qRT‐PCR) assay and virus isolation. There was serological evidence of previous exposure to bovine viral diarrhoea virus, bovine respiratory syncytial virus and bovine parainfluenza virus type 3, but not to bovine herpesvirus type 1. None of these viral pathogens or Mycoplasma bovis was identified by qRT‐PCR.
Conclusion
This is believed to be the first report of BCoV in association with bovine respiratory disease complex in Australia.
Keywords: bovine coronavirus, bovine respiratory disease complex, cattle, feedlots
Abbreviations
- AGID
agar gel immunodiffusion
- BCoV
bovine coronavirus
- BHV‐1
bovine herpesvirus type 1
- BRSV
bovine respiratory syncytial virus
- BVDV
bovine viral diarrhoea virus
- BALT
bronchiole‐associated lymphoid tissue
- BPIV‐3
bovine parainfluenza virus type 3
- Ct
cycle‐threshold
- FBS
fetal bovine serum
- PBGS
phosphate‐buffered gelatin saline
The bovine respiratory disease complex is a significant cause of morbidity, mortality and production losses in cattle, particularly in feedlot situations.1, 2, 3 Bovine respiratory disease is a multi‐factorial disease in which a range of adverse host and environmental factors predispose animals to infection with a variety of viral and bacterial pathogens.4 Outbreaks typically follow entry into a feedlot during which cattle are subjected to stress from transport, commingling and a rapid change in husbandry. Under these circumstances, the animals are exposed to new pathogens under conditions that are conducive to their transmission and at a time when their immune defences are reduced. Several viruses, including bovine herpes virus type 1 (BHV‐1), bovine parainfluenza virus type 3 (BPIV‐3), bovine respiratory syncytial virus (BRSV) and bovine viral diarrhoea virus (BVDV) can act as primary respiratory pathogens.3, 5 The most frequently identified bacterial pathogens are Mannheimia haemolytica, Pasteurella multocida and Histophilus somni, although a variety of other organisms, including Mycoplasma spp., may be involved.4, 6 These bacteria are ubiquitous in cattle as commensal nasopharyngeal flora, but can exacerbate disease through secondary infection in the lower respiratory tract.6
Bovine coronavirus (BCoV) infection is most commonly associated with calf diarrhoea and winter dysentery, but evidence implicating BCoV as a respiratory pathogen of cattle has recently been reviewed.7, 8 BCoV is a member of the genus Coronavirus within the family Coronaviridae, which are large (65–200 nm), pleomorphic, enveloped viruses with a single‐stranded RNA genome.9 The aim of this report was to document the role of BoCV as the probable primary aetiological agent in an outbreak of bovine respiratory disease in intensively managed cattle in Australia.
Materials and methods
Production system
An outbreak of bovine respiratory disease occurred in autumn 2010 in a semi‐intensive, paddock‐based feedlot production system located on the south coast of New South Wales, Australia. The 156‐ha property consisted of improved pastures with a total population of 300 Angus and Murray Grey purebred cows or their crosses with Limousin and Charolais bulls. The cattle were segregated in groups of 25 head in 4‐ha paddocks with a central feed trough, but shared water troughs on the fence lines. The production system was characterised by frequent turnover of stock, with weekly slaughter of 15–25 animals after approximately 70 days on a ration of cereal hay, biscuit meal and dried distillers’ grain. Replacement cattle were sourced at approximately 9 months of age, predominantly from saleyards, but also directly from breeder's holdings, and were transported up to 450 km. The newly purchased cattle were held in a nearby holding, in groups of 40–60 head, for 1 week prior to introduction to the feedlot. During this time they were vaccinated with a 5‐in‐1 clostridial vaccine (Ultravac, Pfizer, West Ryde, NSW, Australia), drenched with triclabendazole (Fasinex 240, Novartis, West Ryde, NSW, Australia) and fenbendazole (Panacur 100, Coopers Animal Health, Bendigo, VIC, Australia) and dosed with an oral chelated trace element mixture (Maxi‐Min®, Virbac, Milperra, NSW, Australia). Treatment with the clostridial vaccine and trace element preparation was repeated after 4 weeks.
Collection of samples
Samples were collected in the acute stage of disease from animals with the most severe clinical signs. Swabs were taken from inside both nostrils and placed in 3 mL phosphate‐buffered gelatin saline (PBGS). Blood was collected in clot activator and lithium heparin‐treated evacuated tubes. Convalescent serum samples were collected from the same animals and additional cattle from the same cohorts 2 weeks after collection of the acute samples. Lung and tracheal tissues were collected from animals with the most severe lesions identified at slaughter and were transported fresh on ice or fixed in 10% neutral‐buffered formalin. These animals were slaughtered 2 months after the onset of disease.
Serology
An agar gel immunodiffusion (AGID) assay was performed to detect antibodies to BVDV.10 Commercial enzyme‐linked immunosorbent assay (ELISA) kits were used to test for antibodies to BHV‐1 (ELISA IBR Verification kit, Pourquier, France), BPIV‐3 (Svanovir PIV3‐Ab ELISA kit, Svanova, Uppsala, Sweden) and BRSV (Svanovir BRSV‐Ab ELISA kit, Svanova).
Real‐time polymerase chain reaction (qPCR) and reverse transcriptase qPCR (qRT‐PCR) assays
Nucleic acids were purified from a 50‐μL aliquot of swab fluid (PBGS) or a 20% suspension of tissues homogenised in PBGS, using the MagMax‐96 Viral RNA extraction kit (Ambion, Austin, TX, USA) with a Kingfisher‐96 magnetic particle handling system (Thermo Fisher Scientific, Vantaa, Finland). A 5‐μL aliquot of purified nucleic acid was used as template for each qPCR and qRT‐PCR assay.
Assays for BCoV, BVDV, BHV‐1 and M. bovis were performed in 25‐μL reactions using the AgPathID one‐step RT‐PCR kit (Ambion) with previously described oligonucleotide primers and linear hydrolysis probes.11, 12, 13, 14 Thermocycling and collection of fluorescence data was performed with an ABI 7500‐Fast Real Time PCR system (Applied Biosystems, Foster City, CA, USA) with the following program: 95°C for 10 min, then 45 cycles at 95°C for 15 s and 60°C for 45 s. The thermocycling program was preceded by incubation at 45°C for 10 min for reverse transcription for BVDV and BCoV assays. The fluorescence threshold was set manually at 0.05 and results were expressed as cycle‐threshold (Ct) values, the point at which the fluorescence amplification curve crossed the threshold. A positive result was defined by a logarithmic increase in fluorescence that produced a Ct value <36. The Ct values between 36 and 40 were considered inconclusive and values >40 as negative. The TaqVet Triplex BRSV and PI3 qRT‐PCR kit (Laboratoire Service International, Lissieu, France) were used to test for BRSV and PIV‐3, respectively.
Bacteriology
Tracheal swabs and lung tissue were tested by routine bacterial culture methods at the Elizabeth Macarthur Agricultural Institute.15 Briefly, samples were inoculated in parallel onto MacConkey agar no. 3 (CM0115 Oxoid, Adelaide, SA, Australia) and sheep blood agar (7% v/v ovine blood in blood agar base, CM0271 Oxoid) plates. MacConkey agar plates were incubated at 37°C in air for 48 h and sheep blood agar plates in an atmosphere containing 5% CO2 for up to 72 h. For detection of Mycoplasma spp., the samples were inoculated onto culture plates prepared from Mycoplasma Agar Base (CM0401 Oxoid) with Mycoplasma Selective Supplement‐G (SR0059C Oxoid) and 0.2% w/v DNA (calf thymus) (Sigma‐Aldrich D1501, Castle Hill, NSW, Australia) and incubated at 37°C in an atmosphere containing 5% CO2 for 7 days.
Virus isolation
BCoV was isolated using human rectal tumour (HRT‐18) cells.16 Viral transport medium from swabs, or a 20% suspension of tissues homogenised in PBGS, was centrifuged at 560g for 15 min. The supernatant was aspirated and diluted 1 : 5 in a cell culture medium: RPMI‐1640 medium (Gibco, Life Technologies, Melbourne, VIC, Australia) supplemented with penicillin (1000 IU/mL), streptomycin (1000 μg/mL), trypsin (5 μg/mL) and pancreatin (5 μg/mL). This preparation was passed through a 0.45‐μm filter (Minisart, Sartorius, Germany) and 2 mL was adsorbed onto a freshly washed near‐confluent monolayer of HRT‐18 cells in 25 cm2 tissue culture flasks (Falcon, CA, USA). The cells were incubated at 37°C with 5% CO2 for 2 h and then for a further 4 h after the addition of 8 mL of cell culture medium supplemented with 1% fetal bovine serum (FBS). The inoculum was then discarded and 10 mL of fresh cell culture medium with 1% FBS was added. The cultures were incubated at 37°C with 5% CO2 for 5 days. The cells were then scraped from the plastic surface and mixed with the culture medium in the flask. The resulting supernatant was used to inoculate a near‐confluent HRT‐18 cell monolayer. Sub‐cultivation was repeated twice and replication of BCoV monitored by qRT‐PCR performed on the culture supernatant.
Virus isolation for BVDV and BHV‐1 was performed in primary bovine testis cells with immunostaining where appropriate.10, 17 After three passages, in the absence of cytopathology the cultures of testis cells were also screened for BPIV‐3 by haemadsorption with a 0.5% suspension of guinea pig red blood cells in phosphate‐buffered saline solution on the monolayers, followed by examination under a light microscope.
Results
Clinical signs
Respiratory disease was first observed in a group of 45 recently introduced cattle (cohort 1) and approximately 80 of the 300 cattle on the property were affected over the ensuing month. Affected cattle showed reduced exercise tolerance when being yarded, were anorexic, pyrexic (rectal temperature, 39.2–40.9°C), had an infrequent dry cough and an elevated respiratory rate. A serous nasal discharge was evident in some cases, predominantly in the animals with the highest rectal temperature. This discharge was occasionally flecked with mucopurulent material. Diarrhoea was not a feature of the disease outbreak. In most cases, the duration of clinical signs was 7–10 days, but signs persisted for 2 months in some cattle. More severely affected animals were treated by injection of 20 mg/kg long‐acting oxytetracycline (Bivatop LA Injectable, Boehringer Ingelheim, New Zealand). Clinical signs resolved in most animals, but two chronically affected animals died.
After the initial disease outbreak, similar clinical signs were observed in cohorts 2 and 3, which were introduced 1 and 2 months after the initial outbreak, respectively. All introduced cattle exhibited clinical signs within 1 week of introduction to the property. Similar clinical signs were also reported in cattle on neighbouring properties, but these cases were not investigated. The effect of disease on production parameters was not formally quantified.
Pathology
Gross lesions in the respiratory tract were identified in 15 of 49 cattle that were slaughtered during the study period. These cattle represented the heavier individuals that were suitable for slaughter, but a proportion of those slaughtered were still coughing occasionally. Multifocal, coalescent regions of consolidation up to 5–10 cm in diameter were present in the ventral areas of the cranial lung lobes (Figure 1a). Tags of fibrin were present on the thoracic pleura of six of the animals with pulmonary lesions, but adhesions were rarely present. The tracheas of some of the cattle with pulmonary lesions had petechial haemorrhage along the length of the mucosa and variable quantities of mucopurulent discharge, which extended into the bronchi of the worst affected animals (Figure 1b).
Figure 1.
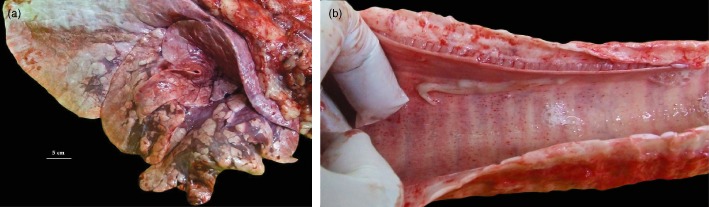
(a) Lung of a steer infected with bovine coronavirus, showing consolidation of the cranioventral lobes with multifocal to coalescing bronchopneumonia. (b) Trachea from a steer infected with bovine coronavirus showing petechial haemorrhages on the mucosa and mucopurulent exudate.
Histopathology
Examination of the lungs of five animals demonstrated multifocal to diffuse thickening of the pulmonary interstitium by infiltrates of macrophages, lymphocytes and fewer plasma cells. There were exudates of neutrophils admixed with mucus in the lumina of bronchi, bronchioles and some alveoli. Multinucleate syncytial epithelial cells were present in the lumina of some bronchioles (Figure 2a). There was moderate to marked hyperplasia of the bronchiole‐associated lymphoid tissue (BALT) (Figure 2b), together with atelectasis of surrounding alveoli and partial occlusion of some bronchiolar lumina. There was segmental hyperplasia and mild focal disruption of the bronchiolar epithelium by infiltrating lymphocytes. Oedema was evident in alveoli, as well as mild exudates of macrophages and occasional neutrophils. Mild, diffuse infiltrates of macrophages, lymphocytes and plasma cells were present in the lamina propria of the trachea, with mild exocytosis of lymphocytes in the epithelium. There was moderate segmental hyperplasia of the tracheal epithelium, with replacement by cuboidal to flattened epithelium.
Figure 2.
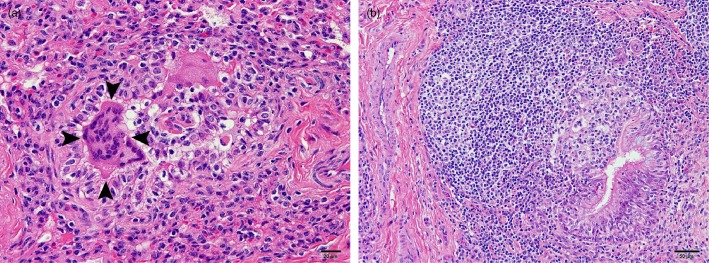
(a) Sample of the lung of a steer infected with bovine coronavirus showing multinucleate epithelial syncytial cells in a collapsed bronchiole (arrowheads). (b) Sample of the lung of a calf infected with bovine coronavirus showing marked hyperplasia of the bronchiole‐associated lymphoid tissue, with hyperplasia and focal disruption of the bronchiolar epithelium by infiltrating lymphocytes. (Haematoxylin and eosin staining.)
Virology
A combination of serology, virus isolation and qRT‐PCR was used to detect viral pathogens involved in the outbreak. Serology was performed with paired acute and convalescent samples from cohorts 2 and 3, but convalescent sera were the only samples available from cohort 1. Acute serum samples from newly introduced cattle indicated that BPIV‐3 and BRSV exposure occurred prior to entry (Table 1). On the basis of strong reactivity in the BVDV AGID test, transmission of BVDV had occurred recently, but prior to recruitment of cohorts 1 and 2. Transmission of BVDV after entry to the property was only detected in cohort 3. There was no serological evidence of infection with BHV‐1.
Table 1.
Results of laboratory assays performed on samples collected from affected animals: respiratory disease occurred in three cohorts that were introduced to the property at different times
| Cohort | Date | Sample | Antibody detection (positive results/number tested) | qPCR or qRT‐PCR (positive results/no. tested) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BVDV | BHV‐1 | BPIV‐3 | BRSV | BVDV | BHV‐1 | BPIV‐3 | BRSV | BoCV | Mycoplasma bovis | |||
| 1 | 23/4/10 | Convalescent serum | 0/9 | 0/9 | 5/9 | 9/9 | ||||||
| 01/6/10 |
Lung tissue Tracheal swab |
0/5 0/5 |
0/5 0/5 |
0/5 0/5 |
0/5 0/5 |
2/5 0/5 |
0/5 0/5 |
|||||
| 2 | 27/4/10 | Acute serum | 8/10 | 0/10 | 10/10 | 10/10 | ||||||
| Nasal swabs | 0/10 | 0/10 | 0/10 | 0/10 | 4/10 | 0/10 | ||||||
| 13/5/10 | Convalescent serum | 4/5a | 0/5 | Not tested (already positive) | ||||||||
| 21/6/10 | Lung tissue | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | |||||
| 3 | 01/6/10 | Acute serum | 1/5 | 0/5 | 5/5 | 5/5 | ||||||
| Nasal swabs | 0/5 | 0/5 | 0/10 | 0/10 | 2/10 | 0/5 | ||||||
| 10/6/10 | Convalescent serum | 4/10b | 0/10 | 10/10 | 10/10 | |||||||
| Nasal swabs | 0/10 | 0/10 | 0/10 | 0/10 | 4/10c | 0/10 | ||||||
One of the animals with no BVDV antibodies was resampled. Neither of the seronegative animals sampled from this cohort was persistently infected with BVDV.
Samples included the four seronegative animals identified by the acute samples; these animals did not seroconvert and did not have persistent BVDV infection.
Five of the 10 swabs taken on 10/6/2010 were from the same animals sampled on the 1/6/2010. The two animals that were initially positive for BCoV were also positive 9 days later.
BCoV, bovine coronavirus; BHV‐1, bovine herpesvirus type 1; BRSV, bovine respiratory syncytial virus; BVDV, bovine viral diarrhoea virus; BPIV‐3, bovine parainfluenza virus type 3; qPCR, real‐time polymerase chain reaction assay; qRT‐PCR, real‐time reverse transcriptase–polymerase chain reaction assay.
Bovine coronavirus was the only viral pathogen detected, with positive qRT‐PCR results in nasal swabs from 10 of 30 acutely infected animals and in the lungs of 2 of 15 animals with pulmonary lesions at slaughter (Table 1). BCoV was isolated from the nasal swab with the highest viral load, as determined by qRT‐PCR. Virus isolation and qRT‐PCR assays for BHV‐1, BRSV, BVDV, and BPIV‐3 were all negative.
Bacteriology
Pasteurella multocida was cultured from one of five lung lesions and one tracheal swab from cohort 1. A sparse pure growth of H. somni was obtained from another tracheal swab. No other bacteria, including Mycoplasma spp., were cultured and all pulmonary and tracheal samples tested negative for M. bovis by qPCR.
Discussion
Detailed laboratory investigation of this outbreak of bovine respiratory disease identified BCoV in the absence of more common viral respiratory pathogens. Detection of BCoV in nasal swabs and pulmonary tissue supported a primary viral aetiology with exacerbation by secondary bacterial infection in some cases, which is typical of the bovine respiratory disease complex.4, 10 Histopathology and bacteriology suggested that BCoV also contributed substantially to morbidity in this outbreak. BCoV has recently been described as a cause of bovine respiratory disease in Italy.18 A study in the USA demonstrated that BCoV infection was involved in two high‐mortality outbreaks of shipping fever in calves.19 Two other studies have identified an association between infection with BCoV and increased incidence of respiratory disease in feedlot cattle, but decreased weight gain was identified in only one of those studies.20, 21
Previous exposure to BPIV‐3 and BRSV was common among the weaner cattle recruited to this feedlot, which is consistent with previous studies of Australian feedlot cattle indicating that these agents, as well as BVDV and BHV‐1, contributed to the bovine respiratory disease complex.3 Tests for antibodies in paired serum samples, together with sensitive molecular and culture‐based pathogen detection assays, indicated that these agents were not involved in the disease outbreak described here. Exposure to BVDV occurred prior to entry to the feedlot in cohorts 1 and 2, but active transmission in cohort 3, may have contributed to the course of disease through immunosuppression.22 An assay to detect antibodies to BCoV was not available during this study. Such an assay could provide a sensitive herd test for BCoV infection because a study in feedlots in the USA indicated that seroconversion occurred at higher prevalence and persisted for a greater period of time compared with viral shedding.23 Detailed analysis of production and morbidity data in combination with serology could assist in determining the effect of BCoV infection, but pathogen detection is required to discriminate between respiratory and enteric infections.24
Real‐time PCR assays have recently become a standard method in veterinary virology because of their potential to provide rapid, sensitive and specific results.25 The assay used in this study appeared to provide superior sensitivity compared with virus isolation, which did not detect coronavirus in every qRT‐PCR positive sample (unpubl. data). A similar qRT‐PCR assay for BCoV detected as few as 10 copies of viral RNA in clinical specimens.26 However, the detection of BCoV by qRT‐PCR can be affected by differences in the RNA sequence in the target region of the genome.27 The qRT‐PCR assay used in the present study was designed using nucleotide sequence from a recent isolate of BCoV that caused neonatal diarrhoea in Australian calves.13 The assay amplified a target in the ORF1b region, which is a relatively conserved portion of the coronavirus genome.27 It is unclear if different strains of BCoV are responsible for enteric and respiratory syndromes. Antigenic and phylogenetic differences between isolates of BCoV from enteric and respiratory disease have been identified28, 29 and the quasispecies structure of BCoV produces nucleotide sequence variation within a single isolate.30 However, in other cases, isolates of BoCV with >98% phylogenetic similarity have been identified in enteric and respiratory disease31 and an isolate of BCoV from a case of winter dysentery demonstrated both respiratory and enteric tropism after experimental infection.32 Phylogenetic differences between BCoV isolates from different geographical locations are greater than the differences between isolates from different clinical syndromes.29, 33 Further characterisation of BCoV isolates from Australian cattle is required and the potential for variation in the genome sequence needs to be considered when interpreting qRT‐PCR assays.
Histological observations suggested that BCoV contributed to morbidity in this outbreak. Multinucleated syncytial epithelial cells have been observed previously in the lungs of cattle with viral infection.18 In the present cases, the syncytia were confined to the bronchioles, whereas syncytia associated with BSRV may be found in both airways and alveoli.34Syncytial cells have been also observed in humans infected with severe acute respiratory syndrome coronavirus (SARS‐CoV).35 Hyperplasia of BALT in the lungs of some cattle is suggestive of infection with Mycoplasma spp.,36 but these organisms were not cultured and M. bovis was not detected by qPCR in the present investigation. Interestingly, hyperplasia of BALT was observed after experimental infection of ferrets with SARS‐CoV.37
The effect of bovine respiratory disease on production can be minimised by reducing stress during adjustment to the feedlot environment.38 Walker et al. also demonstrated that morbidity was reduced by administration of BVDV, BHV‐1, BPIV‐3 and Pasteurella vaccines to animals that did not have antibodies to these agents.38 Although not currently available in Australia, evidence suggests that use of a vaccine for BCoV prior to entry into a feedlot may be beneficial. In Canada, calves with low neutralising titres for BCoV were more likely to require treatment for respiratory disease after commingling in a feedlot.39 Furthermore, intranasal administration of an infectious, modified BoCV vaccine reduced the risk of clinical signs of respiratory disease after feedlot entry in an experimental trial in the USA.40
Following the outbreak described here, BCoV has been further implicated as a component of the bovine respiratory disease complex in Australia during the investigation of shipping fever in cattle being exported from Australia (P Kluver and P Kirkland, unpubl. obs.) and also during diagnostic investigations of sporadic outbreaks of respiratory disease in adult cattle at Elizabeth Macarthur Agriculture Institute. BCoV should be considered in the diagnosis of respiratory disease in intensively managed cattle, but infection trials and detailed risk factor analyses are required to further elucidate the contribution of BCoV to the bovine respiratory disease complex in Australian cattle.
Acknowledgments
We are indebted to the staff of the Virology Laboratory at Elizabeth Macarthur Agriculture Institute for technical assistance during these investigations, particularly Elizabeth Moane (virus isolation) and Rodney Davis (qRT‐PCR). Adrian Philbey provided valuable assistance in preparing the manuscript.
References
- 1. Cusack PMV, McMeniman NP, Lean IJ. Feedlot entry characteristics and climate: their relationship with cattle growth rate, bovine respiratory disease and mortality. Aust Vet J 2007;85:311–316. [DOI] [PubMed] [Google Scholar]
- 2. Smith RA. Impact of disease on feedlot performance: a review. J Anim Sci 1998;76:272–274. [DOI] [PubMed] [Google Scholar]
- 3. Dunne SE, Godwin J, Hoare RJT, Kirkland PD. Diseases of feedlot cattle . Final report to Meat Research Council of Australia on Project DAN 064, 1994, 1–79.
- 4. Cusack PMV, McMeniman N, Lean IJ. The medicine and epidemiology of bovine respiratory disease in feedlots. Aust Vet J 2003;81:480–487. [DOI] [PubMed] [Google Scholar]
- 5. Panciera RJ, Confer AW. Pathogenesis and pathology of bovine pneumonia. Vet Clin North Am Food Anim Pract 2010;26:191–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Griffin D. Bovine pasteurellosis and other bacterial infections of the respiratory tract. Vet Clin North Am Food Anim Pract 2010;26:57–71. [DOI] [PubMed] [Google Scholar]
- 7. Saif LJ. Bovine respiratory coronavirus. Vet Clin North Am Food Anim Pract 2010;26:349–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boileau MJ, Kapil S. Bovine coronavirus associated syndromes. Vet Clin North Am Food Anim Pract 2010;26:123–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. International Committee on Taxonomy of Viruses (ICTV) . Management. 03.019.0.01 Coronavirus. In: ICTVdb – The Universal Virus Database, version 4. 2006. http://www.ncbi.nlm.nih.gov/ICTVdb/ICTV/dB. Accessed December 2011.
- 10. Kirkland PD, MacKintosh SG. Ruminant pestivirus infections. Australian and New Zealand Standard Diagnostic Procedures. Sub‐committee on Animal Health Laboratory Standards, Animal Health Australia. 2006:1–30. http://www.scahls.org.au/%20procedures/anzsdps. Accessed November 2011. [Google Scholar]
- 11. Wang J, O'Keefe J, Orr D. Validation of a real‐time PCR assay for detection of bovine herpesvirus 1 in bovine semen. J Virol Methods 2007;144:103–108. [DOI] [PubMed] [Google Scholar]
- 12. Hoffman B, Depner K, Schirrmeier H, Beer M. A universal heterologous internal control system for duplex real‐time RT‐PCR assays used in a detection system for pestiviruses. J Virol Methods 2006;136:200–209. [DOI] [PubMed] [Google Scholar]
- 13. Izzo MM, Kirkland PD, Mohler VL, Gunn AA, House JK. Comparison of three diagnostic techniques for detection of rotavirus and coronavirus in calf faeces in Australia. Aust Vet J 2012;90:122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sachse K, Salam HSH, Diller R et al. Use of a novel real‐time PCR technique to monitor and quantitate Mycoplasma bovis infection in cattle herds with mastitis and respiratory disease. Vet J 2010;186:299–303. [DOI] [PubMed] [Google Scholar]
- 15. Hum S, Kessell A, Djordjevic S et al. Mastitis, polyarthritis and abortion caused by Mycoplasma species bovine group 7 in dairy cattle. Aust Vet J 2000;78:744–750. [DOI] [PubMed] [Google Scholar]
- 16. Kapil S, Richardson KL, Radi C, Chard‐Bergstrom C. Factors affecting isolation and propagation of bovine coronavirus in human rectal tumor‐18 cell line. J Vet Diagn Invest 1996;8:96–99. [DOI] [PubMed] [Google Scholar]
- 17. Kirkland PD, Gu X. Infectious bovine rhinotracheitis. Australian and New Zealand Standard Diagnostic Procedures. Sub‐committee on Animal Health Laboratory Standards, Animal Health Australia. 2008:1–18. http://www.scahls.org.au/%20procedures/anzsdps. Accessed November 2011. [Google Scholar]
- 18. Decaro N, Campolo M, Desario C et al. Respiratory disease associated with bovine coronavirus infection in cattle herds in southern Italy. J Vet Diagn Invest 2008;20:28–32. [DOI] [PubMed] [Google Scholar]
- 19. Storz J, Lin X, Purdy CW et al. Coronavirus and Pasteurella infections in bovine shipping fever pneumonia and Evans’ criteria for causation. J Clin Microbiol 2000;38:3291–3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lathrop SL, Wittum TE, Brock KV et al. Association between infection of the respiratory tract attributable to bovine coronavirus and health and growth performance of cattle in feedlots. Am J Vet Res 2000;61:1062–1066. [DOI] [PubMed] [Google Scholar]
- 21. Thomas CJ, Hoet AR, Sreevatsan S et al. Transmission of bovine coronavirus and serological responses in feedlot calves under field conditions. Am J Vet Res 2006;67:1412–1420. [DOI] [PubMed] [Google Scholar]
- 22. Chase CCL, Elmowalis G, Yousif AAA. The immune response to bovine viral diarrhea virus: a constantly changing picture. Vet Clin North Am Food Anim Pract 2004;20:95–114. [DOI] [PubMed] [Google Scholar]
- 23. Lathrop SL, Wittum TE, Loerch SC, Perino LJ, Saif LJ. Antibody titres against bovine coronavirus and shedding of the virus via the respiratory tract in feedlot cattle. Am J Vet Res 2000;61:1057–1061. [DOI] [PubMed] [Google Scholar]
- 24. Cho KO, Hoet AE, Loerch SC, Wittum TE, Saif LJ. Evaluation of concurrent shedding of bovine coronavirus via the respiratory tract and enteric route in feedlot cattle. Am J Vet Res 2001;62:1436–1441. [DOI] [PubMed] [Google Scholar]
- 25. Hoffman B, Beer M, Reid SM et al. A review of RT‐PCR technologies used in veterinary virology and disease control: sensitive and specific diagnosis of five livestock diseases notifiable to the World Organisation for Animal Health. Vet Microbiol 2009;139:1–23. [DOI] [PubMed] [Google Scholar]
- 26. Decaro N, Elia G, Campolo M et al. Detection of bovine coronavirus using a TaqMan‐based real‐time RT‐PCR assay. J Virol Methods 2008;151:167–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Escutenaire S, Mohamed N, Isaksson M et al. SYBR Green real‐time reverse transcriptase‐polymerase chain reaction assay for generic detection of coronaviruses. Arch Virol 2007;152:41–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gelinas A, Boutin M, Sasseville AM, Dea S. Bovine coronaviruses associated with enteric and respiratory diseases in Canadian dairy cattle display different reactivities to anti‐HE monoclonal antibodies and distinct amino acid changes in their HE, S and ns4.9 protein. Virus Res 2001;76:43–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hasoksuz M, Hoet AE, Loerch SC et al. Detection of respiratory and enteric shedding of bovine coronaviruses in cattle in an Ohio feedlot. J Vet Diagn Invest 2002;14:308–313. [DOI] [PubMed] [Google Scholar]
- 30. Zhang X, Hasoksuz M, Spiro D et al. Quasispecies of bovine enteric and respiratory coronaviruses based on complete genome sequences and genetic changes after tissue culture adaptation. Virology 2007;363:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang X, Herbst K, Kousoulas KG, Storz J. Comparison of the S genes and the biological properties of respiratory and enteropathogenic bovine coronaviruses. Arch Virol 1994;134:421–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Park SJ, Kim GY, Choy HE et al. Dual enteric and respiratory tropisms of winter dysentery bovine coronavirus in calves. Arch Virol 2007;152:1885 – 1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jeong J, Kim GY, Yoon S et al. Detection and isolation of winter dysentery bovine coronavirus circulated in Korea during 2002–2004. Virology 2005;67:187–189. [DOI] [PubMed] [Google Scholar]
- 34. Valarcher J, Taylor G. Bovine respiratory syncytial virus infection. Vet Res 2007;38:153–180. [DOI] [PubMed] [Google Scholar]
- 35. Franks TJ, Chong PY, Chui P et al. Lung pathology of severe acute respiratory syndrome (SARS): a study of 8 autopsy cases from Singapore. Hum Pathol 2003;34:743–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Radaelli E, Luini M, Loria GR, Nicholas RAJ, Scanziani E. Bacteriological, serological, pathological, and immunohistochemical studies of Mycoplasma bovis respiratory infection in veal calves and adult cattle at slaughter. Res Vet Sci 2008;85:282–290. [DOI] [PubMed] [Google Scholar]
- 37. Van den Brand J, Haagmans B, Leijten L et al. Pathology of experimental SARS coronavirus infection in cats and ferrets. Vet Pathol 2008;45:551–562. [DOI] [PubMed] [Google Scholar]
- 38. Walker P, Fell LR, Reddacliff LA et al. Effects of yard weaning and training in the behavioural adaptation of cattle to a feedlot. Livestock Sci 2007;106:210–217. [Google Scholar]
- 39. Fulton RW, Step DL, Wahrmund J et al. Bovine coronavirus (BCV) infections in transported commingled beef cattle and sole‐source ranch calves. Can J Vet Res 2011;25:191–199. [PMC free article] [PubMed] [Google Scholar]
- 40. Plummer PJ, Rohrbach BW, Daugherty RA et al. Effect of intranasal vaccination against bovine enteric coronavirus on the occurrence of respiratory tract disease in a commercial backgrounding feedlot. J Am Vet Med Assoc 2004;225:726–731. [DOI] [PubMed] [Google Scholar]


